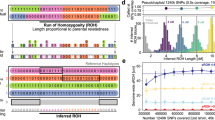
Abstract
Recent studies of human populations suggest that the genome consists of chromosome segments that are ancestrally conserved ('haplotype blocks'; refs. 1–3) and have discrete boundaries defined by recombination hot spots4,5. Using publicly available genetic markers6, we have constructed a first-generation haplotype map of chromosome 19. As expected for this marker density7, approximately one-third of the chromosome is encompassed within haplotype blocks. Evolutionary modeling of the data indicates that recombination hot spots are not required to explain most of the observed blocks, providing that marker ascertainment and the observed marker spacing are considered. In contrast, several long blocks are inconsistent with our evolutionary models, and different mechanisms could explain their origins.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Daly, M.J., Rioux, J.D., Schaffner, S.F., Hudson, T.J. & Lander, E.S. High-resolution haplotype structure in the human genome. Nat. Genet. 29, 229–232 (2001).
Gabriel, S.B. et al. The structure of haplotype blocks in the human genome. Science 296, 2225–2229 (2002).
Reich, D.E. et al. Human genome sequence variation and the influence of gene history, mutation and recombination. Nat. Genet. 32, 135–142 (2002).
Goldstein, D.B. Islands of linkage disequilibrium. Nat. Genet. 29, 109–111 (2001).
Jeffreys, A.J., Kauppi, L. & Neumann, R. Intensely punctate meiotic recombination in the class II region of the major histocompatibility complex. Nat. Genet. 29, 217–222 (2001).
Sachidanandam, R. et al. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature 409, 928–933 (2001).
Dawson, E. et al. A first generation linkage disequilibrium map of human chromosome 22. Nature 418, 544–548 (2002).
Roses, A.D. Pharmacogenetics and the practice of medicine. Nature 405, 857–865 (2000).
Patil, N. et al. Blocks of limited haplotype diversity revealed by high-resolution scanning of human chromosome 21. Science 294, 1719–1723 (2001).
Casci, T. Haplotype mapping: shortcut around the block. Nat. Rev. Genet. 3, 573 (2002).
Weiss, K.M. & Clark, A.G. Linkage disequilibrium and the mapping of complex human traits. Trends Genet. 18, 19–24 (2002).
Couzin, J. Genomics. New mapping project splits the community. Science 296, 1391–1393 (2002).
Ardlie, K.G., Kruglyak, L. & Seielstad, M. Patterns of linkage disequilibrium in the human genome. Nat. Rev. Genet. 3, 299–309 (2002).
Jeffreys, A.J., Ritchie, A. & Neumann, R. High resolution analysis of haplotype diversity and meiotic crossover in the human TAP2 recombination hotspot. Hum. Mol. Genet. 9, 725–733 (2000).
Johnson, G.C. et al. Haplotype tagging for the identification of common disease genes. Nat. Genet. 29, 233–237 (2001).
Subrahmanyan, L., Eberle, M.A., Clark, A.G., Kruglyak, L. & Nickerson, D.A. Sequence variation and linkage disequilibrium in the human T-cell receptor β (TCRB) locus. Am. J. Hum. Genet. 69, 381–395 (2001).
Dausset, J. et al. Centre d'etude du polymorphisme humain (CEPH): collaborative genetic mapping of the human genome. Genomics 6, 575–577 (1990).
Petes, T.D. Meiotic recombination hot spots and cold spots. Nat. Rev. Genet. 2, 360–369 (2001).
Hudson, R.R. Properties of a neutral allele model with intragenic recombination. Theor. Popul. Biol. 23, 183–201 (1983).
Griffiths, R.C. & Marjoram, P. Ancestral inference from samples of DNA sequences with recombination. J. Comput. Biol. 3, 479–502 (1996).
Kong, A. et al. A high-resolution recombination map of the human genome. Nat. Genet. 31, 242–247 (2002).
Smith, R.A., Ho, P.J., Clegg, J.B., Kidd, J.R. & Thein, S.L. Recombination breakpoints in the human β-globin gene cluster. Blood 92, 4415–4421 (1998).
Hartl, D.L. & Clark, A.G. Principles of Population Genetics (Sinauer Associates, Sunderland, Massachusetts, 1997).
Thompson, E. Pedigree Analysis in Human Genetics (Johns Hopkins University Press, Baltimore, Maryland, 1986).
McKeigue, P.M. Mapping genes underlying ethnic differences in disease risk by linkage disequilibrium in recently admixed populations. Am. J. Hum. Genet. 60, 188–196 (1997).
Bell, P.A. et al. SNPstream UHT: ultra-high throughput SNP genotyping for pharmacogenomics and drug discovery. Biotechniques (Suppl.) 70–72, 74, 76–77 (2002).
Fan, J.B. et al. Parallel genotyping of human SNPs using generic high-density oligonucleotide tag arrays. Genome Res. 10, 853–860 (2000).
Weir, B.S. Genetic Data Analysis II. 113–138 (Sinauer Associates, Sunderland, Massachusetts, 1996).
Abecasis, G.R., Cherny, S.S., Cookson, W.O. & Cardon, L.R. Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat. Genet. 30, 97–101 (2002).
Eberle, M.A. & Kruglyak, L. An analysis of strategies for discovery of single-nucleotide polymorphisms. Genet. Epidemiol. 19, S29–S35 (2000).
Acknowledgements
The authors thank J. Marcella, J. Ball and R. Tomacelli for advice and guidance during the development of this project at Orchid. R.S. thanks the cancer center at Cold Spring Harbor Laboratory for support. R.L., A.P.M., J.B. and L.R.C. were supported by the Wellcome Trust.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Authors affiliated with Orchid Biosciences are employed by or have personal financial interests in the company, which specializes in high-throughput genotyping technologies.
Rights and permissions
About this article
Cite this article
Phillips, M., Lawrence, R., Sachidanandam, R. et al. Chromosome-wide distribution of haplotype blocks and the role of recombination hot spots. Nat Genet 33, 382–387 (2003). https://doi.org/10.1038/ng1100
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng1100
This article is cited by
-
Genotype imputation for soybean nested association mapping population to improve precision of QTL detection
Theoretical and Applied Genetics (2022)
-
Exploring novel single nucleotide polymorphisms and haplotypes of the diacylglycerol O-acyltransferase 1 (DGAT1) gene and their effects on protein structure in Iranian buffalo
Genes & Genomics (2019)
-
Analysis of tomato meiotic recombination profile reveals preferential chromosome positions for NB-LRR genes
Euphytica (2017)
-
Linkage disequilibrium and haplotype block structure in a composite beef cattle breed
BMC Genomics (2014)
-
Evaluation of the long-term storage stability of saliva as a source of human DNA
Clinical Oral Investigations (2013)