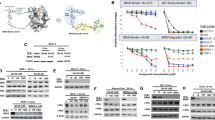
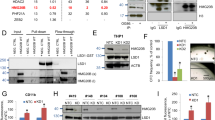
Abstract
Targeted therapies against disruptor of telomeric silencing 1-like (DOT1L) and bromodomain-containing protein 4 (BRD4) are currently being evaluated in clinical trials. However, the mechanisms by which BRD4 and DOT1L regulate leukemogenic transcription programs remain unclear. Using quantitative proteomics, chemoproteomics and biochemical fractionation, we found that native BRD4 and DOT1L exist in separate protein complexes. Genetic disruption or small-molecule inhibition of BRD4 and DOT1L showed marked synergistic activity against MLL leukemia cell lines, primary human leukemia cells and mouse leukemia models. Mechanistically, we found a previously unrecognized functional collaboration between DOT1L and BRD4 that is especially important at highly transcribed genes in proximity to superenhancers. DOT1L, via dimethylated histone H3 K79, facilitates histone H4 acetylation, which in turn regulates the binding of BRD4 to chromatin. These data provide new insights into the regulation of transcription and specify a molecular framework for therapeutic intervention in this disease with poor prognosis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Stratton, M.R., Campbell, P.J. & Futreal, P.A. The cancer genome. Nature 458, 719–724 (2009).
Dawson, M.A. & Kouzarides, T. Cancer epigenetics: from mechanism to therapy. Cell 150, 12–27 (2012).
Patel, J.P. et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N. Engl. J. Med. 366, 1079–1089 (2012).
Cancer Genome Atlas Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 368, 2059–2074 (2013).
Cai, S.F., Chen, C.W. & Armstrong, S.A. Drugging chromatin in cancer: recent advances and novel approaches. Mol. Cell 60, 561–570 (2015).
Dawson, M.A., Kouzarides, T. & Huntly, B.J. Targeting epigenetic readers in cancer. N. Engl. J. Med. 367, 647–657 (2012).
Rathert, P. et al. Transcriptional plasticity promotes primary and acquired resistance to BET inhibition. Nature 525, 543–547 (2015).
Prebet, T. et al. Prolonged administration of azacitidine with or without entinostat for myelodysplastic syndrome and acute myeloid leukemia with myelodysplasia-related changes: results of the US Leukemia Intergroup trial E1905. J. Clin. Oncol. 32, 1242–1248 (2014).
Issa, J.P. et al. Results of phase 2 randomized study of low-dose decitabine with or without valproic acid in patients with myelodysplastic syndrome and acute myelogenous leukemia. Cancer 121, 556–561 (2015).
Yu, B.D., Hess, J.L., Horning, S.E., Brown, G.A. & Korsmeyer, S.J. Altered Hox expression and segmental identity in Mll-mutant mice. Nature 378, 505–508 (1995).
McMahon, K.A. et al. Mll has a critical role in fetal and adult hematopoietic stem cell self-renewal. Cell Stem Cell 1, 338–345 (2007).
Jude, C.D. et al. Unique and independent roles for MLL in adult hematopoietic stem cells and progenitors. Cell Stem Cell 1, 324–337 (2007).
Neff, T. & Armstrong, S.A. Recent progress toward epigenetic therapies: the example of mixed lineage leukemia. Blood 121, 4847–4853 (2013).
Dawson, M.A. et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature 478, 529–533 (2011).
Daigle, S.R. et al. Selective killing of mixed lineage leukemia cells by a potent small-molecule DOT1L inhibitor. Cancer Cell 20, 53–65 (2011).
Grembecka, J. et al. Menin-MLL inhibitors reverse oncogenic activity of MLL fusion proteins in leukemia. Nat. Chem. Biol. 8, 277–284 (2012).
Harris, W.J. et al. The histone demethylase KDM1A sustains the oncogenic potential of MLL-AF9 leukemia stem cells. Cancer Cell 21, 473–487 (2012).
Mohan, M., Lin, C., Guest, E. & Shilatifard, A. Licensed to elongate: a molecular mechanism for MLL-based leukaemogenesis. Nat. Rev. Cancer 10, 721–728 (2010).
