Abstract
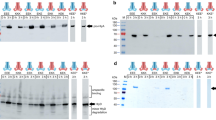
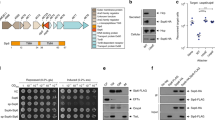
Bacteria have mechanisms to export proteins for diverse purposes, including colonization of hosts and pathogenesis. A small number of archetypal bacterial secretion machines have been found in several groups of bacteria and mediate a fundamentally distinct secretion process. Perhaps erroneously, proteins called 'autotransporters' have long been thought to be one of these protein secretion systems. Mounting evidence suggests that autotransporters might be substrates to be secreted, not an autonomous transporter system. We have discovered a new translocation and assembly module (TAM) that promotes efficient secretion of autotransporters in proteobacteria. Functional analysis of the TAM in Citrobacter rodentium, Salmonella enterica and Escherichia coli showed that it consists of an Omp85-family protein, TamA, in the outer membrane and TamB in the inner membrane of diverse bacterial species. The discovery of the TAM provides a new target for the development of therapies to inhibit colonization by bacterial pathogens.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
References
Gerlach, R.G. & Hensel, M. Protein secretion systems and adhesins: the molecular armory of Gram-negative pathogens. Int. J. Med. Microbiol. 297, 401–415 (2007).
Durand, E. et al. Structural biology of bacterial secretion systems in gram-negative pathogens–potential for new drug targets. Infect. Disord. Drug Targets 9, 518–547 (2009).
Holland, I.B. The extraordinary diversity of bacterial protein secretion mechanisms. Methods Mol. Biol. 619, 1–20 (2010).
Bernstein, H.D. Are bacterial 'autotransporters' really transporters? Trends Microbiol. 15, 441–447 (2007).
Ieva, R., Tian, P., Peterson, J.H. & Bernstein, H.D. Sequential and spatially restricted interactions of assembly factors with an autotransporter β-domain. Proc. Natl. Acad. Sci. USA 108, E383–E391 (2011).
Dautin, N. & Bernstein, H.D. Protein secretion in gram-negative bacteria via the autotransporter pathway. Annu. Rev. Microbiol. 61, 89–112 (2007).
Gentle, I.E., Burri, L. & Lithgow, T. Molecular architecture and function of the Omp85 family of proteins. Mol. Microbiol. 58, 1216–1225 (2005).
Jacob-Dubuisson, F., Villeret, V., Clantin, B., Delattre, A.S. & Saint, N. First structural insights into the TpsB/Omp85 superfamily. Biol. Chem. 390, 675–684 (2009).
Tommassen, J. Biochemistry. Getting into and through the outer membrane. Science 317, 903–904 (2007).
Bos, M.P., Robert, V. & Tommassen, J. Biogenesis of the gram-negative bacterial outer membrane. Annu. Rev. Microbiol. 61, 191–214 (2007).
Gentle, I., Gabriel, K., Beech, P., Waller, R. & Lithgow, T. The Omp85 family of proteins is essential for outer membrane biogenesis in mitochondria and bacteria. J. Cell Biol. 164, 19–24 (2004).
Pfanner, N., Wiedemann, N., Meisinger, C. & Lithgow, T. Assembling the mitochondrial outer membrane. Nat. Struct. Mol. Biol. 11, 1044–1048 (2004).
Schleiff, E. & Soll, J. Membrane protein insertion: mixing eukaryotic and prokaryotic concepts. EMBO Rep. 6, 1023–1027 (2005).
Hagan, C.L., Silhavy, T.J. & Kahne, D. β-Barrel membrane protein assembly by the Bam complex. Annu. Rev. Biochem. 80, 189–210 (2011).
Hagan, C.L. & Kahne, D. The reconstituted Escherichia coli Bam complex catalyzes multiple rounds of β-barrel assembly. Biochemistry 50, 7444–7446 (2011).
Gatzeva-Topalova, P.Z., Walton, T.A. & Sousa, M.C. Crystal structure of YaeT: conformational flexibility and substrate recognition. Structure 16, 1873–1881 (2008).
Kim, S. et al. Structure and function of an essential component of the outer membrane protein assembly machine. Science 317, 961–964 (2007).
Knowles, T.J. et al. Fold and function of polypeptide transport-associated domains responsible for delivering unfolded proteins to membranes. Mol. Microbiol. 68, 1216–1227 (2008).
Arnold, T., Zeth, K. & Linke, D. Omp85 from the thermophilic cyanobacterium Thermosynechococcus elongatus differs from proteobacterial Omp85 in structure and domain composition. J. Biol. Chem. 285, 18003–18015 (2010).
Clantin, B. et al. Structure of the membrane protein FhaC: a member of the Omp85-TpsB transporter superfamily. Science 317, 957–961 (2007).
Burall, L.S. et al. Proteus mirabilis genes that contribute to pathogenesis of urinary tract infection: identification of 25 signature-tagged mutants attenuated at least 100-fold. Infect. Immun. 72, 2922–2938 (2004).
Frickey, T. & Lupas, A. CLANS: a Java application for visualizing protein families based on pairwise similarity. Bioinformatics 20, 3702–3704 (2004).
Luirink, J., von Heijne, G., Houben, E. & de Gier, J.W. Biogenesis of inner membrane proteins in Escherichia coli. Annu. Rev. Microbiol. 59, 329–355 (2005).
Clements, A. et al. The reducible complexity of a mitochondrial molecular machine. Proc. Natl. Acad. Sci. USA 106, 15791–15795 (2009).
