Abstract
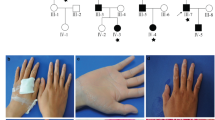
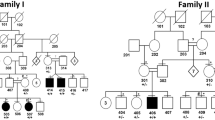
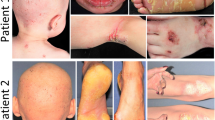
Molecular diagnosis of rare inherited palmoplantar keratoderma (PPK) is still challenging. We investigated at the clinical and genetic level a consanguineous Tunisian family presenting an autosomal dominant atypical form of transgrediens and progrediens PPK to better characterize this ultrarare disease and to identify its molecular etiology. Whole-exome sequencing (WES), filtering strategies, and bioinformatics analysis have been achieved. Clinical investigation and follow up over 13 years of this Tunisian family with three siblings formerly diagnosed as an autosomal recessive form of Mal de Melela-like conducted us to reconsider its initial phenotype. Indeed, the three patients presented clinical features that overlap both Mal de Meleda and progressive symmetric erythrokeratoderma (PSEK). The mode of inheritance was also reconsidered, since the mother, initially classified as unaffected, exhibited a similar expression of the disease. WES analysis showed the absence of potentially functional rare variants in known PPKs or PSEK-related genes. Results revealed a novel heterozygous nonsynonymous variant in cadherin-12 gene (CDH12, NM_004061, c.1655C > A, p.Thr552Asn) in all affected family members. This variant is absent in dbSNP and in 50 in-house control exomes. In addition, in silico analysis of the mutated 3D domain structure predicted that this variant would result in cadherin-12 protein destabilization and thermal instability. Functional annotation and biological network construction data provide further supporting evidence for the potential role of CDH12 in the maintenance of skin integrity. Taken together, these results suggest that CDH12 gene is a potential candidate gene for an atypical presentation of an autosomal dominant form of transgrediens and progrediens PPK.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Botstein D, Risch N. Discovering genotypes underlying human phenotypes: past successes for mendelian disease, future approaches for complex disease. Nat Genet. 2003;33:228–37. http://www.ncbi.nlm.nih.gov/pubmed/12610532.
Lai-Cheong JE, McGrath JA. Next-generation diagnostics for inherited skin disorders. J Investig Dermatol. 2011;131:1971–3. https://linkinghub.elsevier.com/retrieve/pii/S0022202X15350557.
Hsu TM, Kwok PY. Advances in molecular medicine. J Am Acad Dermatol. 2001;44:847–55. http://linkinghub.elsevier.com/retrieve/pii/S0190962201537731.
Kimyai-Asadi A, Kotcher LB, Jih MH. The molecular basis of hereditary palmoplantar keratodermas. J Am Acad Dermatol. 2002;47:327–43. http://www.ncbi.nlm.nih.gov/pubmed/12196741.
Bchetnia M, Charfeddine C, Kassar S, Zribi H, Guettiti HT, Ellouze F, et al. Clinical and mutational heterogeneity of Darier disease in Tunisian Families. Arch Dermatol. 2009;145. https://doi.org/10.1001/archdermatol.2009.52.
Bchetnia M, Laroussi N, Youssef M, Charfeddine C, Ben Brick AS, Boubaker MS, et al. Particular Mal de Meleda phenotypes in Tunisia and mutations founder effect in the Mediterranean region. Biomed Res Int. 2013;2013:1–7. http://www.hindawi.com/journals/bmri/2013/206803/.
Charfeddine C, Mokni M, Kassar S, Zribi H, Bouchlaka C, Boubaker S, et al. Further evidence of the clinical and genetic heterogeneity of recessive transgressive PPK in the Mediterranean region. J Hum Genet. 2006;51:841–5. https://doi.org/10.1007/s10038-006-0002-8.
Castellana S, Mazza T. Congruency in the prediction of pathogenic missense mutations: state-of-the-art web-based tools. Brief Bioinform. 2013;14:448–59. http://www.ncbi.nlm.nih.gov/pubmed/23505257.
Castellana S, Fusilli C, Mazza T. A broad overview of computational methods for predicting the pathophysiological effects of non-synonymous variants. 2016; 423–40. http://link.springer.com/10.1007/978-1-4939-3572-7_22.
Desmet F-O, Hamroun D, Lalande M, Collod-Béroud G, Claustres M, Béroud C. Human splicing finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009;37:e67. https://doi.org/10.1093/nar/gkp215.
Has C, Technau-Hafsi K. Palmoplantar keratodermas: clinical and genetic aspects. J der Dtsch Dermatologischen Ges. 2016;14:123–40. http://www.ncbi.nlm.nih.gov/pubmed/26819106.
Stelzer G, Plaschkes I, Oz-Levi D, Alkelai A, Olender T, Zimmerman S, et al. VarElect: the phenotype-based variation prioritizer of the GeneCards Suite. BMC Genom. 2016;17 Suppl 2:444. http://bmcgenomics.biomedcentral.com/articles/10.1186/s12864-016-2722-2.
Chivian D, Kim DE, Malmström L, Bradley P, Robertson T, Murphy P, et al. Automated prediction of CASP-5 structures using the Robetta server. Proteins. 2003;53 Suppl 6:524–33. https://doi.org/10.1002/prot.10529.
Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–13.
Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38 Suppl 2: (Web Server issue) W214–W220.
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504.
Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, et al. ClueGO: a cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–3. http://www.ncbi.nlm.nih.gov/pubmed/19237447.
Bindea G, Galon J, Mlecnik B. CluePedia cytoscape plugin: pathway insights using integrated experimental and in silico data. Bioinformatics. 2013;29:661–3. http://www.ncbi.nlm.nih.gov/pubmed/23325622.
