Abstract
Stress responses are controlled by the hypothalamus pituitary adrenal (HPA)-axis and maladaptive stress responses are associated with the onset and maintenance of stress-related disorders such as major depressive disorder (MDD). Genes that play a role in the HPA-axis regulation may likely contribute to the relation between relevant neurobiological substrates and stress-related disorders. Therefore, we performed gene-wide analyses for 30 a priori literature-based genes involved in HPA-axis regulation in 2014 subjects (34% male; mean age: 42.5) to study the relations with lifetime MDD diagnosis, cortisol awakening response, and dexamethasone suppression test (DST) levels (subsample N=1472) and hippocampal and amygdala volume (3T MR images; subsample N=225). Additionally, gene by childhood maltreatment (CM) interactions were investigated. Gene-wide significant results were found for dexamethasone suppression (CYP11A1, CYP17A1, POU1F1, AKR1D1), hippocampal volume (CYP17A1, CYP11A1, HSD3B2, PROP1, AVPRA1, SRD5A1), amygdala volume (POMC, CRH, HSD3B2), and lifetime MDD diagnosis (FKBP5 and CRH), all permutation p-values<0.05. Interactions with CM were found for several genes; the strongest interactions were found for NR3C2, where the minor allele of SNP rs17581262 was related to smaller hippocampal volume, smaller amygdala volume, higher DST levels, and higher odds of MDD diagnosis only in participants with CM. As hypothesized, several HPA-axis genes are associated with stress-related endophenotypes including cortisol response and reduced brain volumes. Furthermore, we found a pleiotropic interaction between CM and the mineralocorticoid receptor gene, suggesting that this gene plays an important moderating role in stress and stress-related disorders.
Similar content being viewed by others
Introduction
Stress in the form of childhood maltreatment (CM), stressful life events or taking care of a sick child has been related to several detrimental health outcomes (Gouin et al, 2012; Steptoe and Kivimaki, 2012), including cardiovascular disease (Steptoe and Kivimaki, 2012), immune disorders (Bauer et al, 2009), and neuropsychiatric disorders, among which post-traumatic stress disorder (Liu et al, 2017)and major depressive disorder (MDD) (Nelson et al, 2017). The human stress response involves a complex succession of signals; when a stressor (either physiological or psychological) is encountered the hypothalamus releases corticotropin-releasing hormone (CRH) and vasopressin. CRH stimulates the anterior pituitary to release corticotropin and corticotropin activates the adrenal cortex to upregulate the production of glucocorticoids (GCs). Glucocorticoids are the primary stress hormones and their main function is to restore homeostasis following exposure to stress.
A maladaptive stress response or dysregulation of the hypothalamus pituitary adrenal (HPA)-axis is a frequent finding in MDD; a meta-analysis showed that MDD patients overall have higher levels of cortisol (Stetler and Miller, 2011). Further, it has been shown that high cortisol at the start of a psychological intervention is related to poorer outcomes (Fischer et al, 2017) and that the dexamethasone/CRH test could serve as a diagnostic test for MDD (Mokhtari et al, 2013), suggesting that dysregulation of the HPA-axis in MDD plays a clinically relevant role. Moreover, some studies showed that genetic variation in the GC receptor (GR) and mineralocorticoid receptor (MR) genes play a role in the etiology of MDD as well (Keller et al, 2017; Kuningas et al, 2007), although hypothesis-free genome-wide studies have not yet confirmed a role for these genes (Bond, 2015; Ripke et al, 2013). Additionally, interactions between some HPA-axis genes and exposure to stressors have repeatedly been found to play a role in the onset of MDD, for instance with the MR gene (Vinkers et al, 2015), the GR gene (Bet et al, 2009), and the FKBP5 gene (Zannas and Binder, 2014).
Particularly severe stressors experienced early in life, such as CM increase the risk of MDD onset (Kendler et al, 2004) and early-life stress has also been shown to have long-lasting effects on HPA-axis regulation and the brain (Teicher et al, 2016). Two limbic brain regions that are thought to be vulnerable to early-life stressors are the hippocampus (McEwen, 2007) and the amygdala (Roozendaal et al, 2009). Moreover, in humans smaller hippocampal volume is a consistent finding in MDD and both enlarged and decreased amygdala volumes have been observed in MDD (Kempton et al, 2011; Price and Drevets, 2012; Schmaal et al, 2015; van Eijndhoven et al, 2009).
Given the strong associations of HPA-axis regulation with stress-related disorders, brain structure and function, genes that play a role in HPA-axis regulation seem likely candidates to be investigated further. This current study therefore examines the relation between genetic variation in an literature-based a priori selection of 30 HPA-axis genes on one hand and cortisol levels (cortisol awakening response levels, a low-dose dexamethasone suppression test (DST) cortisol levels), brain volumes (amygdala and hippocampus) and clinical MDD diagnosis on the other hand, in a large cohort consisting of patients with MDD and healthy controls. We will do a look-up of the most clear pleiotropic genetic findings (that is, genes or SNPs that affect multiple phenotypes). Additionally, to investigate whether genetic variation in the HPA-axis would specifically play a role in interaction with stressors, we will also examine gene by CM interactions.
We will use gene-based methods, which will enable us to include jointly the total genetic variation, and by doing so we can increase our statistical power (Franke et al, 2010), opposed to looking solely at candidate single nucleotide polymorphisms.