Biswas, D. et al. Function of leukemogenic mixed lineage leukemia 1 (MLL) fusion proteins through distinct partner protein complexes. Proc. Natl. Acad. Sci. USA 108, 15751–15756 (2011).
Bitoun, E., Oliver, P.L. & Davies, K.E. The mixed-lineage leukemia fusion partner AF4 stimulates RNA polymerase II transcriptional elongation and mediates coordinated chromatin remodeling. Hum. Mol. Genet. 16, 92–106 (2007).
Lin, C. et al. AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol. Cell 37, 429–437 (2010).
Mohan, M. et al. Linking H3K79 trimethylation to Wnt signaling through a novel Dot1-containing complex (DotCom). Genes Dev. 24, 574–589 (2010).
Mueller, D. et al. A role for the MLL fusion partner ENL in transcriptional elongation and chromatin modification. Blood 110, 4445–4454 (2007).
Mueller, D. et al. Misguided transcriptional elongation causes mixed lineage leukemia. PLoS Biol. 7, e1000249 (2009).
Yokoyama, A., Lin, M., Naresh, A., Kitabayashi, I. & Cleary, M.L. A higher-order complex containing AF4 and ENL family proteins with P-TEFb facilitates oncogenic and physiologic MLL-dependent transcription. Cancer Cell 17, 198–212 (2010).
Yu, W. et al. Catalytic site remodelling of the DOT1L methyltransferase by selective inhibitors. Nat. Commun. 3, 1288 (2012).
Zuber, J. et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature 478, 524–528 (2011).
Chen, C.W. et al. DOT1L inhibits SIRT1-mediated epigenetic silencing to maintain leukemic gene expression in MLL-rearranged leukemia. Nat. Med. 21, 335–343 (2015).
Shi, J. et al. Discovery of cancer drug targets by CRISPR-Cas9 screening of protein domains. Nat. Biotechnol. 33, 661–667 (2015).
Bernt, K.M. et al. MLL-rearranged leukemia is dependent on aberrant H3K79 methylation by DOT1L. Cancer Cell 20, 66–78 (2011).
Fong, C.Y. et al. BET inhibitor resistance emerges from leukaemia stem cells. Nature 525, 538–542 (2015).
Nguyen, A.T. & Zhang, Y. The diverse functions of Dot1 and H3K79 methylation. Genes Dev. 25, 1345–1358 (2011).
Lovén, J. et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 153, 320–334 (2013).
Pott, S. & Lieb, J.D. What are super-enhancers? Nat. Genet. 47, 8–12 (2015).
Yang, L. et al. lncRNA-dependent mechanisms of androgen-receptor-regulated gene activation programs. Nature 500, 598–602 (2013).
LeRoy, G. et al. Proteogenomic characterization and mapping of nucleosomes decoded by Brd and HP1 proteins. Genome Biol. 13, R68 (2012).
Zhao, R., Nakamura, T., Fu, Y., Lazar, Z. & Spector, D.L. Gene bookmarking accelerates the kinetics of post-mitotic transcriptional re-activation. Nat. Cell Biol. 13, 1295–1304 (2011).
Lee, J.S., Smith, E. & Shilatifard, A. The language of histone crosstalk. Cell 142, 682–685 (2010).
Shankar, D.B. et al. The role of CREB as a proto-oncogene in hematopoiesis and in acute myeloid leukemia. Cancer Cell 7, 351–362 (2005).
Henry, R.A., Kuo, Y.M. & Andrews, A.J. Differences in specificity and selectivity between CBP and p300 acetylation of histone H3 and H3/H4. Biochemistry 52, 5746–5759 (2013).
Roe, J.S., Mercan, F., Rivera, K., Pappin, D.J. & Vakoc, C.R. BET bromodomain inhibition suppresses the function of hematopoietic transcription factors in acute myeloid leukemia. Mol. Cell 58, 1028–1039 (2015).
Lavallée, V.P. et al. The transcriptomic landscape and directed chemical interrogation of MLL-rearranged acute myeloid leukemias. Nat. Genet. 47, 1030–1037 (2015).
Jo, S.Y., Granowicz, E.M., Maillard, I., Thomas, D. & Hess, J.L. Requirement for Dot1l in murine postnatal hematopoiesis and leukemogenesis by MLL translocation. Blood 117, 4759–4768 (2011).
