Abstract
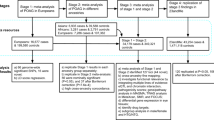
Primary angle closure glaucoma (PACG) is a major cause of blindness worldwide. We conducted a genome-wide association study (GWAS) followed by replication in a combined total of 10,503 PACG cases and 29,567 controls drawn from 24 countries across Asia, Australia, Europe, North America, and South America. We observed significant evidence of disease association at five new genetic loci upon meta-analysis of all patient collections. These loci are at EPDR1 rs3816415 (odds ratio (OR) = 1.24, P = 5.94 × 10−15), CHAT rs1258267 (OR = 1.22, P = 2.85 × 10−16), GLIS3 rs736893 (OR = 1.18, P = 1.43 × 10−14), FERMT2 rs7494379 (OR = 1.14, P = 3.43 × 10−11), and DPM2–FAM102A rs3739821 (OR = 1.15, P = 8.32 × 10−12). We also confirmed significant association at three previously described loci (P < 5 × 10−8 for each sentinel SNP at PLEKHA7, COL11A1, and PCMTD1–ST18)1, providing new insights into the biology of PACG.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Vithana, E.N. et al. Genome-wide association analyses identify three new susceptibility loci for primary angle closure glaucoma. Nat. Genet. 44, 1142–1146 (2012).
Thylefors, B., Négrel, A.D., Pararajasegaram, R. & Dadzie, K.Y. Global data on blindness. Bull. World Health Organ. 73, 115–121 (1995).
Hysi, P.G. et al. Genome-wide analysis of multi-ancestry cohorts identifies new loci influencing intraocular pressure and susceptibility to glaucoma. Nat. Genet. 46, 1126–1130 (2014).
Gharahkhani, P. et al. Common variants near ABCA1, AFAP1 and GMDS confer risk of primary open-angle glaucoma. Nat. Genet. 46, 1120–1125 (2014).
Wiggs, J.L. et al. Common variants at 9p21 and 8q22 are associated with increased susceptibility to optic nerve degeneration in glaucoma. PLoS Genet. 8, e1002654 (2012).
Aung, T. et al. A common variant mapping to CACNA1A is associated with susceptibility to exfoliation syndrome. Nat. Genet. 47, 387–392 (2015).
Foster, P.J., Buhrmann, R., Quigley, H.A. & Johnson, G.J. The definition and classification of glaucoma in prevalence surveys. Br. J. Ophthalmol. 86, 238–242 (2002).
Cheng, J.W., Zong, Y., Zeng, Y.Y. & Wei, R.L. The prevalence of primary angle closure glaucoma in adult Asians: a systematic review and meta-analysis. PLoS One 9, e103222 (2014).
Quigley, H.A. & Broman, A.T. The number of people with glaucoma worldwide in 2010 and 2020. Br. J. Ophthalmol. 90, 262–267 (2006).
Foster, P.J. & Johnson, G.J. Glaucoma in China: how big is the problem? Br. J. Ophthalmol. 85, 1277–1282 (2001).
Foster, P.J. et al. The prevalence of glaucoma in Chinese residents of Singapore: a cross-sectional population survey of the Tanjong Pagar district. Arch. Ophthalmol. 118, 1105–1111 (2000).
Dandona, L. et al. Angle-closure glaucoma in an urban population in southern India. The Andhra Pradesh eye disease study. Ophthalmology 107, 1710–1716 (2000).
Quigley, H.A., Congdon, N.G. & Friedman, D.S. Glaucoma in China (and worldwide): changes in established thinking will decrease preventable blindness. Br. J. Ophthalmol. 85, 1271–1272 (2001).
Purdue, M.P. et al. Genome-wide association study of renal cell carcinoma identifies two susceptibility loci on 2p21 and 11q13.3. Nat. Genet. 43, 60–65 (2011).