Material and methods
Sample
The Netherlands Study of Depression and Anxiety (NESDA) is a large multicenter cohort study examining the course of depressive and anxiety disorders, including in total 2981 respondents recruited from the community, general practice care, and specialized mental health care. The study sample included persons with depressive or anxiety disorders as well as control subjects without a lifetime psychiatric diagnosis.
For NESDA’s objectives and methods see Penninx et al (2008). The research protocol was approved by the ethical committees of the participating universities and all participants provided written informed consent. Genome-wide data were available for 2327 participants (with North European ancestry and consent for genetic research) and these constitute the basic sample size for these analyses.
Genotyping
Whole blood was collected and DNA extracted as previously described (Boomsma et al, 2008) Genotyping was performed on Affymetrix Human Genome-Wide SNP 6.0 array (Affymetrix, Santa Clara, CA, USA). More details on genotyping can be found in the Supplementary Material.
We selected 30 genes which are known to play a role in HPA-axis regulation (Supplementary Table S1 in Supplementary Material), which were divided into three functional sets: GC biosynthesis (6 genes), HPA-axis control (15 genes), and GC metabolism (9 genes), as was defined in the paper by Velders et al (2011). Gene start and end positions were derived from UCSC genome browser gateway (NCBI37/HG19)±20 kB. In total 3437 SNPs in these 30 HPA-axis genes met the post imputation QC standards (MAF>0.05, INFO>0.8, INFO<1.1 and HWE<1e-3).
Phenotypes
Salivary cortisol measurement
At the baseline interview, respondents were instructed to collect saliva samples at home on a regular (preferably working) day.
Two cortisol indicators were used: the cortisol awakening response and cortisol suppression on the DST. We calculated the area under the curve with respect to the increase (AUCi) of the CAR using the formulas by Pruessner et al (2003). The AUCi is a measure of the dynamic of the CAR, more related to the sensitivity of the system and emphasizing changes over time.
In all, 1472 subjects (96.3%) had taken 0.5 mg of dexamethasone after 2300 hours on the first sampling day and were available for the DST analyses. To assess the magnitude of suppression we calculated the ratio between basal awakening cortisol and DST awakening cortisol. Because DST levels had a skewed distribution we log-transformed the DST data to obtain normally distributed data.
Hippocampal and amygdala volumes
A subset of the 301 NESDA participants (both patients and controls) participated in the NESDA-MRI study. The imaging protocol is described in the Supplementary Material.
Hippocampal and amygdala volumes were obtained using automatic subcortical volumetric segmentation with the FreeSurfer software. This fully automated process includes motion correction, removal of non-brain tissue, automated Talairach transformation, segmentation of the subcortical white matter and deep gray-matter volumetric structures, intensity normalization, and cortical reconstruction. This segmentation procedure assigns a neuroanatomical label to every voxel in the MR image volume. The method is based on probabilistic information estimated from a manually labeled training set. For 225 participants we had data available on both brain volumes and genotypes.
MDD diagnosis
The lifetime presence of MDD was assessed according to DSM-IV criteria using the Composite International Depression Interview (CIDI, version 2.1) (Robins et al, 1988), which was administered by trained research staff members. The CIDI is a structured interview that is applicable for use by trained non-clinicians and has shown good reliability and validity (Wittchen, 1994). Patients with an MDD diagnosis were contrasted against subjects without any lifetime psychiatric disorder as assessed using the CIDI.
Childhood Maltreatment
Childhood maltreatment was assessed with the Nemesis Trauma Interview (Spijker et al, 2002). Participants were asked whether they had experienced emotional neglect, psychological abuse, physical abuse, and/or sexual trauma before age 16. The number of different traumata encountered was combined with their frequencies, resulting in a sum score ranging from 0 to 8, as has been defined before (Peyrot et al, 2014). More details are described in the Supplementary Material.
Statistical Analysis
For each phenotype (MDD lifetime diagnosis, cortisol awakening response, DST cortisol, amygdala, and hippocampal volumes), we performed analyses within the three functional gene sets and subsequently performed gene-wide analyses. All analyses were adjusted for age, gender, three ancestry-informative principal components capturing population stratification in the Netherlands (Abdellaoui et al, 2013) and in case of brain volumes we additionally adjusted for intracranial volume.
Because there are several methods to perform gene-wide analyses (Mooney et al, 2014), we decided to use three frequently used methods (Gamma method, VEGAS, and Plink set-based test). Given that we already made a selection of genes for which we hypothesized that they are involved in cortisol levels, brain volumes, and MDD, we chose only self-contained gene-wide methods, opposed to competitive methods that are more suitable for hypothesis-free genome-wide data.
First, gene-set analysis within the three functional gene-sets was performed using the Plink Set-based tests and the Gamma method.
Second, we performed gene-wide analyses using the total genetic coverage within each gene using Plink Set-based tests, the Gamma method test, and VEGAS2 method (see Supplementary Material for further description of each method). To control for multiple comparison we first applied 10 000 permutations that controlled for the total number of SNP tests within each gene and second we applied FDR correction on gene-wide level using a q of 0.05; for each gene-wide and interaction test we used an FDR-corrected p-value of 0.008; based on the fact that we have 5 phenotypes × 30 genes.
In case of significant gene-wide findings, we looked for top SNPs and also for overlapping SNPs across all phenotypes. We expect that several genes will indeed play a role in more than one phenotype and therefore we particularly looked for overlapping top SNPs and their pleiotropic effects on multiple phenotypes.
We used online available data from the ENIGMA consortium (Hibar et al, 2015) to look up top hits for hippocampal and amygdala volume, data available from previous GWAS on serum morning cortisol (Bolton et al, 2014) to look up top hits of cortisol awakening response findings, and we used the online available Psychiatric Genomics Consortium (Ripke et al, 2013) data to look up our top findings on MDD.