Nguyen, A.T., Taranova, O., He, J. & Zhang, Y. DOT1L, the H3K79 methyltransferase, is required for MLL-AF9-mediated leukemogenesis. Blood 117, 6912–6922 (2011).
Bannister, A.J. & Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 21, 381–395 (2011).
Dawson, M.A. et al. Recurrent mutations, including NPM1c, activate a BRD4-dependent core transcriptional program in acute myeloid leukemia. Leukemia 28, 311–320 (2014).
McLean, C.M., Karemaker, I.D. & van Leeuwen, F. The emerging roles of DOT1L in leukemia and normal development. Leukemia 28, 2131–2138 (2014).
Shi, J. & Vakoc, C.R. The mechanisms behind the therapeutic activity of BET bromodomain inhibition. Mol. Cell 54, 728–736 (2014).
Sarkaria, S.M., Christopher, M.J., Klco, J.M. & Ley, T.J. Primary acute myeloid leukemia cells with IDH1 or IDH2 mutations respond to a DOT1L inhibitor in vitro. Leukemia 28, 2403–2406 (2014).
Jonkers, I. & Lis, J.T. Getting up to speed with transcription elongation by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 16, 167–177 (2015).
Smith, E. & Shilatifard, A. Transcriptional elongation checkpoint control in development and disease. Genes Dev. 27, 1079–1088 (2013).
Herait, P.E. et al. BET-bromodomain inhibitor OTX015 shows clinically meaningful activity at nontoxic doses: interim results of an ongoing phase I trial in hematologic malignancies. Cancer Res. 74, CT231 (2014).
Stein, E.M. et al. The DOT1L inhibitor EPZ-5676: safety and activity in relapsed/refractory patients with mll-rearranged leukemia. Blood 124, 387 (2014).
Kruse, U. et al. Chemoproteomics-based kinome profiling and target deconvolution of clinical multi-kinase inhibitors in primary chronic lymphocytic leukemia cells. Leukemia 25, 89–100 (2011).
Savitski, M.M. et al. Tracking cancer drugs in living cells by thermal profiling of the proteome. Science 346, 1255784 (2014).
Werner, T. et al. High-resolution enabled TMT 8-plexing. Anal. Chem. 84, 7188–7194 (2012).
Savitski, M.M. et al. Delayed fragmentation and optimized isolation width settings for improvement of protein identification and accuracy of isobaric mass tag quantification on Orbitrap-type mass spectrometers. Anal. Chem. 83, 8959–8967 (2011).
Savitski, M.M. et al. Targeted data acquisition for improved reproducibility and robustness of proteomic mass spectrometry assays. J. Am. Soc. Mass Spectrom. 21, 1668–1679 (2010).
Savitski, M.M. et al. Measuring and managing ratio compression for accurate iTRAQ/TMT quantification. J. Proteome Res. 12, 3586–3598 (2013).
Robinson, M.D., McCarthy, D.J. & Smyth, G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Stat. Methodol. 57, 289–300 (1995).
Anders, S. & Huber, W. Differential expression analysis for sequence count data. Genome Biol. 11, R106 (2010).
Wu, D. et al. ROAST: rotation gene set tests for complex microarray experiments. Bioinformatics 26, 2176–2182 (2010).
Langmead, B. & Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008).
Robinson, J.T. et al. Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 (2011).
Quinlan, A.R. & Hall, I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550 (2005).
Wickham, H. ggplot2: Elegant Graphics for Data Analysis (Springer, 2009).
Tsirigos, A., Haiminen, N., Bilal, E. & Utro, F. GenomicTools: a computational platform for developing high-throughput analytics in genomics. Bioinformatics 28, 282–283 (2012).
Rädle, B. et al. Metabolic labeling of newly transcribed RNA for high resolution gene expression profiling of RNA synthesis, processing and decay in cell culture. J. Vis. Exp. 78, e50195 (2013).
Bailey, T.L. & Machanick, P. Inferring direct DNA binding from ChIP-seq. Nucleic Acids Res. 40, e128 (2012).