International Parkinson Disease Genomics Consortium. Imputation of sequence variants for identification of genetic risks for Parkinson's disease: a meta-analysis of genome-wide association studies. Lancet 377, 641–649 (2011).
Ellinor, P.T. et al. Meta-analysis identifies six new susceptibility loci for atrial fibrillation. Nat. Genet. 44, 670–675 (2012).
Gudbjartsson, D.F. et al. A sequence variant in ZFHX3 on 16q22 associates with atrial fibrillation and ischemic stroke. Nat. Genet. 41, 876–878 (2009).
Pomerantz, M.M. et al. The 8q24 cancer risk variant rs6983267 shows long-range interaction with MYC in colorectal cancer. Nat. Genet. 41, 882–884 (2009).
Tuupanen, S. et al. The common colorectal cancer predisposition SNP rs6983267 at chromosome 8q24 confers potential to enhanced Wnt signaling. Nat. Genet. 41, 885–890 (2009).
Gregorio-King, C.C. et al. MERP1: a mammalian ependymin-related protein gene differentially expressed in hematopoietic cells. Gene 286, 249–257 (2002).
Dolmans, G.H. et al. Wnt signaling and Dupuytren's disease. N. Engl. J. Med. 365, 307–317 (2011).
Barone, R. et al. DPM2-CDG: a muscular dystrophy–dystroglycanopathy syndrome with severe epilepsy. Ann. Neurol. 72, 550–558 (2012).
Wang, D.Y., Fulthorpe, R., Liss, S.N. & Edwards, E.A. Identification of estrogen-responsive genes by complementary deoxyribonucleic acid microarray and characterization of a novel early estrogen-induced gene: EEIG1. Mol. Endocrinol. 18, 402–411 (2004).
Shi, L. et al. Overexpression of PIP5KL1 suppresses cell proliferation and migration in human gastric cancer cells. Mol. Biol. Rep. 37, 2189–2198 (2010).
Lachkar, Y. & Bouassida, W. Drug-induced acute angle closure glaucoma. Curr. Opin. Ophthalmol. 18, 129–133 (2007).
Mandak, J.S., Minerva, P., Wilson, T.W. & Smith, E.K. Angle closure glaucoma complicating systemic atropine use in the cardiac catheterization laboratory. Cathet. Cardiovasc. Diagn. 39, 262–264 (1996).
Shen, Z. et al. Novel focal adhesion protein kindlin-2 promotes the invasion of gastric cancer cells through phosphorylation of integrin β1 and β3. J. Surg. Oncol. 108, 106–112 (2013).
Kim, Y.S., Nakanishi, G., Lewandoski, M. & Jetten, A.M. GLIS3, a novel member of the GLIS subfamily of Krüppel-like zinc finger proteins with repressor and activation functions. Nucleic Acids Res. 31, 5513–5525 (2003).
Senée, V. et al. Mutations in GLIS3 are responsible for a rare syndrome with neonatal diabetes mellitus and congenital hypothyroidism. Nat. Genet. 38, 682–687 (2006).
Barrett, J.C. et al. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat. Genet. 41, 703–707 (2009).
Cho, Y.S. et al. Meta-analysis of genome-wide association studies identifies eight new loci for type 2 diabetes in east Asians. Nat. Genet. 44, 67–72 (2012).
Dupuis, J. et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat. Genet. 42, 105–116 (2010).
Burdon, K.P. et al. Genome-wide association study identifies susceptibility loci for open angle glaucoma at TMCO1 and CDKN2B-AS1. Nat. Genet. 43, 574–578 (2011).
Chen, Y. et al. Common variants near ABCA1 and in PMM2 are associated with primary open-angle glaucoma. Nat. Genet. 46, 1115–1119 (2014).
Li, Z. et al. A common variant near TGFBR3 is associated with primary open angle glaucoma. Hum. Mol. Genet. 24, 3880–3892 (2015).
