Key Points
-
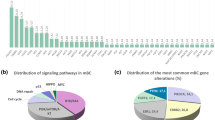
Recent research data defining the genomic landscape of breast cancer have reinforced the notion that this disease is driven by genomic alterations
-
So far very few gene drivers validated in breast cancer have been identified, including ESR1, ERBB 2, PIK3CA and ATK1
-
Identification of drivers, characterization of resistant clones, identification of DNA repair defects and mechanisms of immune suppression are potential uses of genomics to personalize medicine
-
The development of precision medicine for the treatment of breast cancer has several major challenges that include low frequency of targetable molecular alterations, feasibility of high-throughput technologies and availability of targeted therapy
Abstract
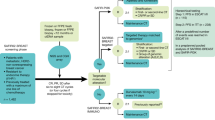
The development of precision medicine for the management of metastatic breast cancer is an appealing concept; however, major scientific and logistical challenges hinder its implementation in the clinic. The identification of driver mutational events remains the biggest challenge, because, with the few exceptions of ER, HER2, PIK3CA and AKT1, no validated oncogenic drivers of breast cancer exist. The development of bioinformatic tools to help identify driver mutations, together with assessment of pathway activation and dependency should help resolve this issue in the future. The occurrence of secondary resistance, such as ESR1 mutations, following endocrine therapy poses a further challenge. Ultra-deep sequencing and monitoring of circulating tumour DNA (ctDNA) could permit early detection of the genetic events underlying resistance and inform on combination therapy approaches. Beside these scientific challenges, logistical and operational issues are a major limitation to the development of precision medicine. For example, the low incidence of most candidate genomic alterations hinders randomized trials, as the number of patients to be screened would be too high. We discuss these limitations and the solutions, which include scaling-up the number of patients screened for identifying a genomic alteration, the clustering of genomic alterations into pathways, and the development of personalized medicine trials.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Jensen, E. V. & DeSombre, E. R. Oestrogen-receptor interaction. Science 182, 126–134 (1973).
Slamon, D. J. et al. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science 235, 177–182 (1987).
Osborne, C. K. Tamoxifen in the treatment of breast cancer. N. Engl. J. Med. 339, 1609–1618 (1998).
Dawood, S., Broglio, K., Buzdar, A. U., Hortobagyi, G. N. & Giordano, S. H. Prognosis of women with metastatic breast cancer by HER2 status and trastuzumab treatment: an institutional-based review. J. Clin. Oncol. 28, 92–98 (2010).
Kandoth, C. et al. Mutational landscape and significance across 12 major cancer types. Nature 502, 333–339 (2013).
Stephens, P. J. et al. The landscape of cancer genes and mutational processes in breast cancer. Nature 486, 400–404 (2012).
Hortobagyi, G. N. et al. Correlation of molecular alterations with efficacy of everolimus in hormone receptor–positive, HER2-negative advanced breast cancer: results from BOLERO-2 [abstract]. J. Clin. Oncol. 31 (Suppl.), LBA509 (2013).
Treilleux, I. et al. Translational studies within the TAMRAD randomized GINECO trial: evidence for mTORC1 activation marker as a predictive factor for everolimus efficacy in advanced breast cancer. Ann. Oncol. 26, 120–125 (2015).
Shah, S. P. et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature 486, 395–399 (2012).
Amir, E. et al. Prospective study evaluating the impact of tissue confirmation of metastatic disease in patients with breast cancer. J. Clin. Oncol. 30, 587–592 (2012).
Niikura, N. et al. Loss of human epidermal growth factor receptor 2 (HER2) expression in metastatic sites of HER2-overexpressing primary breast tumours. J. Clin. Oncol. 30, 593–599 (2012).
Toy, W. et al. ESR1 ligand-binding domain mutations in hormone-resistant breast cancer. Nat. Genet. 45, 1439–1445 (2013).
Robinson, D. R. et al. Activating ESR1 mutations in hormone-resistant metastatic breast cancer. Nat. Genet. 45, 1446–1451 (2013).
Arnedos, M. et al. Genomic and immune characterization of metastatic breast cancer (mBC): And ancillary study of the SAFIR01 & MOSCATO trials [abstract]. Ann. Oncol. 25, a351O (2014).