Interaction with childhood maltreatment
Interactions with CM were examined on gene-wide level, using Plink and VEGAS2 methods. The summed CM score was added as a covariate and SNP by CM interaction terms were created. For further details on this analysis see the Supplementary Material.
Results
Data on age, gender, and CM was available for 2327 participants, including 1615 with a lifetime MDD disorder, 313 patients with an anxiety disorder, and 399 healthy controls. Table 1 shows the sample characteristics in further detail. Correlations among all phenotypes are shown in Supplementary Table S3.
Gene-Set Results
On HPA-axis function set level, only Set 1, consisting of genes that play a role in GC biosynthesis, had a set-wide nominally significant association with hippocampal volume (PLINK p-value 0.07 and Gamma method p-value=0.01). None of the other sets had a significant association with any of the phenotypes (Table 2; all p-values>0.05).
Gene-Wide Results
Table 2 also shows the results of the gene-wide analyses, organized by functional set and separate for each method.
Cortisol awakening response was not significantly related to any of the selected genes, although there was a borderline significant association with the SERPINA6 gene.
DST cortisol levels showed significant gene-wide findings for the genes LEP and AKR1D1. For hippocampal volume there were gene-wide significant associations with the genes CYP17A1, CYP11A1, and AVPRA1. Amygdala volume showed significant gene-wide associations with the genes POMC and CRH.
For lifetime diagnosis of MDD, there was a gene-wide significant finding for the gene FKBP5.
For all gene-wide analyses we examined SNP-wise tests; Supplementary Table S3 shows the top 20 most significant SNPs for each phenotype.
Two genes (CYP17A1 and CYP11A1) showed significant gene-wide associations with multiple phenotypes. For each of these significant gene-wide findings, we looked for overlapping significant SNPs within a significant gene among the phenotypes (Supplementary Table S4). Overlapping SNPs were found in the CYP17A1 (rs6162, rs6163, and rs743572) and CYP11A1 (rs80047157 and rs11072479) genes; these SNPs were associated with both DST cortisol and hippocampal volume. For the SNPs in the CYP11A1 gene, the minor allele was associated with higher DST cortisol levels and smaller hippocampal volume and for the SNPs in the CYP17A1 gene, the minor allele was associated with lower DST cortisol levels and larger hippocampal volume.
After adding smoking habits and alcohol use as covariates to these gene-wide analyses none of the results changed substantially (data not shown).
Look-Up of Gene-Wide Hits
Using the ENIGMA consortium data (on N=12 826 (Hibar et al, 2015)), we could replicate gene-wide significance for hippocampal volume in the gene AVPR1 (gene-wide P-value=0.03), but not for any of the other genes. Top SNP in AVPR1A, rs11174811 was associated with decreased hippocampal volume (NESDA: B=−258.8, p-value=0.01; ENIGMA: B=−10.6, p-value=0.03). For amygdala volume we could replicate the gene-wide significant finding in the CRH gene (gene-wide p=0.006), but not for any of the other hits. Top SNP rs12721510, in CRH was related with increased amygdala volume (NESDA: B=191.6, p-value=0.02 and ENIGMA: B=16.22, p-value=0.002). For more details please see Supplementary Table S5 in Supplementary Material.
Also for MDD we could replicate the gene-wide finding for FKBP5, in PGC (based on N=18 759 (Ripke et al, 2013)) (gene-wide p-value=0.002) and the overlapping top SNP rs4173904 was related to lower risk for MDD in both (NESDA: b=0.17, p-value=0.003 and PGC: B=−0.05, p-value=0.002).
For the cortisol awakening response we could not find any overlapping SNPs nor SNPs that are in LD; however, in both the GWAS (Bolton et al, 2014) and our study the SERPINA6 gene showed significant associations.
Interactions with CM
Gene-wide level
On gene-wide level we found several significant adjusted interactions with CM; Table 3 shows the gene-wide interaction p-values and Z-values stratified for CM− and CM+ for the most significant SNPs in each gene. For the NR3C2 gene we found interactions in all phenotypes that reached (borderline) significance considering an FDR-corrected p-value of 0.008.
NR3C2 gene
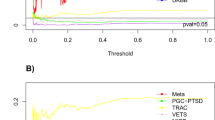
For four of our phenotypes there was a nominal significant interaction between CM and the NR3C2 gene; amygdala volume (p-value=0.001), hippocampal volume (p-value=0.012), DST cortisol (p-value=0.004) and MDD lifetime diagnosis (p-value=0.036). For each of these phenotypes, Figure 1 shows all p-values for interactions between each SNP in the NR3C2 gene and CM. We decided to focus on this result in more detail, because it showed overlapping effects in more than two phenotypes.
Interaction between NR3C2 gene and childhood maltreatment on four different phenotypes (MDD status, DST ratio cortisol, amygdala, and hippocampal volume). For every phenotype a different symbol was used (see legend in the figure) and each single symbol represents an association on that specific base pair locus (for which location in gene is depicted on x axis).
