Abstract
Avoiding destruction by immune cells is a hallmark of cancer, yet how tumors ultimately evade control by natural killer (NK) cells remains incompletely defined. Using global transcriptomic and flow-cytometry analyses and genetically engineered mouse models, we identified the cytokine-TGF-β-signaling-dependent conversion of NK cells (CD49a−CD49b+Eomes+) into intermediate type 1 innate lymphoid cell (intILC1) (CD49a+CD49b+Eomes+) populations and ILC1 (CD49a+CD49b−Eomesint) populations in the tumor microenvironment. Strikingly, intILC1s and ILC1s were unable to control local tumor growth and metastasis, whereas NK cells favored tumor immunosurveillance. Experiments with an antibody that neutralizes the cytokine TNF suggested that escape from the innate immune system was partially mediated by TNF-producing ILC1s. Our findings provide new insight into the plasticity of group 1 ILCs in the tumor microenvironment and suggest that the TGF-β-driven conversion of NK cells into ILC1s is a previously unknown mechanism by which tumors escape surveillance by the innate immune system.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Change history
21 March 2024
A Correction to this paper has been published: https://doi.org/10.1038/s41590-024-01799-9
References
Guillerey, C., Huntington, N.D. & Smyth, M.J. Targeting natural killer cells in cancer immunotherapy. Nat. Immunol. 17, 1025–1036 (2016).
Yang, L., Pang, Y. & Moses, H.L. TGF-beta and immune cells: an important regulatory axis in the tumor microenvironment and progression. Trends Immunol. 31, 220–227 (2010).
Wrzesinski, S.H., Wan, Y.Y. & Flavell, R.A. Transforming growth factor-β and the immune response: implications for anticancer therapy. Clin. Cancer Res. 13, 5262–5270 (2007).
Smyth, M.J., Strobl, S.L., Young, H.A., Ortaldo, J.R. & Ochoa, A.C. Regulation of lymphokine-activated killer activity and pore-forming protein gene expression in human peripheral blood CD8+ T lymphocytes. Inhibition by transforming growth factor-beta. J. Immunol. 146, 3289–3297 (1991).
Donatelli, S.S. et al. TGF-β-inducible microRNA-183 silences tumor-associated natural killer cells. Proc. Natl. Acad. Sci. USA 111, 4203–4208 (2014).
Wilson, E.B. et al. Human tumour immune evasion via TGF-β blocks NK cell activation but not survival allowing therapeutic restoration of anti-tumour activity. PLoS One 6, e22842 (2011).
Smyth, M.J. et al. CD4+CD25+ T regulatory cells suppress NK cell-mediated immunotherapy of cancer. J. Immunol. 176, 1582–1587 (2006).
Viel, S. et al. TGF-β inhibits the activation and functions of NK cells by repressing the mTOR pathway. Sci. Signal. 9, ra19 (2016).
Sojka, D.K. et al. Tissue-resident natural killer (NK) cells are cell lineages distinct from thymic and conventional splenic NK cells. eLife 3, e01659 (2014).
Constantinides, M.G., McDonald, B.D., Verhoef, P.A. & Bendelac, A. A committed precursor to innate lymphoid cells. Nature 508, 397–401 (2014).
Fuchs, A. ILC1s in tissue inflammation and infection. Front. Immunol. 7, 104 (2016).
Robinette, M.L. et al. Transcriptional programs define molecular characteristics of innate lymphoid cell classes and subsets. Nat. Immunol. 16, 306–317 (2015).
Seillet, C. et al. Differential requirement for Nfil3 during NK cell development. J. Immunol. 192, 2667–2676 (2014).
Gasteiger, G., Fan, X., Dikiy, S., Lee, S.Y. & Rudensky, A.Y. Tissue residency of innate lymphoid cells in lymphoid and nonlymphoid organs. Science 350, 981–985 (2015).
Klose, C.S. et al. Differentiation of type 1 ILCs from a common progenitor to all helper-like innate lymphoid cell lineages. Cell 157, 340–356 (2014).