Nongpiur, M.E. et al. Lack of association between primary angle-closure glaucoma susceptibility loci and the ocular biometric parameters anterior chamber depth and axial length. Invest. Ophthalmol. Vis. Sci. 54, 5824–5828 (2013).
Cheng, C.Y. et al. Nine loci for ocular axial length identified through genome-wide association studies, including shared loci with refractive error. Am. J. Hum. Genet. 93, 264–277 (2013).
Nongpiur, M.E. et al. ABCC5, a gene that influences the anterior chamber depth, is associated with primary angle closure glaucoma. PLoS Genet. 10, e1004089 (2014).
Boyle, A.P. et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 22, 1790–1797 (2012).
Ward, L.D. & Kellis, M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 40, D930–D934 (2012).
ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012).
Westra, H.J. et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat. Genet. 45, 1238–1243 (2013).
GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 45, 580–585 (2013).
Fairfax, B.P. et al. Genetics of gene expression in primary immune cells identifies cell type–specific master regulators and roles of HLA alleles. Nat. Genet. 44, 502–510 (2012).
Okada, Y. et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature 506, 376–381 (2014).
Speliotes, E.K. et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat. Genet. 42, 937–948 (2010).
Dunstan, S.J. et al. Variation at HLA-DRB1 is associated with resistance to enteric fever. Nat. Genet. 46, 1333–1336 (2014).
Verhoeven, V.J. et al. Genome-wide meta-analyses of multiancestry cohorts identify multiple new susceptibility loci for refractive error and myopia. Nat. Genet. 45, 314–318 (2013).
Al Olama, A.A. et al. A meta-analysis of 87,040 individuals identifies 23 new susceptibility loci for prostate cancer. Nat. Genet. 46, 1103–1109 (2014).
Kiryluk, K. et al. Discovery of new risk loci for IgA nephropathy implicates genes involved in immunity against intestinal pathogens. Nat. Genet. 46, 1187–1196 (2014).
Acknowledgements
This research is supported by the Singapore Ministry of Health's National Medical Research Council under its Translational and Clinical Research (TCR) Flagship Programme Grant Stratified Medicine for Primary Angle Closure Glaucoma (NMRC/TCR/008-SERI/2013) and the Singapore Translational Research (STaR) Investigator Award Singapore Angle Closure Glaucoma Program Characterization, Prevention, and Management (NMRC/STAR/0023/2014), as well as the Biomedical Research Council, Agency for Science, Technology and Research (A-STAR), Singapore. A.T.L.-S. gratefully acknowledges support from grants RUI 1001/PPSP/812101 and RUI 1001/PPSP/812152 from the Universiti Sains Malaysia. H.J., C.Q., and N. Wang acknowledge support from the Program of Beijing Scholars (2013), Leading Talents–High-Level Talents of the Health System of Beijing (2009-1-05), and the National Major Scientific and Technological Special Project for 'Significant New Drugs Development' (2011ZX09302-007-05), as well as Project of the National Natural Science Foundation of China (81570837) grants.