Stenberg, F., von Heijne, G. & Daley, D.O. Assembly of the cytochrome bo3 complex. J. Mol. Biol. 371, 765–773 (2007).
Ulett, G.C. et al. Functional analysis of antigen 43 in uropathogenic Escherichia coli reveals a role in long-term persistence in the urinary tract. Infect. Immun. 75, 3233–3244 (2007).
van der Woude, M.W. & Henderson, I.R. Regulation and function of Ag43 (flu). Annu. Rev. Microbiol. 62, 153–169 (2008).
Wells, T.J. et al. EhaA is a novel autotransporter protein of enterohemorrhagic Escherichia coli O157:H7 that contributes to adhesion and biofilm formation. Environ. Microbiol. 10, 589–604 (2008).
Klemm, P., Hjerrild, L., Gjermansen, M. & Schembri, M.A. Structure-function analysis of the self-recognizing Antigen 43 autotransporter protein from Escherichia coli. Mol. Microbiol. 51, 283–296 (2004).
Ieva, R., Skillman, K.M. & Bernstein, H.D. Incorporation of a polypeptide segment into the β-domain pore during the assembly of a bacterial autotransporter. Mol. Microbiol. 67, 188–201 (2008).
Jain, S. & Goldberg, M.B. Requirement for YaeT in the outer membrane assembly of autotransporter proteins. J. Bacteriol. 189, 5393–5398 (2007).
Rossiter, A.E. et al. The essential BAM complex components BamD and BamA are required for autotransporter biogenesis. J. Bacteriol. 193, 4250–4253 (2011).
Struve, C., Forestier, C. & Krogfelt, K.A. Application of a novel multi-screening signature-tagged mutagenesis assay for identification of Klebsiella pneumoniae genes essential in colonization and infection. Microbiology 149, 167–176 (2003).
Pusnik, M. et al. Mitochondrial preprotein translocase of trypanosomatids has a bacterial origin. Curr. Biol. 21, 1738–1743 (2011).
Alcock, F., Clements, A., Webb, C. & Lithgow, T. Evolution. Tinkering inside the organelle. Science 327, 649–650 (2010).
Anwari, K. et al. A modular BAM complex in the outer membrane of the α-proteobacterium Caulobacter crescentus. PLoS ONE 5, e8619 (2010).
Kelly, M. et al. Essential role of the type III secretion system effector NleB in colonization of mice by Citrobacter rodentium. Infect. Immun. 74, 2328–2337 (2006).
Bendtsen, J.D., Nielsen, H., von Heijne, G. & Brunak, S. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340, 783–795 (2004).
McGuffin, L.J., Bryson, K. & Jones, D.T. The PSIPRED protein structure prediction server. Bioinformatics 16, 404–405 (2000).
Owen, P., Meehan, M., de Loughry-Doherty, H. & Henderson, I. Phase-variable outer membrane proteins in Escherichia coli. FEMS Immunol. Med. Microbiol. 16, 63–76 (1996).
Acknowledgements
We thank D. Daley from the Department of Chemistry and Biophysics at Stockholm University for reagents, J. Rossjohn for critical discussions and A. Traven and T. Beilharz for critical comments on the manuscript. This work was made possible by the National Health & Medical Research Council (NHMRC) of Australia through an NHMRC Project grant (508911, to T.L. and A.W.P.), an NHMRC Program grant (606788, to T.L., E.L.H. and R.A.S.), an ARC Super Science grant (to T.L. and R.A.S.) and a BBSRC New Investigator grant to D.W. T.L. is an Australian Research Council Federation Fellow, E.L.H. and M.A.S. are ARC Future Fellows, A.W.P. is an NHMRC Senior Research Fellow, C.T.W., M.J.B. and A.J.P. are NHMRC Biomedical Fellows, D.L.L. is an ARC Super Science Fellow and J.S. is supported by an NHMRC Postgraduate Research Scholarship.
Author information
Authors and Affiliations
Contributions
J.S. initiated the project. J.S., C.T.W., M.-D.P., A.J.P., N.C., K.M. and M.J.B. conducted experiments and data analysis. I.R.H., T.J.W., F.M., M.T., R.M.R.-B., M.D.-P., C.O., E.L.H. D.W. and M.K. engineered bacterial strains and conducted experiments and data analysis. A.W.P. and S.H.R carried out and analyzed data from MS experiments. T.L. supervised the project, and T.L., J.S., C.T.W., M.J.B., R.A.S., I.R.H., D.L.L. and M.A.S. contributed to writing the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–3, Supplementary Table 1 and Supplementary Methods (PDF 418 kb)
Rights and permissions
About this article
Cite this article
Selkrig, J., Mosbahi, K., Webb, C. et al. Discovery of an archetypal protein transport system in bacterial outer membranes. Nat Struct Mol Biol 19, 506–510 (2012). https://doi.org/10.1038/nsmb.2261
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2261
This article is cited by
-
Surveying membrane landscapes: a new look at the bacterial cell surface
Nature Reviews Microbiology (2023)
-
A multipoint guidance mechanism for β-barrel folding on the SAM complex
Nature Structural & Molecular Biology (2023)
-
Mitochondrial sorting and assembly machinery operates by β-barrel switching
Nature (2021)
-
Recent advances in the understanding of trimeric autotransporter adhesins
Medical Microbiology and Immunology (2020)
-
Remote homology searches identify bacterial homologues of eukaryotic lipid transfer proteins, including Chorein-N domains in TamB and AsmA and Mdm31p
BMC Molecular and Cell Biology (2019)