Massard, C. et al. Enriching phase I trials with molecular alterations: Interim analysis of 708 patients enrolled in the MOSCATO 01 trial [abstract]. 13th International Congress on Targeted Anticancer Therapies, Paris, France, aO3.7 (2015).
Lord, C. J. & Ashworth, A. Mechanisms of resistance to therapies targeting BRCA-mutant cancers. Nat. Med. 19, 1381–1388 (2013).
Wagle, N. et al. Whole exome sequencing (WES) of HER2+ metastatic breast cancer (MBC) from patients with or without prior trastuzumab (T): A correlative analysis of TBCRC003. Cancer Res. 75, PD3–PD5 (2015).
Hammond, M. E. et al. American Society of Clinical Oncology/College Of American Pathologists guideline recommendations for immunohistochemical testing of oestrogen and progesterone receptors in breast cancer. J. Clin. Oncol. 28, 2784–2795 (2010).
Wolff, A. C. et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J. Clin. Oncol. 31, 3997–4013 (2013).
Paik, S. et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N. Engl. J. Med. 351, 2817–2826 (2004).
van de Vijver, M. J. et al. A gene-expression signature as a predictor of survival in breast cancer. N. Engl. J. Med. 347, 1999–2009 (2002).
Sorlie, T. et al. Repeated observation of breast tumour subtypes in independent gene expression data sets. Proc. Natl Acad. Sci. USA 100, 8418–8423 (2003).
Loi, S. et al. PIK3CA genotype and a PIK3CA mutation-related gene signature and response to everolimus and letrozole in oestrogen receptor positive breast cancer. PLoS ONE 8, e53292 (2013).
Balko, J. M. et al. A gene expression signature of MEK pathway activation to predict survival in triple-negative breast cancer [abstract]. J. Clin. Oncol. 30 (Suppl.), a1024 (2012).
Andre, F. et al. Comparative genomic hybridisation array and DNA sequencing to direct treatment of metastatic breast cancer: a multicentre, prospective trial (SAFIR01/UNICANCER). Lancet Oncol. 15, 267–274 (2014).
Arnedos, M. et al. Array CGH and PIK3CA/AKT1 mutations to drive patients to specific targeted agents: a clinical experience in 108 patients with metastatic breast cancer. Eur. J. Cancer 48, 2293–2299 (2012).
Van Allen, E. M. et al. Whole-exome sequencing and clinical interpretation of formalin-fixed, paraffin-embedded tumour samples to guide precision cancer medicine. Nat. Med. 20, 682–688 (2014).
Watkins, J. A., Irshad, S., Grigoriadis, A. & Tutt, A. N. Genomic scars as biomarkers of homologous recombination deficiency and drug response in breast and ovarian cancers. Breast Cancer Res. 16, 211 (2014).
Dawson, S. J. et al. Analysis of circulating tumour DNA to monitor metastatic breast cancer. N. Engl. J. Med. 368, 1199–1209 (2013).
Murtaza, M. et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature 497, 108–112 (2013).
Garnett, M. J. et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature 483, 570–575 (2012).
Cancer Genome Atlas, N. Comprehensive molecular portraits of human breast tumours. Nature 490, 61–70 (2012).
Janku, F. et al. PI3K/AKT/mTOR inhibitors in patients with breast and gynecologic malignancies harbouring PIK3CA mutations. J. Clin. Oncol. 30, 777–782 (2012).
Andre, F. et al. Targeting FGFR with dovitinib (TKI258): preclinical and clinical data in breast cancer. Clin. Cancer Res. 19, 3693–3702 (2013).
Soria, J. C. et al. Phase I/IIa study evaluating the safety, efficacy, pharmacokinetics, and pharmacodynamics of lucitanib in advanced solid tumours. Ann. Oncol. 25, 2244–2251 (2014).
Bose, R. et al. Activating HER2 mutations in HER2 gene amplification negative breast cancer. Cancer Discov. 3, 224–237 (2013).
Finn, R. S. et al. The cyclin-dependent kinase 4/6 inhibitor palbociclib in combination with letrozole versus letrozole alone as first-line treatment of oestrogen receptor-positive, HER2-negative, advanced breast cancer (PALOMA-1/TRIO-18): a randomised phase 2 study. Lancet Oncol. 16, 25–35 (2015).