Author information
Authors and Affiliations
Contributions
T.A. and C.C. Khor conceived the project. T. Do., H.J., M.N., R.G., K.A.-A., R. Duvesh., L.J.C., Z.L., M.E.N., S.A.P., C.Q., H.-T.W., H. Sakai., M.B.d.M., Y.A., T.L.H.D., Y.I., R.A.P.-G., T.Z., A.C.D., J.B.J., P.O.S.T., T.A.T., H.A., F.A., S.M., P.T.K.C., L.A.A., T. Dada., T.T.L., M.S.A., N.K., B.W., Y.Y.A., J.M.-N., S.V., S.S., R.H., A.J., M.B., D.G., D.H.S., H.W., V.K.Y., L.W.Y., T.B.T., M.M., T.T.N., E.U.L., K.-H.P., W.A.W., R.S.K., C.T., Y.K., S.S.T., K.P., J.F.S., Y.H.S., A.F., M.O., J.S.M.L., V.T., C.C. Khaing., T.M., S.N., C.-Y.K., G.T., S.F., R.W., H.M., T.T.G.N., T.D.T., M.U., J.M.M., N.R., Y.M.A., R.D.R., S.A.V., S.K.F., Z.X., X.Y.C., J.N.F., K.S.S., T.T.W., D.T.Q., R.V., S. Kavitha., S.R.K., N. Soumittra., B.S., B.-A.L., J.O., J.P.C.d.V., V.P.C., R.Y.A., B.B.d.S., C.C.S., M.C.A., E.K.-J., G.B.F., V.C.T., R.F., Y.J., N. Waseem., S.L., H.N.P., S.A.-S., E.R.C., M.I.K., R. Dada., K. Mohanty., M.A.F., A.W.H., K.P.B., E.H.G., A.P., T.P., M.A.T.C., I.R.F., C.S.L., E.R., V.W.I., G.C., G.P., C.L., P.R., L.M., S.C., J.C.H.C., B.N.K.C., J.W.H.S., H.M.T., K.T.O., A.T.H., V.H.Y., X.-Y.N., F.P., D.P., P.F., J.J.W., P.M., J.H.F., R.R.A., M.A.H., R. Stead., R. Quino., S.N.Z., U.L., R. Shetty., M.Z., H.W.B., N.L.O., T.K., A.M., W.L.H., L.D., Y.H.H., C.A.K., M.K., Z.E.D., J.I.M.P., A.G., J.W.J., T.S.H., N. Srisamran, T.S., S.H.S., V.H.D., S.S.B., C.-L.H., D.T.T., R. Sihota., S.-C.L., K. Mori., S. Kinoshita., A.I.d.H., R. Qamar., Y.-X.W., Y.Y.T., E.-S.T., C.H.-M., D.L.-G., S.M.S., C.-Y.C., J.C.Z., C.P.P., H.T.T.B., O.H., J.E.C., D.P.E., M.Y., J.M.N., M.L.G.-F., L.X., R.R., A.T.L.-S., T.Y.W., S.A.-O., N.H.D., P.S., C.C.T., P.J.F., L.V., K.T., E.N.V., and N. Wang were responsible for sample collection, data analysis, and sample administration. M.L.H., M.S., E.P., S.J.D., N.V.V.C., J.B., Y.X.Z., A.K., S.T.L., S.H.C., R.P.E., A.S., K.H.P., J.A., G.B., H. Snippe., and B.B. contributed control data sets with genome-wide genotyping data. M.-C.L., A.S.C., S.R.G., Y.F.C., and E.N.V. were responsible for molecular biology work and pathological tissue staining and interpretation. The manuscript was written by C.C. Khor, with approval from all authors. All authors read and approved the final version of the manuscript for publication. T.A. was responsible for obtaining financial support for the study.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Regional association analysis for the five newly identified loci surpassing genome-wide significance.
The horizontal axis represents physical distance in megabases on hg37 coordinates. The vertical axis of the left represents –log10 (P values) for association with PACG. The vertical axis on the right represents the recombination rate in centiMorgans and is taken from the HapMap panels.
Supplementary Figure 2 Ocular expression of genes at PACG-associated loci.