For all four phenotypes there was a significant interaction around SNP rs17581262 (Supplementary Figure S1 showing LD blocks). Figure 2 shows the stratified results for the AA genotype vs G-allele carriers; compared to individuals carrying one or more major alleles (genotypes AG and GG) homozygotes for the minor allele (genotype AA) of this SNP had smaller hippocampal volume (B=−403; p=0.006), smaller amygdala volume (B=−221; p-value=0.02), and higher levels of cortisol after DST (B=0.04; p=0.03) in case of CM, but no SNP effect was found in participants without CM on hippocampal volume (B=177; p=0.18), amygdala volume (B=97; p=0.13), or DST (B=0.001; p=0.91). Also we found that participants who had the AA genotype had a higher likelihood of having a lifetime diagnosis of MDD after experiencing CM (odds ratio= 1.95; 95%CI=1.59–2.41; p=0.0003) than participants who were carriers of one or two G-alleles (odds ratio= 1.49; 95%CI=1.25−1.65; p=0.008). The X2 for the difference in odds ratio between participants with CM and without CM for the effect of AA genotype on MDD lifetime diagnosis was a Z-score of 1.97, p-value=0.05.
Interaction between rs17581262 and childhood maltreatment (CM) on different phenotypes (hippocampal volume, amygdala volume, and DST ratio cortisol). *p-value<0.05. G-allele consists of GG and AG genotype. Adjusted for age, sex (hippocampal and amygdala volume also adjusted for ICV).
Discussion
Using gene-wide analyses, we showed that several genes known to be involved in HPA-axis regulation play a role in cortisol levels and also in hippocampal volume, amygdala volume, and MDD. More specifically, we found gene-wide effects for genes that have previously been shown to play a role in stress-related disorders, such as CRH on amygdala volume, AVPR1A on hippocampal volume, and FKBP5 on MDD. But we also found new hits of genes playing a role in the biosynthesis of GCs (CYP11A1 and CYP17A1) on hippocampal volume and a pleiotropic interaction with CM and the gene coding for the MR (NR3C2) on DST cortisol, hippocampal and amygdala volume and MDD status. The NR3C2 gene was more strongly associated with amygdala volume, hippocampal volume, and MDD status in participants with CM than in participants without CM.
When we focused on the top overlapping SNP in a locus that showed most significant interactions, we found that the minor allele of rs17581262 in participants with CM was related to lower amygdala volume, lower hippocampal volume, higher DST cortisol levels, and more lifetime MDD diagnosis, while there was no effect in participants without CM. The overlapping top SNP, rs17581262, is located in the intron region of NR3C2 and has not previously been associated with any neuropsychiatric phenotype. A recent study on genetic risk for preterm birth found two SNPs in this same locus that are in high LD (R2>0.7) with rs17581261 (Christiaens et al, 2015), suggesting that this region plays a role in multiple phenotypes.
The NR3C2 gene, coding for the MR, is widely distributed in the hippocampus and amygdala and it is thought that particularly the receptors in the limbic system play a role in HPA-axis inhibition. There is increasing evidence that the MR is also involved in susceptibility for stress-related disorders (Klok et al, 2011; Otte et al, 2007; van Leeuwen et al, 2011) and animal studies have shown the MR located in the amygdala and hippocampus mediates rapid cortisol effects and influences stress appraisal (Karst et al, 2010). As such, the MR is thought to play a role in the susceptibility for psychiatric disorders, particularly following exposure to severe stress.
Preceding studies on genetic variation in the NR3C2 gene have mostly focused on the well-described haplotype consisting of the 2G/C (rs2070951) and I180V (rs5522) SNPs in the NR3C2 promoter region that displays differential activity at the transcriptional, translational, and transactivational level (Joels et al, 2008). This haplotype has been associated with MDD risk in women (Klok et al, 2011), changes in amygdala activation (Bogdan et al, 2012) and increased subjective stress and cortisol stress response (van Leeuwen et al, 2011). Moreover, in the NESDA study it was previously found that there was no main effect of this haplotype on MDD and course of MDD (Hardeveld et al, 2015), but that in participants with a history of CM, the CA haplotype was related to depressive symptoms in men, but not in women (Vinkers et al, 2015). In the current study we found that for MDD, the most significant interaction with CM was found for the rs5520 SNP, which is in high LD with rs5522, but for all other phenotypes the most significant findings were around rs17581262, which has very low LD with the well-described haplotype.
Unlike previous studies we found little support for previously reported candidate SNPs in the GC gene NR3C1 and the cortico-tropin releasing hormone receptor genes CRHR1 and CRHR2. Previous studies that did report associations with MDD consisted of either different populations (eg psychotic depression (Keller et al, 2017)) or reported indirect effects via epigenetics (Bakusic et al, 2017). Furthermore another recent study like ours did not find support for any of the HPA-axis candidate genes and a role in MDD (Buttenschon et al, 2017). Owing to generally smaller sample sizes candidate SNP studies can be more prone to publication bias and biased estimates (Duncan and Keller, 2011).
We did however find support for the FKBP5 gene, which is in line with preceding studies, using candidate SNPs, that reported a role for the FKBP5 gene in onset of depression (Zobel et al, 2010), treatment response, and recurrence of depressive episodes (Binder et al, 2004). This gene-wide result could also be looked up in the PGC data and interestingly the SNP that reached significance in both data sets is a well-known SNP, rs4173904, which has been shown to play a role in the stress response (Binder et al, 2004).