Acknowledgements
A postdoctoral Fellowship awarded to O.G. from Leukaemia Foundation Australia partly supported this work. A mid-career Fellowship awarded to E.Y.N.L. from the Victoria Cancer Agency partly supported this work. A Senior Leukaemia Foundation Australia Fellowship and VESKI Innovation Fellowship currently support M.A.D. The National Health and Medical Research Council of Australia (1066545 (M.A.D.), 1085015 (M.A.D.), 1106444 (M.A.D.)) and Leukaemia Foundation Australia fund M.A.D.'s laboratory. This work was also funded in part by the Structural Genomics Consortium, a registered charity (no. 1097737) that received funds from AbbVie; Bayer; Boehringer Ingelheim; Genome Canada through the Ontario Genomics Institute (OGI-055); GlaxoSmithKline; Janssen; Lilly Canada; the Novartis Research Foundation; the Ontario Ministry of Economic Development and Innovation; Pfizer; Takeda; and the Wellcome Trust (092809/Z/10/Z).
Author information
Authors and Affiliations
Contributions
O.G., E.Y.N.L. and M.A.D. designed the research, interpreted data and wrote the manuscript. O.G., E.Y.N.L., I.B., D.L., E.C., G.J., A.W., M.W., C.Y.F., S.F., D.T., K.S., L.M., C.-F.W., Y.-C.C. and N.G. performed experiments and analyzed data. M.G., D.S., C.C., P.B., M.E.B., A.J.B., T.K., B.J.P.H., R.W.J., G.D., S.-J.D., C.H.A., P.G. and R.K.P. provided critical reagents and aided in manuscript preparation.
Corresponding author
Ethics declarations
Competing interests
I.B., D.L., G.J., A.W., C.C., N.G., G.D., P.G. and R.K.P. are employees of GlaxoSmithKline. D.S., P.B. and C.A. are employed by the Structural Genomics Consortium, a registered charity (no. 1097737) that received funds from AbbVie; Bayer; Boehringer Ingelheim; Genome Canada through the Ontario Genomics Institute (OGI-055); GlaxoSmithKline; Janssen; Lilly Canada; the Novartis Research Foundation; the Ontario Ministry of Economic Development and Innovation; Pfizer; Takeda; and the Wellcome Trust (092809/Z/10/Z).
Integrated supplementary information
Supplementary Figure 1 Characterization of SGC0946 and the BRD4- or DOT1L-containing protein complexes.
(a) Co-immunoprecipitation (Co-IP) of endogenous BRD4, DOT1L, MLLT1, and LEO1 in MOLM-13 cells. IgG was used as a control. Co-IP was followed by western blot analysis with antibodies against BRD4. (b) Size exclusion chromatography followed by western blot analysis of BRD4 and DOT1L, in MV4;11 cells. (c) Dose response to SGC0946 treatment in MV4;11 cells by western blot analysis using H3K79me2 antibodies, total H3 was used as control. (d) Time course of SGC0946 treatment in MV4;11 cells followed by western blot analysis for H3K79me2. (e) Western blot analysis of various histone lysine methylations (H3K79me/2/3, H3K36me3, and H3K4me3) following SGC0946 treatment in MV4;11 cells. (f) MV4;11 cells were treated with vehicle (DMSO), SGC0946, I-BET or combination followed by western blot analysis with antibodies against H3K79me2 and total H3.
Supplementary Figure 2 Chemical probes and proliferation assays.
(a) Chemical structure of the DOT1L inhibitors used in this study. (b) Chemical structure of DOT1L inhibitor compounds used for bead immobilization. (c) Pull downs with DOT1L inhibitor compounds immobilized on beads at 2 different coupling densities (CD) in HL60 nuclear extracts. 10µM FED1 (+) or DMSO (–) was used as soluble competitor. The eluates from the beads were analysed on Western blot using DOT1L antibodies. In vitro sub-therapeutic doses of (d) I-BET and (e) SGC0946 in MV4;11 cells. (f-h) Proliferation assay of the combination of low dose (LD, 100nM) I-BET plus low dose (LD, 1μM) SGC0946 in (f) SKM-1, (g) NB-4 (APML) and (h) HL-60 cells. Mean, error bars, s.d. (n = 2 cell culture replicates), representative graph from experiments done on 3 separate occasions.
Supplementary Figure 3 Phenotypic and pharmacokinetic analysis of single and combination therapies.