Spits, H., Bernink, J.H. & Lanier, L. NK cells and type 1 innate lymphoid cells: partners in host defense. Nat. Immunol. 17, 758–764 (2016).
Seillet, C. & Belz, G.T. Differentiation and diversity of subsets in group 1 innate lymphoid cells. Int. Immunol. 28, 3–11 (2016).
Vallentin, B. et al. Innate Lymphoid Cells in Cancer. Cancer Immunol. Res. 3, 1109–1114 (2015).
Morvan, M.G. & Lanier, L.L. NK cells and cancer: you can teach innate cells new tricks. Nat. Rev. Cancer 16, 7–19 (2016).
Cortez, V.S. et al. Transforming growth factor-β signaling guides the differentiation of innate lymphoid cells in salivary glands. Immunity 44, 1127–1139 (2016).
Keskin, D.B. et al. TGFbeta promotes conversion of CD16+ peripheral blood NK cells into CD16− NK cells with similarities to decidual NK cells. Proc. Natl. Acad. Sci. USA 104, 3378–3383 (2007).
Narni-Mancinelli, E. et al. Fate mapping analysis of lymphoid cells expressing the NKp46 cell surface receptor. Proc. Natl. Acad. Sci. USA 108, 18324–18329 (2011).
Ruijtenberg, S. & van den Heuvel, S. Coordinating cell proliferation and differentiation: antagonism between cell cycle regulators and cell type-specific gene expression. Cell Cycle 15, 196–212 (2016).
Liberzon, A. et al. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 1, 417–425 (2015).
Smyth, M.J. et al. Differential tumor surveillance by natural killer (NK) and NKT cells. J. Exp. Med. 191, 661–668 (2000).
Smyth, M.J., Crowe, N.Y. & Godfrey, D.I. NK cells and NKT cells collaborate in host protection from methylcholanthrene-induced fibrosarcoma. Int. Immunol. 13, 459–463 (2001).
Sathe, P. et al. Innate immunodeficiency following genetic ablation of Mcl1 in natural killer cells. Nat. Commun. 5, 4539 (2014).
Knight, D.A. et al. Host immunity contributes to the anti-melanoma activity of BRAF inhibitors. J. Clin. Invest. 123, 1371–1381 (2013).
Krasnova, Y., Putz, E.M., Smyth, M.J. & Souza-Fonseca-Guimaraes, F. Bench to bedside: NK cells and control of metastasis. Clin. Immunol. 177, 50–59 (2017).
Chaput, N. et al. Phase I clinical trial combining imatinib mesylate and IL-2: HLA-DR+ NK cell levels correlate with disease outcome. OncoImmunology 2, e23080 (2013).
Ménard, C. et al. Natural killer cell IFN-gamma levels predict long-term survival with imatinib mesylate therapy in gastrointestinal stromal tumor-bearing patients. Cancer Res. 69, 3563–3569 (2009).
Fuchs, A. et al. Intraepithelial type 1 innate lymphoid cells are a unique subset of IL-12- and IL-15-responsive IFN-γ-producing cells. Immunity 38, 769–781 (2013).
Crome, S.Q. et al. A distinct innate lymphoid cell population regulates tumor-associated T cells. Nat. Med. 23, 368–375 (2017).
Dadi, S. et al. Cancer immunosurveillance by tissue-resident innate lymphoid cells and innate-like T cells. Cell 164, 365–377 (2016).
Pikovskaya, O. et al. Cutting edge: eomesodermin is sufficient to direct type 1 innate lymphocyte development into the conventional NK lineage. J. Immunol. 196, 1449–1454 (2016).
Cortez, V.S. et al. SMAD4 impedes the conversion of NK cells into ILC1-like cells by curtailing non-canonical TGF-β signaling. Nat. Immunol. doi:10.1038/ni.3809 (2016).