Two other genes from the gene-set that are related to HPA-axis regulation showed significant associations, namely the gene coding for cortico-tropin releasing hormone (CRH) and the gene coding for the vasopressin receptor 1A (AVPR1A). The CRH gene was related to amygdala volume, which we could also replicate using the ENIGMA data. CRH stimulates pituitary ACTH secretion and thereby controls the activity of the HPA axis. Moreover, CRH containing neurons in the central nucleus of the amygdala innervate the locus coeruleus, whereby activating noradrenergic and sympathetic nervous systems. Furthermore, effects of CRH in limbic brain regions have been associated with increased fear, alertness, decreased appetite, and libido, all functions relevant in the fight or flight response and dysregulated in depression and anxiety disorders (Binder and Nemeroff, 2010). Given that the CRH gene is more expressed in the amygdala than in the hippocampus (Flandreau et al, 2012), it makes sense that we found a gene-wide result for the CRH gene with amygdala volume but not with hippocampal volume. The minor allele of the top SNP (rs12721510) was found to be related to increased amygdala volume. In preceding studies this SNP was found to be part of a functional haplotype that is associated with increased stress-induced alcohol use and aggressive temperament in primates (Barr et al, 2009, 2008).
The top SNP in AVPR1A gene that was related to smaller hippocampal volume is a known functional SNP that has previously been found to be associated with increased expression levels of AVPR1A in prefrontal cortex tissue (Maher et al, 2011) and another study demonstrated that this SNP disrupts a microRNA binding site (Nossent et al, 2011). We could also replicate this finding using the ENIGMA consortium, suggesting that genetic control of the vasopressin neurohumoral system also plays a role in hippocampal volume variation.
In our study, most genetic associations were found for hippocampal volume. Two of the genes (CYP17A1 and CYP11A1) associated with hippocampal volume code for enzymes that play a role in the biosynthesis of GCs and other neurosteroids and have not been previously linked to neuropsychiatric outcomes. These two genes are expressed throughout the brain (Munetomo et al, 2015). In our study, minor alleles of the CYP17A1 gene were related to smaller hippocampal volume and increased cortisol levels after the DST. The hippocampus plays an important role in the feedback loop of the HPA-axis and smaller hippocampal volume has previously been found to be related to higher DST cortisol levels (Knoops et al, 2010). Unfortunately no other (large) data set was available to look up the gene-wide hits we found on. DST cortisol is considered to be a proxy for stress-response cortisol and other gene-wide hits we found for DST were for the leptin gene LEP and the aldo-keto reductase family 1 gene (AKR1D1). Leptin has previously been linked to play a role in endocrine and neuropsychiatric disorders (Stieg et al, 2015) while AKR1D1 catalyzes the metabolism of steroid hormones (Penning, 2015), but has otherwise never been related to any stress-related disorder.
Unlike DST cortisol we found no gene-wide results on cortisol awakening response; however, there was a borderline significant effect with the SERPINA6 gene, which has also been reported in a recent genome-wide study on morning cortisol levels (Bolton et al, 2014). A major difference is that the GWAS was performed on one sample of serum cortisol, whereas we looked at the cortisol awakening response in saliva. Nevertheless our results do suggest that genetic variations in SERPINA6 can explain variation in cortisol awakening response levels and thus more research on this gene in cortisol and stress response research is warranted.
Strengths and Limitations
The largest strength of this study is that we applied gene-wide analyses using all available genetic variation in a selection of 30 genes relevant to HPA-axis functioning on various phenotypes. This method combines the strength of genome-wide association studies by looking at the total gene and the strength of candidate SNP analysis, by looking at a selection of genes of which we hypothesized that they would be related to the chosen phenotypes. Furthermore, by looking specifically for genetic findings that were present across several phenotypes we decreased the chance of false positive findings even more.
Ideally we would have had an independent sample to replicate all our findings; however, only few cohorts have similar wealth of data on MDD, CM, cortisol, and brain volumes. We could only look for replication of the genetic associations and not the interaction with CM. However the fact that we found significant interactions in the same locus for four out of five phenotypes could be seen as internal validation of the finding.
Cortisol is known to be influenced by a multitude of external factors, including age, lifestyle factors, and disease. In a previous study from our group we found that the cortisol measures as used in the current study (AUCi morning cortisol, DST, and evening cortisol) were not affected by lifestyle, somatic disease, or sampling factors (eg time of awakening or workday vs weekend day) (Vreeburg et al, 2009). It has also been suggested that multiple days of sampling are necessary to reliably measure salivary cortisol levels, especially directly after awakening (Hellhammer et al, 2007) but this was not an option in our large-scale study. However, these limitations regarding reliability of individual measures are likely to be compensated by the large sample size in this study.
Apart from MDD it would also be interesting to investigate to which extent genetic variation and the interaction with CM can explain other stress-related disorders, such as post-traumatic stress disorder and generalized anxiety disorder. Unfortunately our data were not suitable to investigate the relation with (specific) anxiety disorders, due to too small number of cases with anxiety disorder.
In conclusion, here we report that several genes that play a role in HPA-axis regulation are related to cortisol levels and other stress-related endophenotypes. More specifically, we found strong support that genetic variation in the genes CRH, AVPR1A and FKBP5 are related to amygdala hippocampal volume and MDD status, respectively. In addition, there was a pleiotropic interaction between CM and the MR gene on MDD, DST cortisol levels, and amygdala and hippocampal volumes, suggesting that the MR plays an important role in stress and stress-related disorders.
