Abstract
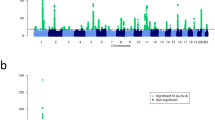
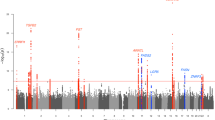
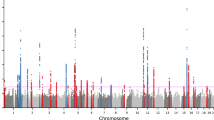
Genetic association studies have identified 21 loci associated with atopic dermatitis risk predominantly in populations of European ancestry. To identify further susceptibility loci for this common, complex skin disease, we performed a meta-analysis of >15 million genetic variants in 21,399 cases and 95,464 controls from populations of European, African, Japanese and Latino ancestry, followed by replication in 32,059 cases and 228,628 controls from 18 studies. We identified ten new risk loci, bringing the total number of known atopic dermatitis risk loci to 31 (with new secondary signals at four of these loci). Notably, the new loci include candidate genes with roles in the regulation of innate host defenses and T cell function, underscoring the important contribution of (auto)immune mechanisms to atopic dermatitis pathogenesis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
Accession codes
References
Weidinger, S. & Novak, N. Atopic dermatitis. Lancet 10.1016/S0140-6736(15)00149-X (13 September 2015).
Bataille, V., Lens, M. & Spector, T.D. The use of the twin model to investigate the genetics and epigenetics of skin diseases with genomic, transcriptomic and methylation data. J. Eur. Acad. Dermatol. Venereol. 26, 1067–1073 (2012).
Irvine, A.D., McLean, W.H. & Leung, D.Y. Filaggrin mutations associated with skin and allergic diseases. N. Engl. J. Med. 365, 1315–1327 (2011).
Palmer, C.N.A. et al. Common loss-of-function variants of the epidermal barrier protein filaggrin are a major predisposing factor for atopic dermatitis. Nat. Genet. 38, 441–446 (2006).
Rodríguez, E. et al. Meta-analysis of filaggrin polymorphisms in eczema and asthma: robust risk factors in atopic disease. J. Allergy Clin. Immunol. 123, 1361–1370 (2009).
Weidinger, S. et al. A genome-wide association study of atopic dermatitis identifies loci with overlapping effects on asthma and psoriasis. Hum. Mol. Genet. 22, 4841–4856 (2013).
Ellinghaus, D. et al. High-density genotyping study identifies four new susceptibility loci for atopic dermatitis. Nat. Genet. 45, 808–812 (2013).
Hirota, T. et al. Genome-wide association study identifies eight new susceptibility loci for atopic dermatitis in the Japanese population. Nat. Genet. 44, 1222–1226 (2012).
Paternoster, L. et al. Meta-analysis of genome-wide association studies identifies three new risk loci for atopic dermatitis. Nat. Genet. 44, 187–192 (2012).
Sun, L.-D. et al. Genome-wide association study identifies two new susceptibility loci for atopic dermatitis in the Chinese Han population. Nat. Genet. 43, 690–694 (2011).
Esparza-Gordillo, J. et al. A common variant on chromosome 11q13 is associated with atopic dermatitis. Nat. Genet. 41, 596–601 (2009).
Esparza-Gordillo, J. et al. A functional IL-6 receptor (IL6R) variant is a risk factor for persistent atopic dermatitis. J. Allergy Clin. Immunol. 132, 371–377 (2013).
Agarwala, V., Flannick, J., Sunyaev, S., Go, T.D.C. & Altshuler, D. Evaluating empirical bounds on complex disease genetic architecture. Nat. Genet. 45, 1418–1427 (2013).
Hinds, D.A. et al. A genome-wide association meta-analysis of self-reported allergy identifies shared and allergy-specific susceptibility loci. Nat. Genet. 45, 907–911 (2013).
Ferreira, M.A.R. et al. Identification of IL6R and chromosome 11q13.5 as risk loci for asthma. Lancet 378, 1006–1014 (2011).
Himes, B.E. et al. Genome-wide association analysis identifies PDE4D as an asthma-susceptibility gene. Am. J. Hum. Genet. 84, 581–593 (2009).
Noguchi, E. et al. Genome-wide association study identifies HLA-DP as a susceptibility gene for pediatric asthma in Asian populations. PLoS Genet. 7, e1002170 (2011).
Moffatt, M.F. et al. A large-scale, consortium-based genomewide association study of asthma. N. Engl. J. Med. 363, 1211–1221 (2010).
Sleiman, P.M. et al. Variants of DENND1B associated with asthma in children. N. Engl. J. Med. 362, 36–44 (2010).