Hayakawa, Y. et al. IFN-gamma-mediated inhibition of tumor angiogenesis by natural killer T-cell ligand, α-galactosylceramide. Blood 100, 1728–1733 (2002).
Ikeda, H., Old, L.J. & Schreiber, R.D. The roles of IFNγ in protection against tumor development and cancer immunoediting. Cytokine Growth Factor Rev. 13, 95–109 (2002).
Balkwill, F. Tumour necrosis factor and cancer. Nat. Rev. Cancer 9, 361–371 (2009).
Baluk, P. et al. TNF-α drives remodeling of blood vessels and lymphatics in sustained airway inflammation in mice. J. Clin. Invest. 119, 2954–2964 (2009).
Sainson, R.C. et al. TNF primes endothelial cells for angiogenic sprouting by inducing a tip cell phenotype. Blood 111, 4997–5007 (2008).
Gill, S. et al. Rapid development of exhaustion and down-regulation of eomesodermin limit the antitumor activity of adoptively transferred murine natural killer cells. Blood 119, 5758–5768 (2012).
Doisne, J.M. et al. Composition, development, and function of uterine innate lymphoid cells. J. Immunol. 195, 3937–3945 (2015).
Levi, I. et al. Characterization of tumor infiltrating natural killer cell subset. Oncotarget 6, 13835–13843 (2015).
Bruno, A. et al. The proangiogenic phenotype of natural killer cells in patients with non-small cell lung cancer. Neoplasia 15, 133–142 (2013).
Lima, P.D., Zhang, J., Dunk, C., Lye, S.J. & Croy, B.A. Leukocyte driven-decidual angiogenesis in early pregnancy. Cell. Mol. Immunol. 11, 522–537 (2014).
Vacca, P. et al. Crosstalk between decidual NK and CD14+ myelomonocytic cells results in induction of Tregs and immunosuppression. Proc. Natl. Acad. Sci. USA 107, 11918–11923 (2010).
Arteaga, C.L. et al. Anti-transforming growth factor (TGF)-β antibodies inhibit breast cancer cell tumorigenicity and increase mouse spleen natural killer cell activity. Implications for a possible role of tumor cell/host TGF-β interactions in human breast cancer progression. J. Clin. Invest. 92, 2569–2576 (1993).
Terabe, M. et al. Synergistic enhancement of CD8+ T cell-mediated tumor vaccine efficacy by an anti-transforming growth factor-β monoclonal antibody. Clin. Cancer Res. 15, 6560–6569 (2009).
Morris, J.C. et al. Phase I study of GC1008 (fresolimumab): a human anti-transforming growth factor-β (TGFβ) monoclonal antibody in patients with advanced malignant melanoma or renal cell carcinoma. PLoS One 9, e90353 (2014).
Akhurst, R.J. & Hata, A. Targeting the TGFβ signalling pathway in disease. Nat. Rev. Drug Discov. 11, 790–811 (2012).
Viant, C. et al. Transforming growth factor-β and Notch ligands act as opposing environmental cues in regulating the plasticity of type 3 innate lymphoid cells. Sci. Signal. 9, ra46 (2016).
Kara, E.E. et al. CCR2 defines in vivo development and homing of IL-23-driven GM-CSF-producing Th17 cells. Nat. Commun. 6, 8644 (2015).
Johnstone, C.N. et al. Functional and molecular characterisation of EO771.LMB tumours, a new C57BL/6-mouse-derived model of spontaneously metastatic mammary cancer. Dis. Model. Mech. 8, 237–251 (2015).
Blake, S.J. et al. Suppression of metastases using a new lymphocyte checkpoint target for cancer immunotherapy. Cancer Discov. 6, 446–459 (2016).
Souza-Fonseca-Guimaraes, F. et al. NK cells require IL-28R for optimal in vivo activity. Proc. Natl. Acad. Sci. USA 112, E2376–E2384 (2015).
Delconte, R.B. et al. CIS is a potent checkpoint in NK cell-mediated tumor immunity. Nat. Immunol. 17, 816–824 (2016).