Malumbres, M. & Barbacid, M. Cell cycle, CDKs and cancer: a changing paradigm. Nat. Rev. Cancer 9, 153–166 (2009).
Zoncu, R., Efeyan, A. & Sabatini, D. M. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat. Rev. Mol. Cell. Biol. 12, 21–35 (2011).
Dickler, M. et al. A first-in-human phase I study to evaluate the oral selective oestrogen receptor degrader GDC-0810 (ARN-810) in postmenopausal women with oestrogen receptor+ HER2−, advanced/metastatic breast cancer. AACR [abstract CT231] (2015).
Juric, D. et al. Convergent loss of PTEN leads to clinical resistance to a PI(3)Kalpha inhibitor. Nature 518, 240–244 (2015).
Alexandrov, L. B. et al. Signatures of mutational processes in human cancer. Nature 500, 415–421 (2013).
Henderson, S., Chakravarthy, A., Su, X., Boshoff, C. & Fenton, T. R. APOBEC-mediated cytosine deamination links PIK3CA helical domain mutations to human papillomavirus-driven tumour development. Cell Rep. 7, 1833–1841 (2014).
Swisher, E. et al. ARIEL2: A phase 2 study to prospectively identify ovarian cancer patients likely to respond to rucaparib [abstract]. 26th EORTC-NCI-AACR Symposium on Molecular Targets and Cancer Therapeutics, a215 (2014).
Tutt, A. et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: a proof-of-concept trial. Lancet 376, 235–244 (2010).
Cui, Y., Palii, S. S., Innes, C. L. & Paules, R. S. Depletion of ATR selectively sensitizes ATM-deficient human mammary epithelial cells to ionizing radiation and DNA-damaging agents. Cell Cycle 13, 541–550 (2014).
Weber, A. M. & Ryan, A. J. ATM and ATR as therapeutic targets in cancer. Pharmacol. Ther. 149, 124–138 (2015).
Bartlett, J. M. et al. Predicting anthracycline benefit: TOP2A and CEP17-not only but also. J. Clin. Oncol. 33, 1680–1687 (2015).
Nanda, R. et al. A phase Ib study of pembrolizumab (MK-3475) in patients with advanced triple-negative breast cancer [abstract]. Cancer Res. 75, S1-09 (2015).
Snyder, A. et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N. Engl. J. Med. 371, 2189–2199 (2014).
Seliger, B. et al. Immune escape of melanoma: first evidence of structural alterations in two distinct components of the MHC class I antigen processing pathway. Cancer Res. 61, 8647–8650 (2001).
Tumeh, P. C. et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 515, 568–571 (2014).
Ignatiadis, M. et al. Gene modules and response to neoadjuvant chemotherapy in breast cancer subtypes: a pooled analysis. J. Clin. Oncol. 30, 1996–2004 (2012).
Vacchelli, E. et al. Loss-of-function alleles of P2RX7 and TLR4 fail to affect the response to chemotherapy in non-small cell lung cancer. Onco-immunology 1, 271–278 (2012).
Bendell, J. C. et al. Phase I, dose-escalation study of BKM120, an oral pan-class I PI3K inhibitor, in patients with advanced solid tumours. J. Clin. Oncol. 30, 282–290 (2012).
Lawrence, M. S. et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499, 214–218 (2013).
Mermel, C. H. et al. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 12, R41 (2011).
Taylor, B. S. et al. Functional copy-number alterations in cancer. PLoS ONE 3, e3179 (2008).
Dees, N. D. et al. MuSiC: identifying mutational significance in cancer genomes. Genome Res. 22, 1589–1598 (2012).
Hua, X. et al. DrGaP: a powerful tool for identifying driver genes and pathways in cancer sequencing studies. Am. J. Hum. Genet. 93, 439–451 (2013).
Gonzalez-Perez, A. et al. IntOGen-mutations identifies cancer drivers across tumour types. Nat. Methods 10, 1081–1082 (2013).
Rubio-Perez, C. et al. In silico prescription of anticancer drugs to cohorts of 28 tumour types reveals targeting opportunities. Cancer Cell 27, 382–396 (2015).