Hirota, T. et al. Genome-wide association study identifies three new susceptibility loci for adult asthma in the Japanese population. Nat. Genet. 43, 893–896 (2011).
Bønnelykke, K. et al. Meta-analysis of genome-wide association studies identifies ten loci influencing allergic sensitization. Nat. Genet. 45, 902–906 (2013).
Jostins, L. et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 491, 119–124 (2012).
Tsoi, L.C. et al. Identification of 15 new psoriasis susceptibility loci highlights the role of innate immunity. Nat. Genet. 44, 1341–1348 (2012).
International Genetics of Ankylosing Spondylitis Consortium. Identification of multiple risk variants for ankylosing spondylitis through high-density genotyping of immune-related loci. Nat. Genet. 45, 730–738 (2013).
International Multiple Sclerosis Genetics Consortium. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature 476, 214–219 (2011).
Okada, Y. et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature 506, 376–381 (2014).
Bradfield, J.P. et al. A genome-wide meta-analysis of six type 1 diabetes cohorts identifies multiple associated loci. PLoS Genet. 7, e1002293 (2011).
Niwa, Y., Sumi, H. & Akamatsu, H. An association between ulcerative colitis and atopic dermatitis, diseases of impaired superficial barriers. J. Invest. Dermatol. 123, 999–1000 (2004).
Jakobsen, C., Paerregaard, A., Munkholm, P. & Wewer, V. Environmental factors and risk of developing paediatric inflammatory bowel disease—a population based study 2007–2009. J. Crohns Colitis 7, 79–88 (2013).
Baron, S. et al. Environmental risk factors in paediatric inflammatory bowel diseases: a population based case control study. Gut 54, 357–363 (2005).
Henseler, T. & Christophers, E. Disease concomitance in psoriasis. J. Am. Acad. Dermatol. 32, 982–986 (1995).
Baurecht, H. et al. Genome-wide comparative analysis of atopic dermatitis and psoriasis gives insight into opposing genetic mechanisms. Am. J. Hum. Genet. 96, 104–120 (2015).
Segrè, A.V. et al. Common inherited variation in mitochondrial genes is not enriched for associations with type 2 diabetes or related glycemic traits. PLoS Genet. 6, e1001058 (2010).
Wellcome Trust Case Control Consortium. Bayesian refinement of association signals for 14 loci in 3 common diseases. Nat. Genet. 44, 1294–1301 (2012).
Grundberg, E. et al. Mapping cis- and trans-regulatory effects across multiple tissues in twins. Nat. Genet. 44, 1084–1089 (2012).
ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012).
Bernstein, B.E. et al. The NIH Roadmap Epigenomics Mapping Consortium. Nat. Biotechnol. 28, 1045–1048 (2010).
Malissen, B., Tamoutounour, S. & Henri, S. The origins and functions of dendritic cells and macrophages in the skin. Nat. Rev. Immunol. 14, 417–428 (2014).
de Jong, M.A. & Geijtenbeek, T.B. Langerhans cells in innate defense against pathogens. Trends Immunol. 31, 452–459 (2010).
Baker, B.S. The role of microorganisms in atopic dermatitis. Clin. Exp. Immunol. 144, 1–9 (2006).
Tang, T.S., Bieber, T. & Williams, H.C. Does “autoreactivity” play a role in atopic dermatitis? J. Allergy Clin. Immunol. 129, 1209–1215 (2012).
Kitabatake, M. et al. Transgenic overexpression of G5PR that is normally augmented in centrocytes impairs the enrichment of high-affinity antigen-specific B cells, increases peritoneal B-1a cells, and induces autoimmunity in aged female mice. J. Immunol. 189, 1193–1201 (2012).
Lundström, W., Fewkes, N.M. & Mackall, C.L. IL-7 in human health and disease. Semin. Immunol. 24, 218–224 (2012).
Gregory, S.G. et al. Interleukin 7 receptor α chain (IL7R) shows allelic and functional association with multiple sclerosis. Nat. Genet. 39, 1083–1091 (2007).
Lundmark, F. et al. Variation in interleukin 7 receptor α chain (IL7R) influences risk of multiple sclerosis. Nat. Genet. 39, 1108–1113 (2007).
Lundström, W. et al. Soluble IL7Rα potentiates IL-7 bioactivity and promotes autoimmunity. Proc. Natl. Acad. Sci. USA 110, E1761–E1770 (2013).
Uehira, M., Matsuda, H., Nakamura, A. & Nishimoto, H. Immunologic abnormalities exhibited in IL-7 transgenic mice with dermatitis. J. Invest. Dermatol. 110, 740–745 (1998).