Rautela, J. et al. Loss of Host Type-I IFN signaling accelerates metastasis and impairs NK-cell antitumor function in multiple models of breast cancer. Cancer Immunol. Res. 3, 1207–1217 (2015).
Ngiow, S.F. et al. A threshold level of intratumor CD8+ T-cell PD1 expression dictates therapeutic response to anti-PD1. Cancer Res. 75, 3800–3811 (2015).
Liao, Y., Smyth, G.K. & Shi, W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 41, e108 (2013).
Law, C.W., Chen, Y., Shi, W. & Smyth, G.K. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15, R29 (2014).
Ritchie, M.E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550 (2005).
Lee, E., Chuang, H.Y., Kim, J.W., Ideker, T. & Lee, D. Inferring pathway activity toward precise disease classification. PLOS Comput. Biol. 4, e1000217 (2008).
Bald, T. et al. Ultraviolet-radiation-induced inflammation promotes angiotropism and metastasis in melanoma. Nature 507, 109–113 (2014).
Acknowledgements
We thank R. Schreiber (Washington University School of Medicine) for MCA1956 fibrosarcoma cells and anti-IFN-γ and anti-TNF hybridomas; the animal house and flow cytometry facilities at QIMR Berghofer Medical Research Institute and Walter and Eliza Hall Institute of Medical Research; E. Loza, K. Elder, L. Town, L. Spencer, T. Camilleri and T. Kratina, for mouse breeding, maintenance and genotyping; and K. MacDonald, D. Smith, A. Kallies, L. Beattie, R. Allan, G. Hill and S. Nutt for discussion, comments and advice on this project. Supported by the National Health and Medical Research Council of Australia (Senior Principal Research Fellowship 1078671 to M.J.S.; Peter Doherty Early Career Fellowship 1088703 to F.S.-F.-G. and 1124690 to T.B.; project grant 1027472 to G.T.B.; Elizabeth Blackburn NHMRC Fellowship to G.T.B.; Independent Research Institute Infrastructure Support scheme grant to G.T.B.; project grants 1066770 & 1057852 N.D.H.; and RD Wright Career development Fellowship 1112113 to N.W.), the Cancer Research Institute Clinical and Laboratory Integration Programs (M.J.S. and N.D.H.), Queensland Institute of Medical Research Berghofer International PhD Scholarship (Y.G. and J.Y.), University of Queensland International Scholarship (Y.G. and J.Y.), the National Breast Cancer Foundation (PF-15-008 to F.S.-F.-G.), Cure Cancer Australia (Priority-Driven Young Investigator Project Grant 1082709 and 1120725 to F.S.-F.-G.), European Molecular Biology Organization (long-term fellowship ALTF 945-2015 to T.B.), the Naito Foundation (K.N.), Cancer Council Queensland (PhD fellowship to A.Y.), Griffith University (PhD scholarships to S.S.N.), Inserm-Avenir-Grant (L.B.), Ligue Nationale Contre le Cancer (L.B.), Fondation ARC Pour la Recherche sur le Cancer (L.B.), the Victorian State Government Operational Infrastructure Scheme (G.T.B.), the Harry J Lloyd Charitable Trust (Melanoma Research Grant to N.D.H.) and the DFG Excellence Cluster Immunosensation (EXC 1023 to M.H.).
Author information
Authors and Affiliations
Contributions
Y.G., F.S.-F.-G., T.B., N.D.H., K.N. and M.J.S. designed research, supervised work and wrote the paper; Y.G., F.S.-F.-G., T.B., A.Y., S.F.N., J.R., S.J.B., J.Y., J.S.L., M.M., L.Z., N.D.H., K.N. and M.J.S. performed research; Y.G., F.S.-F.-G., T.B., S.S.N., A.Y., S.F.N., J.R., J.S., N.W., S.J.B., J.Y., M.M., L.Z., M.W.L.T., G.T.B., C.R.E., N.D.H., K.N., M.H. and M.J.S. analyzed data; and L.B., E.V., K.T. and G.T.B. provided experimental materials.