Suo, C. et al. Integration of somatic mutation, expression and functional data reveals potential driver genes predictive of breast cancer survival. Bioinformatics https://dx.doi.org/10.1093/bioinformatics/btv164 (2015).
Andre, F. et al. Everolimus for women with trastuzumab-resistant, HER2-positive, advanced breast cancer (BOLERO-3): a randomised, double-blind, placebo-controlled phase 3 trial. Lancet Oncol. 15, 580–591 (2014).
Andre, F. et al. Expression patterns and predictive value of phosphorylated AKT in early-stage breast cancer. Ann. Oncol. 19, 315–320 (2008).
Mayer, I. A. et al. Stand up to cancer phase Ib study of pan-phosphoinositide-3-kinase inhibitor buparlisib with letrozole in oestrogen receptor-positive/human epidermal growth factor receptor 2-negative metastatic breast cancer. J. Clin. Oncol. 32, 1202–1209 (2014).
Fritsch, C. et al. Characterization of the novel and specific PI3Kalpha inhibitor NVP-BYL719 and development of the patient stratification strategy for clinical trials. Mol. Cancer Ther. 13, 1117–1129 (2014).
Janku, F. et al. Phase I study of the PI3Kα inhibitor BYL719 plus fulvestrant in patients with PIK3CA-altered and wild type ER+/HER2− locally advanced or metastatic breast cancer [abstract]. Cancer Res. 74, PD5–5 (2014).
Fu, Y., Ferte, C., Soria, J. C., F., A. & Lefebvre, C. Combination of targetable gene alterations decreases anti-cancer drug treatment response in cell lines and patients [abstract P10]. Cancer Pharmacogenomics and Targeted Therapies, Hinxton, Cambridge, UK (2014).
Fontanella, C. et al. Does toxicity predict efficacy? Insight into the mechanism of action of lapatinib. J. Clin. Oncol. 32, 3458–3459 (2014).
Baselga, J. et al. Biomarker analyses in CLEOPATRA: a phase III, placebo-controlled study of pertuzumab in human epidermal growth factor receptor 2-positive, first-line metastatic breast cancer. J. Clin. Oncol. 32, 753–761 (2014).
Schneeweiss, A. et al. Evaluating the predictive value of biomarkers for efficacy outcomes in response to pertuzumab- and trastuzumab-based therapy: an exploratory analysis of the TRYPHAENA study. Breast Cancer Res. 16, R73 (2014).
Lito, P., Rosen, N. & Solit, D. B. Tumour adaptation and resistance to RAF inhibitors. Nat. Med. 19, 1401–1409 (2013).
Choi, Y. L. et al. EML4-ALK mutations in lung cancer that confer resistance to ALK inhibitors. N. Engl. J. Med. 363, 1734–1739 (2010).
Eirew, P. et al. Dynamics of genomic clones in breast cancer patient xenografts at single-cell resolution. Nature 518, 422–426 (2015).
Wang, Y. et al. Clonal evolution in breast cancer revealed by single nucleus genome sequencing. Nature 512, 155–160 (2014).
Kosaka, K. et al. Emergence of resistant variants detected by ultra-deep sequencing after asunaprevir and daclatasvir combination therapy in patients infected with hepatitis C virus genotype 1. J. Viral. Hepat. 22, 158–165 (2015).
De Mattos-Arruda, L. et al. Capturing intra-tumour genetic heterogeneity by de novo mutation profiling of circulating cell-free tumour DNA: a proof-of-principle. Ann. Oncol. 25, 1729–1735 (2014).
O'Reilly, K. E. et al. mTOR inhibition induces upstream receptor tyrosine kinase signalling and activates Akt. Cancer Res. 66, 1500–1508 (2006).
Atzori, F. et al. A phase I pharmacokinetic and pharmacodynamic study of dalotuzumab (MK-0646), an anti-insulin-like growth factor-1 receptor monoclonal antibody, in patients with advanced solid tumours. Clin. Cancer Res. 17, 6304–6312 (2011).
Chen, X. et al. Dual inhibition of PI3K and mTOR mitigates compensatory AKT activation and improves tamoxifen response in breast cancer. Mol. Cancer Res. 11, 1269–1278 (2013).