Funding and disclosure
The infrastructure for the NESDA study (www.nesda.nl) is funded through the Geestkracht program of The Netherlands Organization for Health Research and Development (Zon-Mw, grant number 10-000-1002) and is supported by participating universities and mental health-care organizations (VU University Medical Center, GGZ inGeest, Arkin, Leiden University Medical Center, GGZ Rivierduinen, University Medical Center Groningen, Lentis, GGZ Friesland, GGZ Drenthe, Institute for Quality of Health Care (IQ Healthcare), Netherlands Institute for Health Services Research (NIVEL) and Netherlands Institute of Mental Health and Addiction (Trimbos)). NESDA’s genetic data were supported by the National Institutes of Health (NIH, R01D0042157-01A, MH081802, Grand Opportunity grant 1RC2 MH089951), Center for Medical Systems Biology (CSMB, NWO Genomics) and the Biobanking and Biomolecular Resources Research Infrastructure (BBMRI-NL). CHV is supported by a Netherlands Organisation for Scientific Research VENI grant (451-13-001) and a Netherlands Brain Foundation Fellowship (F2013(1)-216). LG is supported by a Netherlands Organisation for Scientific Research VENI grant (916-14-016). BWJHP, LvV and LS are supported by a Netherlands Organisation for Scientific Research VICI grant (918-11-302), and has received research grant funding from Johnson & Johnson. LS gratefully acknowledges support from The Netherlands Brain Foundation (F2014(1)-24), the Neuroscience Campus Amsterdam (IPB-SE-15-PSYCH-Schmaal) and from the NIH Big Data to Knowledge (BD2K) award (U54 EB020403).
References
Abdellaoui A, Hottenga JJ, de Knijff P, Nivard MG, Xiao X, Scheet P et al (2013). Population structure, migration, and diversifying selection in the Netherlands. Eur J Hum Genet 21: 1277–1285.
Bakusic J, Schaufeli W, Claes S, Godderis L (2017). Stress, burnout and depression: a systematic review on DNA methylation mechanisms. J Psychosom Res 92: 34–44.
Barr CS, Dvoskin RL, Gupte M, Sommer W, Sun H, Schwandt ML et al (2009). Functional CRH variation increases stress-induced alcohol consumption in primates. Proc Natl Acad Sci USA 106: 14593–14598.
Barr CS, Dvoskin RL, Yuan Q, Lipsky RH, Gupte M, Hu X et al (2008). CRH haplotype as a factor influencing cerebrospinal fluid levels of corticotropin-releasing hormone, hypothalamic-pituitary-adrenal axis activity, temperament, and alcohol consumption in rhesus macaques. Arch Gen Psychiatry 65: 934–944.
Bauer ME, Jeckel CM, Luz C (2009). The role of stress factors during aging of the immune system. Ann NY Acad Sci 1153: 139–152.
Bet PM, Penninx BW, Bochdanovits Z, Uitterlinden AG, Beekman AT, van Schoor NM et al (2009). Glucocorticoid receptor gene polymorphisms and childhood adversity are associated with depression: new evidence for a gene-environment interaction. Am J Med Genet B Neuropsychiatr Genet 150B: 660–669.
Binder EB, Nemeroff CB (2010). The CRF system, stress, depression and anxiety-insights from human genetic studies. Mol Psychiatry 15: 574–588.
Binder EB, Salyakina D, Lichtner P, Wochnik GM, Ising M, Putz B et al (2004). Polymorphisms in FKBP5 are associated with increased recurrence of depressive episodes and rapid response to antidepressant treatment. Nat Genet 36: 1319–1325.
Bogdan R, Williamson DE, Hariri AR (2012). Mineralocorticoid receptor Iso/Val (rs5522) genotype moderates the association between previous childhood emotional neglect and amygdala reactivity. Am J Psychiatry 169: 515–522.
Bolton JL, Hayward C, Direk N, Lewis JG, Hammond GL, Hill LA et al (2014). Genome wide association identifies common variants at the SERPINA6/SERPINA1 locus influencing plasma cortisol and corticosteroid binding globulin. PLoS Genet 10: e1004474.
Bond D (2015). Overweight and obesity as risk factors for clinical and neurobiological illness progression in early-stage bipolar disorder. Bipolar Disorders 17 ((Bond D.) University of Minnesota, Laboratory for Neuropsychiatric Imaging, Minneapolis, United States), 15.
Boomsma DI, Willemsen G, Sullivan PF, Heutink P, Meijer P, Sondervan D et al (2008). Genome-wide association of major depression: description of samples for the GAIN Major Depressive Disorder Study: NTR and NESDA biobank projects. Eur J Hum Genet 16: 335–342.
Buttenschon HN, Krogh J, Nielsen MN, Kaerlev L, Nordentoft M, Mors O (2017). Association analyses of depression and genes in the hypothalamus–pituitary–adrenal axis. Acta Neuropsychiatr 29: 59–64.
Christiaens I, Ang QW, Gordon LN, Fang X, Williams SM, Pennell CE et al (2015). Two novel genetic variants in the mineralocorticoid receptor gene associated with spontaneous preterm birth. BMC Med Genet 16: 59.
Duncan LE, Keller MC (2011). A critical review of the first 10 years of candidate gene-by-environment interaction research in psychiatry. Am J Psychiatry 168: 1041–1049.
Fischer S, Strawbridge R, Vives AH, Cleare AJ (2017). Cortisol as a predictor of psychological therapy response in depressive disorders: systematic review and meta-analysis. Br J Psychiatry 210: 105–109.
Flandreau EI, Ressler KJ, Owens MJ, Nemeroff CB (2012). Chronic overexpression of corticotropin-releasing factor from the central amygdala produces HPA axis hyperactivity and behavioral anxiety associated with gene-expression changes in the hippocampus and paraventricular nucleus of the hypothalamus. Psychoneuroendocrinology 37: 27–38.
Franke B, Vasquez AA, Veltman JA, Brunner HG, Rijpkema M, Fernandez G (2010). Genetic variation in CACNA1C, a gene associated with bipolar disorder, influences brainstem rather than gray matter volume in healthy individuals. Biol Psychiatry 68: 586–588.