(a) Expression analysis of EPDR1, GLIS3, DPM2, FAM102A, FERMT2, and PIP5KL1 in human ocular tissues by RT-PCR using gene-specific primers (Supplementary Table 9). RT-PCR products were observed differentially in sclera (S), cornea (Co), lens with lens capsule (L), iris (I), trabecular meshwork (TM), ciliary body (CB), retina (R), choroid (Ch), optic nerve head (ONH), and optic nerve (ON). The ubiquitously expressed gene ACTB was used as the normalizing control. A no-template sample acted as the negative control (NC) to ensure no contamination of the RT-PCR reaction mix. M denotes the molecular weight marker. (b–h) Immunoblots of whole-cell lysates from retinal pigment epithelial (ARPE19), non-pigmented ciliary epithelial (NPCE), human trabecular meshwork (HTM), human cervical adenocarcinoma (Hela S), human breast adenocarcinoma (MCF7), and human embryonic kidney epithelial (HEK 293) cells, probed for CHAT, EPDR1, GLIS3, DPM2, FAM102A, FERMT2, and PIP5KL1 proteins and GAPDH, as a loading control. The protein isoforms detected by the respective antibodies were CHAT (83 kDa), EPDR1 (~25 kDa and ~18 kDa corresponding to two alternatively spliced forms of EPDR1), GLIS3 (~118 kDa and 90 kDa corresponding to two alternatively spliced forms of GLIS3), DPM2 (~17 kDa), FAM102A (37 kDa), FERMT2 (~78 kDa), and PIP5KL1 (22 kDa). The arrowheads indicate specific protein bands.
Supplementary Figure 3 Analysis of CHAT distribution in human ocular tissues.
(a) Strong diffuse immunoreactivity of CHAT was detected in the posterior iris epithelium (PIE) and anterior iris border (AIB). In the iris stroma (IS), the iris stromal cells also show moderate to strong positive staining for CHAT, whereas the iris dilator muscle shows only weak and variable immunoreactivity. (b) In the ciliary body, pronounced yet diffuse staining of CHAT was detected in the pigmented ciliary epithelium (PCE). (c) The ciliary muscles (CM) showed strong and diffuse staining of the CHAT protein, whereas more moderate immunoreactivity was detected in the trabecular meshwork (TM) along the trabecular beams. (d,e) The corneal epithelium (C. Epi) and Descemet’s membrane (Descemet’s) demonstrated positive immunoreactivity for CHAT in contrast to the negative staining in stromal keratocytes, corneal stroma (C. Stroma), and endothelial cells (C. Endo). (f) In the retina, CHAT expression was diffusely and strongly positive in photoreceptors (PR). Negative immunoreactivity was seen in the nerve fiber layer, ganglion cell layer (GCL) inner plexiform layer, inner nuclear layer (INL), outer plexiform layer, and outer nuclear layer (ONL).
Supplementary Figure 4 Analysis of EPDR1 distribution in human ocular tissues.
(a) In the iris, diffuse and strong immunoreactivity for EPDR1 was seen throughout the iris tissue. These areas included the posterior iris epithelium (PIE), iris dilator muscle (IDM), cells within the iris stroma (IS) as well as the iris capillaries (IC), and anterior iris border (AIB). (b) In the ciliary body, EPDR1 protein expression was detected as diffuse and strongly positive immunoreactivity in ciliary body vessels (CBV) and non-pigmented ciliary epithelium (NPCE). Although positive staining was detected in pigmented ciliary epithelium (PCE), the intensity was weaker than those for NPCE and CBV. (c) EPDR1 protein was also expressed throughout the trabecular meshwork (TM) and Schlemm’s canal (SC). (d,e) In the cornea, EPDR1 was detected in the cornea epithelium (C. Epi.) and cornea endothelial (C. Endo) layers. (f) In the retina, EPDR1 staining was detected in the nerve fiber layer (NFL), ganglion cell layer (GCL), inner plexiform layer (IPL), outer plexiform layer (OPL), and photoreceptors (PR).
Supplementary Figure 5 Analysis of FERMT2 distribution in human ocular tissues.