Competing Interests StatementM.J.S. has research agreements with Bristol-Myers Squibb, Corvus Pharmaceuticals and Aduro Biotech; E.V. is a cofounder and shareholder in Innate Pharma; and N.D.H. and J.R. are cofounders and shareholders in oNKo-Innate.
Corresponding author
Ethics declarations
Competing interests
M.J.S. has research agreements with Bristol-Myers Squibb, Corvus Pharmaceuticals and Aduro Biotech; E.V. is a cofounder and shareholder in Innate Pharma; and N.D.H. and J.R. are cofounders and shareholders in oNKo-Innate.
Integrated supplementary information
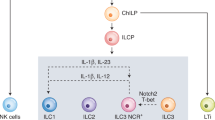
Supplementary Figure 1 Similarities among group 1 ILC subsets from tumor, liver and spleen.
(a) Representative flow cytometric plots illustrating the gating strategy for sorting of group 1 ILC subsets from MCA1956 tumors for transcriptomic analyses. (b) Representative flow cytometric plots illustrating post-sorting purity of tumor NK cells, intILC1s and ILC1s. (c) Heatmap visualizing gene expression profiles from NK cells and ILC1s isolated from liver/spleen (GSE52047) clustered by tumor NK cell and ILC1 gene signatures. (d) Quantification of gene signature expression by combined z-scores. Horizontal bars, median; boxes, 25th to 75th quartile; ‘whiskers’, 10th and 90th quartile. Statistical analysis by pairwise two-sided t-tests with Benjamini & Hochberg (FDR) correction for multiple testing. **P < 0.01, ****P < 0.0001; ns, non-significant. Sp: spleen; Liv: liver.
Supplementary Figure 2 TGF-b promotes NK cell conversion both in vitro and in vivo.
(a) Flow cytometric characterization of group 1 ILC composition based on CD49a and CD49b expression in livers and spleens of indicated transgenic mice. (b) Liver group 1 ILC composition as determined by flow cytometry based on CD49a and Eomes expression in indicated transgenic mice. (c) Corresponding quantification of liver group 1 ILC subsets amongst indicated transgenic mice. Data shown as mean ± s.e.m.; Mann-Whitney U-test; **P < 0.01. (a-c) Data represent n = 5 of one experiment. (d,e) Percentage and phenotype of NK-derived ILC1s (CD49a+Eomes−) in NK cell cultured for 5 days with 25 ng/mL rIL-15/IL-15Rα complex and TGF-β1 at indicated concentration. Statistical comparisons of in vitro NK cell-derived ILC1 percentage (d) and TRAILhiDNAM-1hi cell percentage in in vitro NK cell-derived ILC1s (e) at different rTGF-β1 concentrations. Data shown as mean ± s.e.m. of 5 replicates per group of one experiment. *P < 0.05, ***P < 0.001, ****P < 0.0001 determined by one-way ANOVA and Tukey’s multiple comparison test. (f) Purity of intILC1s and NK cells sorted from pooled spleens of indicated transgenic mice. (g) CD49a and CD49b expression profile of cells as sorted in (f) after cultured in serum-free medium in presence of 50 ng/mL rIL-15 for 4 days (n = 3 of one experiment). (h) Expression of CD49a and CD49b on splenic NK cells cultured in serum-free medium supplemented as indicated over time (n = 3 of one experiment). (i) Corresponding expression of CD49a and CD49b on liver ILC1s (n = 3 of one experiment).
Supplementary Figure 3 Analysis of proliferation-associated gene sets from liver and splenic NK cells and ILC1s.