Steward-Tharp, S.M. et al. A mouse model of HIES reveals pro- and anti-inflammatory functions of STAT3. Blood 123, 2978–2987 (2014).
Milner, J.D. et al. Impaired TH17 cell differentiation in subjects with autosomal dominant hyper-IgE syndrome. Nature 452, 773–776 (2008).
Lupski, J.R., Belmont, J.W., Boerwinkle, E. & Gibbs, R.A. Clan genomics and the complex architecture of human disease. Cell 147, 32–43 (2011).
Blair, D.R. et al. A nondegenerate code of deleterious variants in Mendelian loci contributes to complex disease risk. Cell 155, 70–80 (2013).
Mertens-Talcott, S.U., Chintharlapalli, S., Li, X. & Safe, S. The oncogenic microRNA-27a targets genes that regulate specificity protein transcription factors and the G2-M checkpoint in MDA-MB-231 breast cancer cells. Cancer Res. 67, 11001–11011 (2007).
Ferreira, M.A. et al. Genome-wide association analysis identifies 11 risk variants associated with the asthma with hay fever phenotype. J. Allergy Clin. Immunol. 133, 1564–1571 (2014).
Stritesky, G.L., Jameson, S.C. & Hogquist, K.A. Selection of self-reactive T cells in the thymus. Annu. Rev. Immunol. 30, 95–114 (2012).
Moisan, J., Grenningloh, R., Bettelli, E., Oukka, M. & Ho, I.C. Ets-1 is a negative regulator of Th17 differentiation. J. Exp. Med. 204, 2825–2835 (2007).
Nagarajan, P. et al. Ets1 blocks terminal differentiation of keratinocytes and induces expression of matrix metalloproteases and innate immune mediators. J. Cell Sci. 123, 3566–3575 (2010).
Beck, L.A. et al. Dupilumab treatment in adults with moderate-to-severe atopic dermatitis. N. Engl. J. Med. 371, 130–139 (2014).
Granada, M. et al. A genome-wide association study of plasma total IgE concentrations in the Framingham Heart Study. J. Allergy Clin. Immunol. 129, 840–845 (2012).
Ramasamy, A. et al. A genome-wide meta-analysis of genetic variants associated with allergic rhinitis and grass sensitization and their interaction with birth order. J. Allergy Clin. Immunol. 128, 996–1005 (2011).
Morris, A.P. Transethnic meta-analysis of genomewide association studies. Genet. Epidemiol. 35, 809–822 (2011).
Wang, X. et al. Comparing methods for performing trans-ethnic meta-analysis of genome-wide association studies. Hum. Mol. Genet. 22, 2303–2311 (2013).
Yang, J. et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat. Genet. 44, 369–375 (2012).
Nyholt, D.R. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am. J. Hum. Genet. 74, 765–769 (2004).
Welter, D. et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 42, D1001–D1006 (2014).
Cole, C. et al. Filaggrin-stratified transcriptomic analysis of pediatric skin identifies mechanistic pathways in patients with atopic dermatitis. J. Allergy Clin. Immunol. 134, 82–91 (2014).
Tintle, S. et al. Reversal of atopic dermatitis with narrow-band UVB phototherapy and biomarkers for therapeutic response. J. Allergy Clin. Immunol. 128, 583–593 (2011).
Giambartolomei, C. et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 10, e1004383 (2014).
Maurano, M.T. et al. Systematic localization of common disease-associated variation in regulatory DNA. Science 337, 1190–1195 (2012).
So, H.C., Li, M. & Sham, P.C. Uncovering the total heritability explained by all true susceptibility variants in a genome-wide association study. Genet. Epidemiol. 35, 447–456 (2011).
Acknowledgements
This publication is the work of the authors, and L.P. will serve as guarantor for the contents of this paper. This research was specifically funded by an MRC Population Health Scientist Fellowship awarded to L.P. (MR/J012165/1). D.M.E. is supported by an Australian Research Council Future Fellowship (FT130101709) and a Medical Research Council program grant (MC_UU_12013/4). Individual study acknowledgment and funding statements can be found in the Supplementary Note.