Gouin JP, Glaser R, Malarkey WB, Beversdorf D, Kiecolt-Glaser J (2012). Chronic stress, daily stressors, and circulating inflammatory markers. Health Psychol 31: 264–268.
Hardeveld F, Spijker J, Peyrot WJ, de Graaf R, Hendriks SM, Nolen WA et al (2015). Glucocorticoid and mineralocorticoid receptor polymorphisms and recurrence of major depressive disorder. Psychoneuroendocrinology 55: 154–163.
Hellhammer J, Fries E, Schweisthal OW, Schlotz W, Stone AA, Hagemann D (2007). Several daily measurements are necessary to reliably assess the cortisol rise after awakening: state- and trait components. Psychoneuroendocrinology 32: 80–86.
Hibar DP, Stein JL, Renteria ME, Arias-Vasquez A, Desrivieres S, Jahanshad N et al (2015). Common genetic variants influence human subcortical brain structures. Nature 520: 224–229.
Joels M, Karst H, DeRijk R, de Kloet ER (2008). The coming out of the brain mineralocorticoid receptor. Trends Neurosci 31: 1–7.
Karst H, Berger S, Erdmann G, Schutz G, Joels M (2010). Metaplasticity of amygdalar responses to the stress hormone corticosterone. Proc Natl Acad Sci USA 107: 14449–14454.
Keller J, Gomez R, Williams G, Lembke A, Lazzeroni L, Murphy GM Jr et al (2017). HPA axis in major depression: cortisol, clinical symptomatology and genetic variation predict cognition. Mol Psychiatry 22: 527–536.
Kempton MJ, Salvador Z, Munafo MR, Geddes JR, Simmons A, Frangou S et al (2011). Structural neuroimaging studies in major depressive disorder. Meta-analysis and comparison with bipolar disorder. Arch Gen Psychiatr 68: 675–690.
Kendler KS, Kuhn JW, Prescott CA (2004). Childhood sexual abuse, stressful life events and risk for major depression in women. Psychol Med 34: 1475–1482.
Klok MD, Giltay EJ, Van der Does AJ, Geleijnse JM, Antypa N, Penninx BW et al (2011). A common and functional mineralocorticoid receptor haplotype enhances optimism and protects against depression in females. Transl Psychiatry 1: e62.
Knoops AJ, Gerritsen L, van der GY, Mali WP, Geerlings MI (2010). Basal hypothalamic pituitary adrenal axis activity and hippocampal volumes: the SMART-Medea study. Biol Psychiatry 67: 1191–1198.
Kuningas M, de Rijk RH, Westendorp RG, Jolles J, Slagboom PE, van HD (2007). Mental performance in old age dependent on cortisol and genetic variance in the mineralocorticoid and glucocorticoid receptors. Neuropsychopharmacology 32: 1295–1301.
Liu H, Petukhova MV, Sampson NA, Aguilar-Gaxiola S, Alonso J, Andrade LH et al (2017). Association of DSM-IV posttraumatic stress disorder with traumatic experience type and history in the World Health Organization World mental health surveys. JAMA Psychiatry 74: 270–281.
Maher BS, Vladimirov VI, Latendresse SJ, Thiselton DL, McNamee R, Kang M et al (2011). The AVPR1A gene and substance use disorders: association, replication, and functional evidence. Biol Psychiatry 70: 519–527.
McEwen BS (2007). Physiology and neurobiology of stress and adaptation: central role of the brain. Physiol Rev 87: 873–904.
Mokhtari M, Arfken C, Boutros N (2013). The DEX/CRH test for major depression: a potentially useful diagnostic test. Psychiatry Res 208: 131–139.
Mooney MA, Nigg JT, McWeeney SK, Wilmot B (2014). Functional and genomic context in pathway analysis of GWAS data. Trends Genet 30: 390–400.
Munetomo A, Hojo Y, Higo S, Kato A, Yoshida K, Shirasawa T et al (2015). Aging-induced changes in sex-steroidogenic enzymes and sex-steroid receptors in the cortex, hypothalamus and cerebellum. J Physiol Sci 65: 253–263.
Nelson J, Klumparendt A, Doebler P, Ehring T (2017). Childhood maltreatment and characteristics of adult depression: meta-analysis. Br J Psychiatry 210: 96–104.
Nossent AY, Hansen JL, Doggen C, Quax PH, Sheikh SP, Rosendaal FR (2011). SNPs in microRNA binding sites in 3'-UTRs of RAAS genes influence arterial blood pressure and risk of myocardial infarction. Am J Hypertens 24: 999–1006.
Otte C, Moritz S, Yassouridis A, Koop M, Madrischewski AM, Wiedemann K et al (2007). Blockade of the mineralocorticoid receptor in healthy men: effects on experimentally induced panic symptoms, stress hormones, and cognition. Neuropsychopharmacology 32: 232–238.
Penning TM (2015). The aldo-keto reductases (AKRs): Overview. Chem Biol Interact 234: 236–246.
Penninx BW, Beekman AT, Smit JH, Zitman FG, Nolen WA, Spinhoven P et al (2008). The Netherlands Study of Depression and Anxiety (NESDA): rationale, objectives and methods. Int J Meth Psychiatr Res 17: 121–140.
Peyrot WJ, Milaneschi Y, Abdellaoui A, Sullivan PF, Hottenga JJ, Boomsma DI et al (2014). Effect of polygenic risk scores on depression in childhood trauma. Br J Psychiatry 205: 113–119.