(a) In the iris, strong and diffuse FERMT2 immunoreactivity was detected in the iris dilator muscle (IDM). Moderate to strong immunoreactivity was also seen in cells within the iris stroma (IS) and iris capillaries (IC), whereas the anterior iris border showed only moderate immunoreactivity. (b) In the ciliary body, strong FERMT2 expression was detected in both non-pigmented ciliary epithelium (NPCE) and pigmented ciliary epithelium (PCE). (c) Strong FERMT2 immunoreactivity was detected in the ciliary muscles (CM), trabecular meshwork (TM), and Schlemm’s canal (SC). (d,e) In the cornea, FERMT2 expression was observed in cornea epithelium (C. Epi.) and cornea endothelium (C. Endo.). (f) In the retina, FERMT2 was variably expressed in the retinal layers, with strong and diffuse immunoreactivity detected in the photoreceptors (PR) and moderate immunoreactivity seen in the outer plexiform layer (OPL).
Supplementary Figure 6 Analysis of GLIS3 distribution in human ocular tissues.
(a) In the iris, GLIS3 showed strong and diffuse immunoreactivity in iris dilator muscle (IDM) and the anterior iris border (AIB). A positive punctate membranous staining pattern in addition to weaker cytoplasmic staining was observed in posterior pigment epithelium of the posterior iris epithelium (PIE). The cells in the iris stroma (IS) also demonstrated strong positive GLIS3 immunoreactivity. (b) GLIS3 protein expression was prominently seen in the ciliary muscle (CM). Strong and diffuse immunoreactivity was detected in the non-pigmented ciliary epithelium (NPCE), pigmented ciliary epithelium (PCE), and ciliary muscle (CM). (c) In the trabecular meshwork (TM) and Schlemm’s canal (SC), strong and diffuse immunoreactivity was detected. (d,e) In the cornea, GLIS3 expression was detected in the cornea epithelium (C. Epi.) and cornea endothelium (C. Endo.). (f) In the retina, distinct strong and diffuse GLIS3 immunoreactivity was detected in the photoreceptors (PR) and outer plexiform layer (OPL), with much weaker variable immunoreactivity detected in the ganglion cell layer (GCL).
Supplementary Figure 7 Analysis of DPM2 distribution in human ocular tissues.
(a) In the iris, strong DPM2 expression was detected in the anterior iris border (AIB), cells within the iris stroma (IS), and iris capillaries (IC), with only moderate intensity of DPM2 immunostaining detected in iris dilator muscle (IDM). (b) DPM2 protein was found to be diffusely expressed in the ciliary body. Strong immunoreactivity was detected in both non-pigmented ciliary epithelium (NPCE) and pigmented ciliary epithelium (PCE), ciliary body blood vessels (CBV), and also ciliary muscle (CM). (c) DPM2 immunoreactivity was seen in trabecular meshwork (TM) and Schlemm’s canal (SC). (d,e) In the cornea, DPM2 expression was observed in cornea stromal (C. stroma) collagen fibers but not keratocytes. Immunoreactivity to DPM2 was also seen in the cornea epithelium (C. Epi) and cornea endothelium (C. Endo.) IDM. (f) In the retina, strong and diffuse immunoreactivity of DPM2 was detected in the photoreceptors (PR) in contrast to the much weaker immunoreactivity seen in the outer plexiform layer (OPL) and the ganglion cell layer (GCL).
Supplementary Figure 8 Analysis of PIP5KL1 distribution in human ocular tissues.
(a) In the iris, pronounced and diffuse staining of PIP5KL1 was detected in the iris dilator muscle (IDM). Strong immunoreactivity was also seen in iris stromal cells (IS) and iris capillaries (IC). (b) PIP5KL1 expression was diffuse with strong positive staining observed in the non-pigmented ciliary epithelium (NPCE). A weaker staining profile was observed in pigmented ciliary epithelium (PCE), which is constrained to PCE cell–cell borders. (c) In the trabecular meshwork (TM) and Schlemm’s canal (SC), strong immunoreactivity of PIP5KL1 was detected in TM beams and endothelial cells of the Schlemm’s canal. (d,e) In the cornea, PIP5KL1 was strongly expressed in the nuclei and cytoplasm of the corneal epithelium (C. Epi.). Positive staining of stromal keratocytes was also seen. The corneal endothelium (C. Endo) showed intense immunoreactivity to PIP5KL1. (f) In the retina, PIP5KL1 immunoreactivity in the outer limiting membrane (OLM) was distinctly prominent in comparison to the rest of the retinal layers. Similarly, the ganglion cell layer (GCL) also showed strong nuclear positive staining for PIP5KL1. In contrast, mild to moderate variable expression of PIP5KL1 protein was seen in photoreceptors (PR). Weak and variable immunoreactivity could be seen in the inner plexiform layer (IPL) and the inner nuclear layer (INL). The rest of the retina showed no significant immunoreactivity to PIP5KL1.