Heatmap visualizing E2F gene set expression (a) and G2M checkpoint gene set expression (b) of liver/spleen NK cells and ILC1s (GSE52047) clustered by tumor NK cell and ILC1 gene signatures (left of each panel). Quantifications of gene set expression (right of each panel) by combined z-scores were compared by pairwise two-sided t-tests with Benjamini & Hochberg (FDR) correction for multiple testing. Horizontal lines in whisker boxplots represent quartiles. *P < 0.05; ns, non-significant. Sp: spleen; Liv: liver. Horizontal bars, median; boxes, 25th to 75th quartile; ‘whiskers’, 10th and 90th quartile.
Supplementary Figure 4 Mcl1FL mice are deficient in group 1 ILCs, while anti-asGM1 ‘preferentially’ depletes the tumor microenvironment of NK cells and intILC1s.
(a) Representative flow cytometric plots showing NK1.1+NKp46+ cells (left) and CD49a+CD49b− ILC1s and CD49a−CD49b+ NK cells (right) in the liver (top panel) and spleen (bottom panel) of Mcl1WT and Mcl1FL mice. (b) Corresponding quantifications of cell population in the liver (top panel) and spleen (bottom panel) as indicated. Rag2−/−γc−/− mice were used as negative control (mean ± s.e.m.; n = 5 for liver, n = 4 for spleen and n = 2 for Rag2−/−γc−/− mice from two independent experiments; unpaired two-sided t-tests; *P < 0.05, **P < 0.01). (c) Experimental setup for the treatment of MCA1956 tumor-bearing WT mice with two doses of 50 μg anti-asGM1 antibody or control IgG i.p. for two consecutive days. (d,e) Representative flow cytometric plots showing percentage of group 1 ILCs (in live CD45+Lin− population) (d) and group 1 ILC composition (e) in different tissues from mice treated as indicated (n = 5 for ctrl IgG treated group and n = 6 for anti-asGM1 antibody treated group of two independent experiments). (f-h) Corresponding quantifications of group 1 ILC subset number in the liver (f), spleen (g) and MCA1956 tumor (h) of mice treated as indicated (mean ± s.e.m.; n = 5 for ctrl IgG treated group and n = 6 for anti-asGM1 antibody treated group of two independent experiments; Mann-Whitney U-test; *P < 0.05, **P < 0.01).
Supplementary Figure 5 TGF-b signaling fosters tumor growth and NK cell conversion in SM1WT1 tumors.
(a) Representative flow cytometric plots showing group 1 ILC composition in SM1WT1 melanomas harvested from indicated transgenic mice at day 24 after tumor injection. ND, not determined. (b) Corresponding quantification of tumor group 1 ILC subsets (mean ± s.e.m.; n = 8 for Ncr1cre/wt mice, n = 7 for RIIFL mice and RICA-FL mice of two independent experiments; one-way ANOVA and Tukey’s multiple comparison test; ***P < 0.001, ****P < 0.0001). (c) Tumor growth of SM1WT1 melanomas in indicated transgenic mice (mean ± s.e.m.; n = 10 for Ncr1cre/wt, RICA-FL and RICA-WT mice, n = 5 for Mcl1WT and Mcl1FL mice of two independent experiments; one-way ANOVA and Tukey’s multiple comparison test; **P < 0.01). (d) Tumor growth of SM1WT1 melanomas in RIIFL and RIIWT mice treated with 50 μg control IgG or anti-asGM1 antibody i.p. on day -1, 0, 7, 14 and 21 before or after tumor cell injection at day 0 (mean ± s.e.m.; n = 10 for RIIFL and RIIWT mice of two independent experiments; one-way ANOVA and Tukey’s multiple comparison test; ***P < 0.001, ****P < 0.0001).
Supplementary Figure 6 TGF-β signaling in NKp46+ cells controls lung metastasis.