(a-b) Apoptotic response and cell cycle arrest in the combination treated cells. Annexin V staining of (a) MV4;11 and (b) K562 cells treated with either DMSO, SGC0946, I-BET, or combination. Mean, error bars, s.d. (n = 3 cell culture replicates), representative graph from experiments done on 3 separate occasions. (c-d) Cell cycle analysis by PI staining in (c) MV4;11 and (d) K562 cells treated with the inhibitors. These data show a cell cycle arrest in MV4;11 cells that is most prominent with combination therapy. Mean, error bars, s.d. (n = 3 cell culture replicates), representative graph from experiments done on 3 separate occasions. (e) Mouse pharmacokinetic studies comparing the blood concentration of SGC0946 administered via PO, SC or IP routes at 3 mg/kg (Samples taken post 2h below limit of quantification; 0.02 μM). Mean, error bars, s.d. (n = 3 mice per group). (f) Mouse pharmacokinetic study blood concentration-time profile of SGC0946 administered via surgically implanted mini-pump targeting 6 mg/kg/h for 72 h. Required exposure for in vivo model achieved. Mean, error bars, s.d. (n = 3 mice per group). (g) Western blot analysis of in vivo on-target activity of I-BET and SGC0946 on the bone marrows of three mice per group, using c-MYC, HSP60, H3K79me2 and total H3 antibodies.
Supplementary Figure 4 Inducible knockdown of BRD4 and DOT1L.
(a) Western blot and (b) qRT-PCR analysis of DOT1L expression in inducible RNAi samples. β-ACTIN was used as a loading control. (c) Western blot and (d) qRT-PCR analysis of BRD4 expression in inducible RNAi samples. HSP60 was used as a loading control. shDOT1L #7 and shBRD4 #498 were used in the competition assays (Figure 3c). (e) Schematic overview of the plasmids used. Flow plot examples showing cells transduced with single and combination inducible RNAi constructs expressing two different fluorophores. This strategy allowed for double knockdown within the same cell.
Supplementary Figure 5 Correlation of RNA-seq and ChIP–seq analysis of inhibitor-treated cells.
(a) GSEA using ROAST (Wu, D. et al. Bioinformatics 26, 2176–2182, 2010) of differentially expressed genes from RNA-Seq data of SGC0946 treated cells with previously published up- (red) and down-regulated (blue) genes from microarray analysis of MV4;11 cells treated with EPZ004777 (GSE29828). The shaded area in the centre of the plot shows genes ranked by log fold change in expression in SGC0946 compared with DMSO treated cells. Pink and blue shading represent significantly up- and down- regulated genes, respectively. (b) Heatmap of differential mRNA expression data from RNA-Seq of MV4;11 cells treated with I-BET, SGC0946, or combination in duplicate cell culture experiments. (c) geneGO analysis of the top ten diseases associated with the co-regulated genes. (d) Change in gene expression from RNA-Seq following BRD4 inhibition (I-BET), DOT1L inhibition (SGC0946) or a combination of BRD4 and DOT1L inhibition plotted against the changes in BRD4 binding and H3K79me2 levels in genes from ChIP-Seq. (e) Correlation between POL II and BRD4 binding or H3K79me2 levels at all genes in DMSO, I-BET and SGC0946 treated cells.
Supplementary Figure 6 ChIP–seq analysis of superenhancer-associated genes.
(a) Enhancers ranked by increasing BRD4 ChIP-Seq signal in reads per million (RPM) mapped reads. (b) ChIP-Seq profile of BRD4, H3K27ac, H3K79me2 and POL II at a typical enhancer and superenhancer indicated in Supplementary Fig 6a, in red and blue respectively. (c) Median H3K79me2, BRD4 and POL II coverage in RPKM across the co-regulated genes following I-BET, SGC0946 or combination treatment. (d) Heatmap of the expression of genes in closest proximity to superenhancers with I-BET and SGC0946 combination treatment. (e) qPCR analysis of H3K79me2 and BRD4 ChIP in MOLM-13 cells treated with SGC0946 using primers targeting CDK6, BCL2, MTHFD2 and Neg (negative control region). Mean, error bars, s.d. (n=3 technical replicates), representative graph from experiments done on 3 separate occasions
Supplementary Figure 7 SGC0946 washout experiment.
