Abstract
The GAL4–UAS system is a powerful tool for manipulating gene expression, but its application in Caenorhabditis elegans has not been described. Here we systematically optimize the system's three main components to develop a temperature-optimized GAL4–UAS system (cGAL) that robustly controls gene expression in C. elegans from 15 to 25 °C. We demonstrate this system's utility in transcriptional reporter analysis, site-of-action experiments and exogenous transgene expression; and we provide a basic driver and effector toolkit.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Accession codes
References
Duffy, J.B. Genesis 34, 1–15 (2002).
Traven, A., Jelicic, B. & Sopta, M. EMBO Rep. 7, 496–499 (2006).
Triezenberg, S.J., Kingsbury, R.C. & McKnight, S.L. Genes Dev. 2, 718–729 (1988).
Pfeiffer, B.D. et al. Genetics 186, 735–755 (2010).
Beerli, R.R., Segal, D.J., Dreier, B. & Barbas, C.F. III. Proc. Natl. Acad. Sci. USA 95, 14628–14633 (1998).
Brand, A.H., Manoukian, A.S. & Perrimon, N. Methods Cell Biol. 44, 635–654 (1994).
Salvadó, Z. et al. Appl. Environ. Microbiol. 77, 2292–2302 (2011).
Hittinger, C.T. et al. Nature 464, 54–58 (2010).
Marmorstein, R., Carey, M., Ptashne, M. & Harrison, S.C. Nature 356, 408–414 (1992).
Thomas, J.H. Genetics 124, 855–872 (1990).
Wang, H. et al. Curr. Biol. 23, 746–754 (2013).
Mahoney, T.R. et al. Proc. Natl. Acad. Sci. USA 105, 16350–16355 (2008).
Boyden, E.S., Zhang, F., Bamberg, E., Nagel, G. & Deisseroth, K. Nat. Neurosci. 8, 1263–1268 (2005).
Lee, T. & Luo, L. Neuron 22, 451–461 (1999).
Ma, J. & Ptashne, M. Cell 50, 137–142 (1987).
Wei, X., Potter, C.J., Luo, L. & Shen, K. Nat. Methods 9, 391–395 (2012).
del Valle Rodríguez, A., Didiano, D. & Desplan, C. Nat. Methods 9, 47–55 (2011).
Hoier, E.F., Mohler, W.A., Kim, S.K. & Hajnal, A. Genes Dev. 14, 874–886 (2000).
Voutev, R. & Hubbard, E.J.A.A. Genetics 180, 103–119 (2008).
Davis, M.W., Morton, J.J., Carroll, D. & Jorgensen, E.M. PLoS Genet. 4, e1000028 (2008).
Brenner, S. Genetics 77, 71–94 (1974).
Webster, N., Jin, J.R., Green, S., Hollis, M. & Chambon, P. Cell 52, 169–178 (1988).
Zheng, Y., Brockie, P.J., Mellem, J.E., Madsen, D.M. & Maricq, A.V. Neuron 24, 347–361 (1999).
Chen, T.-W. et al. Nature 499, 295–300 (2013).
Zhang, F. et al. Nature 446, 633–639 (2007).
Pokala, N., Liu, Q., Gordus, A. & Bargmann, C.I. Proc. Natl. Acad. Sci. USA 111, 2770–2775 (2014).
Sweeney, S.T., Broadie, K., Keane, J., Niemann, H. & O'Kane, C.J. Neuron 14, 341–351 (1995).
Mello, C.C., Kramer, J.M., Stinchcomb, D. & Ambros, V. EMBO J. 10, 3959–3970 (1991).
Acknowledgements
We are grateful to A. Fire (Stanford University) for sharing unpublished results and to C.T. Hittinger (University of Wisconsin-Madison), D. Sieburth (University of Southern California), E.M. Jorgensen (University of Utah), C. Bargmann (Rockefeller University) and A. Fire (Stanford University) for reagents. We thank H. Korswagen (Hubrecht Institute) for thoughtful discussion. N.P. thanks C. Bargmann for her support. We thank M. Bao, Y.M. Kim, D. Leighton, J. DeModena and G. Medina for technical assistance; and WormBase for technical support. We also thank M. Kato, H. Schwartz, D. Angeles-Albores and other members of the Sternberg lab for editorial comments on the manuscript. Some strains were provided by the CGC, which is funded by the NIH Office of Research Infrastructure Programs (grant P40 OD010440). Some imaging was performed at the Caltech Biological Imaging Facility with the support of the Caltech Beckman Institute and the Arnold and Mabel Beckman Foundation. H.W. is supported by the Della Martin Fellowship. J.L. was supported by NIH grant T32GM007616. This work is supported by the Howard Hughes Medical Institute, with which P.W.S. is an investigator.