Price JL, Drevets WC (2012). Neural circuits underlying the pathophysiology of mood disorders. Trends Cogn Sci 16: 61–71.
Pruessner JC, Kirschbaum C, Meinlschmid G, Hellhammer DH (2003). Two formulas for computation of the area under the curve represent measures of total hormone concentration versus time-dependent change. Psychoneuroendocrinology 28: 916–931.
Ripke S, Wray NR, Lewis CM, Hamilton SP, Weissman MM, Breen G et al (2013). A mega-analysis of genome-wide association studies for major depressive disorder. Mol Psychiatry 18: 497–511.
Robins LN, Wing J, Wittchen HU, Helzer JE, Babor TF, Burke J et al (1988). The Composite International Diagnostic Interview. An epidemiologic Instrument suitable for use in conjunction with different diagnostic systems and in different cultures. Arch Gen Psychiatry 45: 1069–1077.
Roozendaal B, McEwen BS, Chattarji S (2009). Stress, memory and the amygdala. Nat Rev Neurosci 10: 423–433.
Schmaal L, Veltman DJ, van Erp TG, Samann PG, Frodl T, Jahanshad N et al (2015). Subcortical brain alterations in major depressive disorder: findings from the ENIGMA Major Depressive Disorder working group. Mol Psychiatry 21: 806–812.
Spijker J, de GR, Bijl RV, Beekman AT, Ormel J, Nolen WA (2002). Duration of major depressive episodes in the general population: results from The Netherlands Mental Health Survey and Incidence Study (NEMESIS). Br J Psychiatry 181: 208–213.
Steptoe A, Kivimaki M (2012). Stress and cardiovascular disease. Nat Rev Cardiol 9: 360–370.
Stieg MR, Sievers C, Farr O, Stalla GK, Mantzoros CS (2015). Leptin: A hormone linking activation of neuroendocrine axes with neuropathology. PNEC 51: 47–57.
Stetler C, Miller GE (2011). Depression and hypothalamic–pituitary–adrenal activation: a quantitative summary of four decades of research. Psychosom Med 73: 114–126.
Teicher MH, Samson JA, Anderson CM, Ohashi K (2016). The effects of childhood maltreatment on brain structure, function and connectivity. Nat Rev Neurosci 17: 652–666.
van Eijndhoven P, van Wingen G, van Oijen K, Rijpkema M, Goraj B, Jan VR et al (2009). Amygdala volume marks the acute state in the early course of depression. Biol Psychiatry 65: 812–818.
van Leeuwen N, Bellingrath S, de Kloet ER, Zitman FG, DeRijk RH, Kudielka BM et al (2011). Human mineralocorticoid receptor (MR) gene haplotypes modulate MR expression and transactivation: implication for the stress response. Psychoneuroendocrinology 36: 699–709.
Velders FP, Kuningas M, Kumari M, Dekker MJ, Uitterlinden AG, Kirschbaum C et al (2011). Genetics of cortisol secretion and depressive symptoms: a candidate gene and genome wide association approach. Psychoneuroendocrinology 36: 1053–1061.
Vinkers CH, Joels M, Milaneschi Y, Gerritsen L, Kahn RS, Penninx BW et al (2015). Mineralocorticoid receptor haplotypes sex-dependently moderate depression susceptibility following childhood maltreatment. Psychoneuroendocrinology 54: 90–102.
Vreeburg SA, Kruijtzer BP, van Pelt J, van Dyck R, DeRijk RH, Hoogendijk WJ et al (2009). Associations between sociodemographic, sampling and health factors and various salivary cortisol indicators in a large sample without psychopathology. Psychoneuroendocrinology 34: 1109–1120.
Wittchen HU (1994). Reliability and validity studies of the WHO-Composite International Diagnostic Interview (CIDI): a critical review. J Psychiatr Res 28: 57–84.
Zannas AS, Binder EB (2014). Gene-environment interactions at the FKBP5 locus: sensitive periods, mechanisms and pleiotropism. Genes Brain Behav 13: 25–37.
Zobel A, Schuhmacher A, Jessen F, Hofels S, von Widdern O, Metten M et al (2010). DNA sequence variants of the FKBP5 gene are associated with unipolar depression. Int J Neuropsychopharmacol 13: 649–660.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on the Neuropsychopharmacology website
Supplementary information
PowerPoint slides
Rights and permissions
About this article
Cite this article
Gerritsen, L., Milaneschi, Y., Vinkers, C. et al. HPA Axis Genes, and Their Interaction with Childhood Maltreatment, are Related to Cortisol Levels and Stress-Related Phenotypes. Neuropsychopharmacol 42, 2446–2455 (2017). https://doi.org/10.1038/npp.2017.118
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/npp.2017.118
This article is cited by
-
The cortisol switch between vulnerability and resilience
Molecular Psychiatry (2023)
-
The biobehavioural effectiveness of spiritual-hypnosis-assisted therapy in PTSD with childhood trauma
The Egyptian Journal of Neurology, Psychiatry and Neurosurgery (2022)
-
The interplay between genetic variation and gene expression of the glucocorticoid receptor gene NR3C1 and blood cortisol levels on verbal memory and hippocampal volumes
European Archives of Psychiatry and Clinical Neuroscience (2022)
-
FKBP51 modulates hippocampal size and function in post-translational regulation of Parkin
Cellular and Molecular Life Sciences (2022)
-
Gaining a deeper understanding of social determinants of preterm birth by integrating multi-omics data
Pediatric Research (2021)