Lung metastasis in WT or indicated transgenic mice following i.v. injection of 2 × 105 RM-1 prostate carcinoma cells (a) and 3.5 × 105 EO771-LMB mCherry+ breast cancer cells (b-d). Total fluorescence radiant efficiency (b), relative mCherry mRNA expression (c) and representative fluorescence imaging (d) of lung metastases were shown. Results shown as mean ± s.e.m. and represent 3 mice per group (RICA-WT), 6 mice per group (RIIWT, RIIFL and RICA-FL), and 15 mice per group (WT and Ncr1cre/wt) for (a) and 6 mice per group (Ncr1cre/wt and RICA-FL), 5 mice per group (RIIFL and Mcl1FL) for (b-d) of one experiment. *P < 0.05, **P < 0.01, ****P < 0.0001 determined by one-way ANOVA and Tukey’s multiple comparison test.
Supplementary Figure 7 Expression of inhibitory immune cell receptors on tumor group 1 ILC subsets.
(a) Comparisons of inhibitory immune cell receptor expression among tumor group 1 ILC subsets isolated from s.c. transplanted MCA1956 tumors (mean ± s.e.m.; n = 5 of two experiments; one-way ANOVA and Tukey’s multiple comparison test; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; N.D., not detected). (b) Representative flow cytometric plots showing the expression of indicated receptors in tumor group 1 ILC subsets isolated from primary MCA-induced fibrosarcomas (n = 5 of one experiment). (c) Representative histogram showing NKG2A expression in tumor group 1 ILC subsets isolated from s.c. MCA1956 tumors (n = 5 of two independent experiments).
Supplementary Figure 8 Antibody-mediated neutralization of TNF impairs the growth of SM1WT1 melanomas.
(a) IFN-γ (left) and TNF (middle) production by tumor group 1 ILC subsets from SM1WT1 melanomas after 4 h stimulation by PMA/ionomycin. Percentages of cytokine producing cells were determined by flow cytometry and ratios of IFN-γ/TNF-producing cells (right) were calculated (mean ± s.e.m.; n = 7 tumors from one experiment; one-way ANOVA and Tukey’s multiple comparison; *P < 0.05, **P < 0.01). (b) Experimental setup for antibody-mediated cytokine neutralization in SM1WT1 melanoma-bearing mice. (c) Tumor growth in RIIWT (left) and RIIFL (right) mice treated as indicated (mean ± s.e.m.; n = 5 mice per group of two independent experiments; one-way ANOVA and Tukey’s multiple comparison test; ***P < 0.001, ****P < 0.0001). (d) Representative flow cytometric plots showing the gating strategy for the analyses of CXCR6 expression on CD3−CD56dim and CD3−CD56bright human NK cells from PBMC of healthy volunteers (HV) and GIST patients as well as tumor infiltrating lymphocytes (TILs) from GIST patients. (e) Representative flow cytometric plots showing the gating strategy for the analyses of CXCR6 expression on NK1.1+NKp46+ tumor group 1 ILCs in MCA1956 tumors (n = 5 per group of two independent experiments).
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–8 and Supplementary Tables 1 and 2 (PDF 1237 kb)
Rights and permissions
About this article
Cite this article
Gao, Y., Souza-Fonseca-Guimaraes, F., Bald, T. et al. Tumor immunoevasion by the conversion of effector NK cells into type 1 innate lymphoid cells. Nat Immunol 18, 1004–1015 (2017). https://doi.org/10.1038/ni.3800
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.3800
This article is cited by
-
Real-time ex vivo monitoring of NK cell migration toward obesity-associated oesophageal adenocarcinoma following modulation of CX3CR1
Scientific Reports (2024)
-
Natural killer cell therapies
Nature (2024)
-
NK cell-triggered CCL5/IFNγ-CXCL9/10 axis underlies the clinical efficacy of neoadjuvant anti-HER2 antibodies in breast cancer
Journal of Experimental & Clinical Cancer Research (2024)
-
Revitalizing allicin for cancer therapy: advances in formulation strategies to enhance bioavailability, stability, and clinical efficacy
Naunyn-Schmiedeberg's Archives of Pharmacology (2024)
-
Rapid functional impairment of natural killer cells following tumor entry limits anti-tumor immunity
Nature Communications (2024)