Author information
Authors and Affiliations
Contributions
H.W., J.L. and P.W.S. conceived the project. H.W. and J.L. performed the experiments, analyzed the data and wrote the paper. S.G. helped with molecular cloning and strain handling. E.M.S. devised the idea of trying Gal4p from yeast species with lower growth temperatures. C.M.C. and N.P. contributed reagents.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
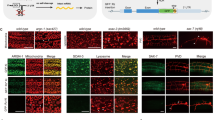
Supplementary Figure 1 Neither driver nor effector alone displays expression of GFP
Comparison of GFP fluorescence in a driver- only strain (a) or an effector-only strain (b) to their driver + effector combination controls. One-tailed t-test with Welch's correction.
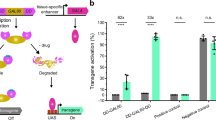
Supplementary Figure 2 Performance of different DBDs from Gal4 proteins at room temperature
Quantification of GFP fluorescence in the pharynx of transgenic worms with either Pmyo‑2::GAL4SC::VP64 or Pmyo‑2::GAL4SK::VP64 drivers injected into a strain carrying an integrated 15xUAS::gfp transgene (syIs300) at room temperature (22‑23°C). The drivers were both injected at 10 ng/μL. Strains with a direct Pmyo‑2::gfp fusion array at 10 ng/μL was measured for comparison. Two independent lines were imaged for each genotype. n = 20 ‑ 30 for each line. Bars are mean ± SEM. * p<0.05. ns, not significant. One‑way ANOVA with Tukey’s post-test. a.u., artificial units.
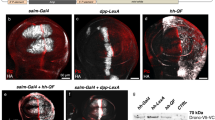
Supplementary Figure 3 Functional verification of integrated effectors
Expression of integrated drivers (left column) or integrated effectors alone (middle column) shows no basal expression. Only the combination (right column) show expression of cytoplasmic or nuclear-localized reporters, or death of appropriate cells. DIC, Differential interference contrast; Green, green filter; Red, red filter. Scale bar is 20 μm.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–3 and Supplementary Notes 1–6. (PDF 2321 kb)
Supplementary Table 1
Integrated cGAL drivers and effectors (XLSX 24 kb)
Supplementary Table 2
Plasmids and oligos used in this study (XLSX 98 kb)
Functional verification of a ChR2 effector with a GABAergic driver
Blue light induces paralysis in transgenic animals carrying a GABAergic driver and a channelrhodopsin (ChR2) effector (MP4 6543 kb)
Negative control of a ChR2 effector without driver
No response to blue light in transgenic animals only carrying a channelrhodopsin (ChR2) effector (MP4 7305 kb)
Functional verification of a GCaMP6s effector with a body wall muscle driver
Calcium imaging in body wall muscles of animals carrying a body wall muscle driver and a GCaMP6s::SL2::mKate2 effector (MP4 2681 kb)
Functional verification of a HisCl1 effector with a body wall muscle driver
Expressing a histamine-gated chloride channel HisCl1 in body wall muscle induces flaccid paralysis on histamine plates. Worms with either the driver or the effector alone fail to respond to histamine (MP4 7339 kb)
Functional verification of a TeTx effector with a GABAergic driver
Expressing a tetanus toxin light chain (TeTx) in GABAergic neurons blocks neurotransmission and leads to the characteristic “shrink” phenotype. Transgenic worms with either the driver or the effector don't display the “shrink” phenotype (MP4 1785 kb)
Rights and permissions
About this article
Cite this article
Wang, H., Liu, J., Gharib, S. et al. cGAL, a temperature-robust GAL4–UAS system for Caenorhabditis elegans. Nat Methods 14, 145–148 (2017). https://doi.org/10.1038/nmeth.4109
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.4109
This article is cited by
-
Neuronal subclass-selective proteomic analysis in Caenorhabditis elegans
Scientific Reports (2020)
-
Development of a Gateway-compatible two-component expression vector system for plants
Transgenic Research (2019)
-
Affinity purification of cell-specific mitochondria from whole animals resolves patterns of genetic mosaicism
Nature Cell Biology (2018)
-
Cellomics approach for high-throughput functional annotation of Caenorhabditis elegans neural network
Scientific Reports (2018)