(a-b) Anti-H3K79me2 and anti-total H3 western blot. Samples are lysates from MV4;11 cells treated with DMSO or SGC0946 for 6, 8 and 10h and (a) harvested at end of treatment or (b) after washout of SGC0946, then harvested 72 hours later. (c) Proliferation Assay. Cell counts after short term treatment of MV4;11 cells with SGC0946 for 6 hours followed by wash-out or continuous treatment with SGC0946. (d-e) qRT-PCR of BCL2 expression from MV4;11 cells treated with DMSO or SGC0946 for 6, 8 and 10h and (d) harvested at end of treatment or (e) after washout of SGC0946, then harvested 72 hours later. ChIP-qPCR analysis for (f) BRD4 and (g) H3K79me2. MV4;11 cells were treated with I-BET for 6 hours, SGC0946 for 8 hours, SGC0946 for 72 hours continuously or SGC0946 for 8 hours followed by washout and assessed 72 hours later. Mean, error bars, s.d. (n=3 technical replicates), representative graph from experiments done on 3 separate occasions. (h), Median BRD4, H3K79me2 and POL II levels in RPKM across genes containing both BRD4 and H3K79me2 including 5kb upstream and downstream regions following short-term SGC0946 or SGC0946 washout treatment.
Supplementary Figure 8 Mechanism of BRD4 displacement by DOT1L inhibition.
(a) Left, Flow chart hierarchy of the numbers of expressed genes with H3K79me2 and BRD4 peaks that change their expression and have decreased H379me2 levels after SGC0946 treatment. Right, scatterplot of the log-fold change in BRD4 and H3K79me2 after SGC0946 treatment in the 175 SGC0946 regulated genes. (b) qRT-PCR of genes identified by RNA-seq as being regulated by both BRD4 depletion and DOT1L inhibition. Mean, error bars, s.d. (n = 3 experiments done on 3 separate occasions). (c) Quantitative proteomic analysis of BRD3 and BRD4 within HL-60 nuclear extracts that are bound to various biotinylated histone H3K79 peptides. BRD3 and BRD4 captured by the histone peptides were differentially quantified by isobaric tagging, as previously performed (Dawson, M.A. et al. Nature 478, 529-33, 2011). (d) ChIP-qPCR analysis of SGC0946 treated MOLM-13 cells using primers against CDK6, BCL2, MTHFD2 and Neg (Negative control region). Mean, error bars, s.d. (n=3 technical replicates), representative graph from 3 experiments done on 3 separate occasions. (e) Scatterplot of the log-fold change in H3K79me2 and H4K5ac levels after DOT1L inhibition in the 175 SGC0946 regulated genes. (f) Log-fold change in the levels of H4K5ac in the subsets of I-BET regulated, SGC0946 regulated and co-regulated genes following SGC0946 treatment. The upper limit, center and lower limit of boxplots denote the upper quartile, median and lower quartile of the data, respectively. Whiskers extend to 1.5 × interquartile range above and below the upper and lower quartiles, respectively. (g) Western blot of CREB1. Lysate from MV4;11 cells transduced with the two independent shRNAs used in Figure 7.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–8 and Supplementary Note (PDF 9045 kb)
Supplementary Table 1
Comparison of I-BET and DOT1L matrix and competition with inhibitor (XLSX 1107 kb)
Supplementary Table 2
Genes co-regulated by I-BET and SGC0946 (XLSX 68 kb)
Supplementary Data Set 1
Uncropped western blots (PDF 344 kb)
Rights and permissions
About this article
Cite this article
Gilan, O., Lam, E., Becher, I. et al. Functional interdependence of BRD4 and DOT1L in MLL leukemia. Nat Struct Mol Biol 23, 673–681 (2016). https://doi.org/10.1038/nsmb.3249
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.3249
This article is cited by
-
Tracking chromatin state changes using nanoscale photo-proximity labelling
Nature (2023)
-
BET inhibition disrupts transcription but retains enhancer-promoter contact
Nature Communications (2021)
-
Histone methyltransferase and drug resistance in cancers
Journal of Experimental & Clinical Cancer Research (2020)
-
HBO1 is required for the maintenance of leukaemia stem cells
Nature (2020)
-
DOT1L inhibition reveals a distinct subset of enhancers dependent on H3K79 methylation
Nature Communications (2019)