Author information
Authors and Affiliations
Consortia
Contributions
Conceived and designed the experiments: L.P., M.S., H. Baurecht, D.P.S., J.A.C., K.B., J.P.T., H.T.d.D., P.M.A.S., F.L.X., M.B., J.Y.T., A.J.H., G.D.S., E.R., J.P., L.L.H., J.C.d.J., F. Rivadeneira, A.H., V.W.V.J., S.G.M.A.P., N.J.E., A.G.U., D.S.P., B.F., A.C., D.A.M., E. Melén, C.O., A.S., B.J., J.W.H., H. Bisgaard, J.S., N.M.P.-H., L.K.W., K.M.G., D.I.B., M. Melbye, G.H.K., Y.-A.L., N.H., D.J., X.J.Z., H.H., L.D., A.L., M.-R.J., M.T., S.J. Brown, J.H., D.M.E., S.W.
Performed the experiments: L.P., K.B., P.M.A.S., F.L.X., M.B., E.K.-M., G.A.L., M. Kubo, W.L.M., J.P.K., J.Z., E.R., F. Rivadeneira, A.G.U., J.L., X.Y.Y., L.D.S., L.E.C., A.M., C.E., D.S.P., C.M.T.T., M.I., S.H., N.V.-T., B.J., H. Bisgaard, N.M.P.-H., L.K.W., K.M.G., G.H.K., A.L., S.J. Brown, D.M.E., S.W.
Performed the statistical analysis: L.P., M.S., J.W., H. Baurecht, M. Hotze, D.P.S., J.A.C., C.T., A.T., A.B., A.C.A., H.T.d.D., M.A.F., E.A., P.M.A.S., J.R.G., I.M., J.E.-G., M.P.-Y., C.-J.X., L.C., M.M.G.-B., C.V., S.J. Barton, A.M.L., I.C., E.K.-M., G.A.L., S.B., R.A.M., F. Rüschendorf, A.K., J.P.K., J.Z., L.L.H., F. Rivadeneira, N.J.E., J.S.R., J.L., X.B.Z., X.D.Z., D.H., B.F., F.G., P.H., C.M.T.T., E.T., B.P., J.J.Y., N.V.-T., R.M., C.A.W., L.D., D.A.H., D.M.E.
Analyzed the data: L.P., M.S., J.W., H. Baurecht, M. Hotze, D.P.S., J.A.C., K.B., C.T., A.B., A.C.A., J.P.T., H.T.d.D., M.A.F., E.A., P.M.A.S., F.L.X., J.R.G., I.M., J.E.-G., M.P.-Y., C.-J.X., L.C., M.M.G.-B., C.V., S.J. Barton, A.M.L., I.C., G.A.L., J.B., S.B., R.A.M., F. Rüschendorf, A.K., A.J.H., M. Horikoshi, S.S., L.L.H., F. Rivadeneira, N.J.E., A.G.U., M.C.M., J.S.R., J.L., X.B.Z., X.D.Z., X.Y.Y., D.H., B.F., F.G., J.J.H., C.M.M., P.H., C.M.T.T., E.T., B.P., J.J.Y., M.I., S.H., N.V.-T., E. Melén, B.J., L.K.W., C.A.W., Y.-A.L., N.H., L.D., A.L., M.T., D.A.H., D.G., S.J. Brown, D.M.E.
Contributed reagents, material and/or analysis tools: L.P., J.W., H. Baurecht, M. Hotze, C.T., H.T.d.D., P.M.A.S., M.P.-Y., C.E.P., A.M.L., M.B., S.B., T.H., M. Kubo, W.L.M., J.Z., G.D.S., M. Macek, M. Kurek, M.A.L.-K., E. Mangold, A.P., A.F., W.L., N.N., R.F.-H., N.G., J.C.d.J., F. Rivadeneira, A.H., V.W.V.J., S.G.M.A.P., N.J.E., A.G.U., G.B.M., P.J.T., C.F.R., J.L., L.D.S., M.A.M., G.M.O'R., C.M.R.F., A.A., G.H., C.O.S., B.K., D.H., C.E., D.S.P., V.B., T.S., B.P., J.J.Y., C.L.R., S.T.W., D.A.M., S.C., T.K., C.S., E. Melén, S.L., C.O., B.A.R., B.J., J.W.H., J.S., L.K.W., K.M.G., M. Melbye, G.H.K., Y.-A.L., N.H., D.J., W.H.I.M., A.D.I., X.J.Z., H.H., C.G., E.G.B., N.G.M., L.D., M.-R.J., M.M.N., M.T., D.A.H., S.J. Brown, J.H., S.W.
Wrote the manuscript: L.P., M.S., J.W., H. Baurecht, M. Hotze, D.P.S., J.A.C., K.B., A.J.H., S.J. Brown, D.M.E., S.W.