US National Library of Medicine. ClinicalTrials.gov[online], (2015).
Baselga, J. et al. Everolimus in postmenopausal hormone-receptor-positive advanced breast cancer. N. Engl. J. Med. 366, 520–529 (2012).
US National Library of Medicine. ClinicalTrials.gov[online], (2015).
Besse, B. et al. Neratinib (N) with or without temsirolimus (TEM) in patients (pts) with non-small cell lung cancer (NSCLC) carrying HER2 somatic mutations: an international randomized phase II study [abstract]. Ann. Oncol. 25, LBA39 (2014).
Hyman, D. M. et al. VE-BASKET, a first-in-kind, phase II, histology-independent “basket” study of vemurafenib (VEM) in nonmelanoma solid tumours harbouring BRAF V600 mutations (V600m) [abstract]. J. Clin. Oncol. 32 (Suppl. 5), a2533 (2014).
US National Library of Medicine. ClinicalTrials.gov[online], (2015).
Zardavas, D. et al. The AURORA initiative for metastatic breast cancer. Br. J. Cancer 111, 1881–1887 (2014).
US National Library of Medicine. ClinicalTrials.gov [online], (2015).
Bedard, P. L., Hansen, A. R., Ratain, M. J. & Siu, L. L. Tumour heterogeneity in the clinic. Nature 501, 355–364 (2013).
Kim, E. S. et al. The BATTLE trial: personalizing therapy for lung cancer. Cancer Discov. 1, 44–53 (2011).
Kaufman, B. et al. Olaparib monotherapy in patients with advanced cancer and a germline BRCA1/2 mutation. J. Clin. Oncol. 33, 244–250 (2015).
Yu, M. et al. Cancer therapy. Ex vivo culture of circulating breast tumour cells for individualized testing of drug susceptibility. Science 345, 216–220 (2014).
Singh, V. M. et al. Analysis of the effect of various decalcification agents on the quantity and quality of nucleic acid (DNA and RNA) recovered from bone biopsies. Ann. Diagn. Pathol. 17, 322–326 (2013).
US National Library of Medicine. ClinicalTrials.gov[online], (2014).
US National Library of Medicine. ClinicalTrials.gov[online], (2015).
US National Library of Medicine. ClinicalTrials.gov[online], (2013).
Acknowledgements
S.L. is supported by Cancer Council Victoria and the National Health and Medical Research Council of Australia (NHMRC). F.A. is supported by Grants from the ARC, Breast cancer Research Foundation, Odyssea and Operation Parrains Chercheurs.
Author information
Authors and Affiliations
Contributions
H.B. and F.A. developed the outline; F.A. wrote the sections related to genomics and modalities of development. All authors made a significant contribution to writing the manuscript, reviewing and/or editing the manuscript and approved the final version before submission.
Corresponding author
Ethics declarations
Competing interests
F.A. receives honourarium and has a research contract with AstraZeneca and Novartis. M.A. receives honourarium from Novartis. The other authors declare no competing interests.
Rights and permissions
About this article
Cite this article
Arnedos, M., Vicier, C., Loi, S. et al. Precision medicine for metastatic breast cancer—limitations and solutions. Nat Rev Clin Oncol 12, 693–704 (2015). https://doi.org/10.1038/nrclinonc.2015.123
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrclinonc.2015.123
This article is cited by
-
Circulating tumor DNA validity and potential uses in metastatic breast cancer
npj Breast Cancer (2024)
-
Survival benefit and biomarker analysis of pyrotinib or pyrotinib plus capecitabine for patients with HER2-positive metastatic breast cancer: a pooled analysis of two phase I studies
Biomarker Research (2023)
-
AKT/mTOR signaling modulates resistance to endocrine therapy and CDK4/6 inhibition in metastatic breast cancers
npj Precision Oncology (2023)
-
PLAGL2 increases adriamycin resistance and EMT in breast cancer cells by activating the Wnt pathway
Genes & Genomics (2023)
-
Subtypes in pancreatic ductal adenocarcinoma based on niche factor dependency show distinct drug treatment responses
Journal of Experimental & Clinical Cancer Research (2022)