Supplementary Figure 9 Analysis of FAM102A distribution in human ocular tissues.
(a) In the iris, strong immunoreactivity of FAM102A protein was seen in the anterior iris border (AIB), iris stromal cells (IS), and the iris dilator muscle (IDM). Moderate and punctate staining of the posterior border of the posterior pigment epithelium portion of the posterior iris epithelium (PIE) was also seen. (b) In the ciliary body, strong immunoreactivity for FAM102A protein was limited to pigmented ciliary epithelium (PCE) and non-pigmented ciliary epithelium (NPCE), with PCE having stronger immunoreactivity. (c) Moderate immunoreactivity of FAM102A was observed in the trabecular meshwork (TM), Schlemm’s canal (SC), and ciliary muscle (CM). (d,e) In cornea, only the basal epithelial layer of the cornea epithelium (C. Epi.) and corneal endothelial cells (C. Endo.) showed moderate FAM102A protein immunoreactivity. (f) In the retina, strong FAM102A immunoreactivity was distinctly localized to the outer limiting membrane (OLM), photoreceptors (PR), and nuclei of the ganglion cell layer (GCL). Patchy moderate immunoreactivity of FAM102A was also noted in the outer plexiform layer (OPL). The rest of the retinal layers showed no significant immunoreactivity to FAM102A.
Supplementary Figure 10 Distribution of PACG cases and controls of East Asian descent from the GWAS discovery stage.
PACG cases and controls are projected onto the top two principal components of genetic stratification, with cases on the left panel and controls on the right panel.
Supplementary Figure 11 Detailed ancestry analysis of PACG cases and controls from the GWAS discovery stage, which encompassed collections from 15 countries.
(a-f) The genetic principal components (PCs) for cases and controls are projected onto PC1 vs. PC3 (a), PC1 vs. PC4 (b), PC1 vs. PC5 (c), PC2 vs. PC3 (d), PC3 vs. PC4 (e), and PC4 vs PC5 (f). For each panel, PACG cases are projected on the left and controls on the right.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–12, Supplementary Note and Supplementary Tables 1–8, 10 and 11. (PDF 5580 kb)
Supplementary Table 9
Variants located within predicted regulatory regions and in LD (r2 >0.8) with the top SNPs in the five newly identified loci. (XLSX 29 kb)
Supplementary Data Set
Summary association statistics of the PACG GWAS SNPs with direct microarray genotyping together with imputation fine-mapping of the genome-wide significant loci. (TXT 29091 kb)
Rights and permissions
About this article
Cite this article
Khor, C., Do, T., Jia, H. et al. Genome-wide association study identifies five new susceptibility loci for primary angle closure glaucoma. Nat Genet 48, 556–562 (2016). https://doi.org/10.1038/ng.3540
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.3540
This article is cited by
-
Impact of rs11024102 PLEKHA7, rs3753841 COL11A1 single nucleotide polymorphisms, and serum levels of oxidative stress markers on the risk of primary angle-closure glaucoma in Egyptians
Journal of Genetic Engineering and Biotechnology (2022)
-
New loci for refractive errors and ocular biometric parameters in young Chinese Han adults
Science China Life Sciences (2022)
-
Haplotype-based genomic analysis reveals novel association of CNTNAP5 genic region with primary angle closure glaucoma
Journal of Biosciences (2021)