Revised and reviewed the paper: L.P., M.S., J.W., H. Baurecht, M. Hotze, D.P.S., J.A.C., K.B., C.T., A.T., A.B., A.C.A., J.P.T., H.T.d.D., M.A.F., E.A., P.M.A.S., F.L.X., J.R.G., I.M., J.E.-G., M.P.-Y., C.-J.X., L.C., M.M.G.-B., C.V., C.E.P., S.J. Barton, A.M.L., I.C., M.B., E.K.-M., G.A.L., S.B., R.A.M., F. Rüschendorf, A.K., J.Y.T., T.H., M. Kubo, W.L.M., A.J.H., J.P.K., J.Z., G.D.S., M. Macek, M. Kurek, M.A.L.-K., E. Mangold, E.R., A.P., A.F., W.L., N.N., R.F.-H., M. Horikoshi, J.P., S.S., L.L.H., N.G., J.C.d.J., F. Rivadeneira, A.H., V.W.V.J., S.G.M.A.P., N.J.E., A.G.U., G.B.M., P.J.T., M.C.M., C.F.R., J.S.R., J.L., X.B.Z., X.D.Z., X.Y.Y., L.D.S., M.A.M., G.M.O'R., C.M.R.F., L.E.C., A.A., G.H., A.M., C.O.S., B.K., D.H., C.E., D.S.P., B.F., F.G., J.J.H., C.M.M., P.H., V.B., T.S., C.M.T.T., E.T., B.P., J.J.Y., M.I., S.H., N.V.-T., C.L.R., R.M., W.N., A.C., S.T.W., D.A.M., S.C., T.K., C.S., E. Melén, S.L., C.O., B.A.R., A.S., B.J., J.W.H., H. Bisgaard, J.S., N.M.P.-H., L.K.W., K.M.G., C.A.W., D.I.B., M. Melbye, G.H.K., Y.-A.L., N.H., D.J., W.H.I.M., A.D.I., X.J.Z., H.H., C.G., E.G.B., N.G.M., L.D., A.L., M.-R.J., M.M.N., M.T., D.A.H., D.G., S.J. Brown, J.H., D.M.E., S.W.
AAGC provided results for the discovery analysis.
Corresponding author
Ethics declarations
Competing interests
C.T., D.A.H. and J.Y.T. are employees of and own stock or stock options in 23andMe, Inc. K.M.G. has received reimbursement for speaking at conferences sponsored by companies selling nutritional and pharmaceutical products. He is part of an academic consortium that has received funding from Abbott Nutrition, Nestec and Danone. The University of Groningen has received money for D.S.P. in the context of an unrestricted educational grant for research from AstraZeneca. Fees for consultancies were given to the University of Groningen by AstraZeneca, Boehringer Ingelheim, Chiesi, GlaxoSmithKline, Takeda and TEVA. A.C. reports personal fees from AstraZeneca, personal fees from Novartis and personal fees from ThermoFisher given outside the context of the current manuscript. A.S. reports personal fees from GlaxoSmithKline and personal fees from ThermoFisher given outside the context of the current manuscript.
Additional information
A full list of members and affiliations is provided in the Supplementary Note.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–8, Supplementary Tables 1–4, 11–16, 22 and 23, and Supplementary Note. (PDF 20319 kb)
Supplementary Tables 5–10
Look-ups of known SNPs for various autoimmune diseases. (XLSX 48 kb)
Supplementary Tables 17–21
Functional investigations of credible SNPs. (XLSX 190 kb)
Rights and permissions
About this article
Cite this article
the EArly Genetics and Lifecourse Epidemiology (EAGLE) Eczema Consortium. Multi-ancestry genome-wide association study of 21,000 cases and 95,000 controls identifies new risk loci for atopic dermatitis. Nat Genet 47, 1449–1456 (2015). https://doi.org/10.1038/ng.3424
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.3424
This article is cited by
-
Mendelian randomization indicates that atopic dermatitis contributes to the occurrence of diabetes
BMC Medical Genomics (2023)
-
Integrated single-cell chromatin and transcriptomic analyses of human scalp identify gene-regulatory programs and critical cell types for hair and skin diseases
Nature Genetics (2023)
-
The Impact of Air Pollution on Atopic Dermatitis
Current Allergy and Asthma Reports (2023)
-
Association between parental autoimmune disease and childhood atopic dermatitis varied by sex: a nationwide case–control study
Archives of Dermatological Research (2023)
-
European and multi-ancestry genome-wide association meta-analysis of atopic dermatitis highlights importance of systemic immune regulation
Nature Communications (2023)