Shapiro L, Weis WI. Structure and biochemistry of cadherins and catenins. Cold Spring Harb Perspect Biol. 2009;1:a003053. https://doi.org/10.1101/cshperspect.a003053.
Zhao L, Vahlquist A, Virtanen M, Wennerstrand L, Lind L, Lundström A, et al. Palmoplantar keratoderma of the Gamborg-Nielsen type is caused by mutations in the SLURP1 gene and represents a variant of Mal de Meleda. Acta Derm Venereol. 2014;94:707–10. http://www.ncbi.nlm.nih.gov/pubmed/24604124.
Kubo A. Nagashima-type palmoplantar keratosis: a common Asian type caused by SERPINB7 protease inhibitor deficiency. J Investig Dermatol. 2014;134:2076–9. https://linkinghub.elsevier.com/retrieve/pii/S0022202X15369499.
Athanikar SB, Inamadar AC, Palit A, Sampagavi VV, Deshmukh NS. Greither’s disease. Indian J Dermatol Venereol Leprol.2003;69:292–3. http://www.ncbi.nlm.nih.gov/pubmed/17642916.
Duchatelet S, Hovnanian A. Erythrokeratodermia variabilis et progressiva allelic to oculo-dento-digital dysplasia. J Investig Dermatol. 2015;135:1475–8. https://linkinghub.elsevier.com/retrieve/pii/S0022202X15372687.
Mahajan V, Khatri G, Chauhan P, Mehta K, Raina R, Gupta M. Progressive symmetric erythrokeratoderma having overlapping features with erythrokeratoderma variabilis and lesional hypertrichosis: is nomenclature “erythrokeratoderma variabilis progressiva” more appropriate? Indian J Dermatol. 2015;60:410 http://www.ncbi.nlm.nih.gov/pubmed/26288417.
Prabhu S, Shenoi SD, Pai SB, Handattu S, Bhattachan B. Progressive and symmetric erythrokeratoderma of adult onset: a rare case. Indian Dermatol Online J. 2010;1:43–5. http://www.idoj.in/text.asp?2010/1/1/43/73261.
Wei S, Zhou Y, Zhang TD, Huang ZM, Zhang XB, Zhu HL, et al. Evidence for the absence of mutations at GJB3, GJB4 and LOR in progressive symmetrical erythrokeratodermia. Clin Exp Dermatol. 2011;36:399–405. https://doi.org/10.1111/j.1365-2230.2010.03974.x.
Common JEA, O’Toole EA, Leigh IM, Thomas A, Griffiths WAD, Venning V, et al. Clinical and genetic heterogeneity of erythrokeratoderma variabilis. J Investig Dermatol. 2005;125:920–7. https://linkinghub.elsevier.com/retrieve/pii/S0022202X15325069.
van Steensel MAM, Oranje AP, van der Schroeff JG, Wagner A, van Geel M. The missense mutation G12D in connexin30.3 can cause both erythrokeratodermia variabilis of Mendes da Costa and progressive symmetric erythrokeratodermia of Gottron. Am J Med Genet A. 2009;149A:657–61. https://doi.org/10.1002/ajmg.a.32744.
Ishida-Yamamoto A. Loricrin keratoderma: a novel disease entity characterized by nuclear accumulation of mutant loricrin. J Dermatol Sci. 2003;31:3–8. http://www.ncbi.nlm.nih.gov/pubmed/12615358.
Bouadjar B, Benmazouzia S, Prud’homme JF, Cure S, Fischer J. Clinical and genetic studies of 3 large, consanguineous, Algerian families with Mal de Meleda. Arch Dermatol. 2000;136:1247–52. http://www.ncbi.nlm.nih.gov/pubmed/11030771.
Schiller S, Seebode C, Hennies HC, Giehl K, Emmert S. Palmoplantar keratoderma (PPK): acquired and genetic causes of a not so rare disease. J Dtsch Dermatol Ges. 2014;12:781–8. https://doi.org/10.1111/ddg.12418.
Sakiyama T, Kubo A. Hereditary palmoplantar keratoderma “clinical and genetic differential diagnosis.”. J Dermatol. 2016;43:264–74. http://www.ncbi.nlm.nih.gov/pubmed/26945534.
Richard G, Smith LE, Bailey RA, Itin P, Hohl D, Epstein EH, et al. Mutations in the human connexin gene GJB3 cause erythrokeratodermia variabilis. Nat Genet. 1998;20:366–9. http://www.nature.com/articles/ng1298_366.
Macari F, Landau M, Cousin P, Mevorah B, Brenner S, Panizzon R, et al. Mutation in the gene for connexin 30.3 in a family with erythrokeratodermia variabilis. Am J Hum Genet. 2000;67:1296–301. https://linkinghub.elsevier.com/retrieve/pii/S0002929707629577.
Boyden LM, Craiglow BG, Zhou J, Hu R, Loring EC, Morel KD, et al. Dominant de novo mutations in GJA1 cause erythrokeratodermia variabilis et progressiva, without features of oculodentodigital dysplasia. J Investig Dermatol. 2015;135:1540–7. https://linkinghub.elsevier.com/retrieve/pii/S0022202X15372833.
Kaliannan K, Hamarneh SR, Economopoulos KP, Nasrin Alam S, Moaven O, Patel P, et al. Intestinal alkaline phosphatase prevents metabolic syndrome in mice. Proc Natl Acad Sci USA. 2013;110:7003–8. https://doi.org/10.1073/pnas.1220180110.
Redies C, Hertel N, Hübner CA. Cadherins and neuropsychiatric disorders. Brain Res. 2012;1470:130–44. https://linkinghub.elsevier.com/retrieve/pii/S0006899312010736.
Gloushankova NA. Changes in regulation of cell-cell adhesion during tumor transformation. Biochemistry. 2008;73:742–50. http://www.ncbi.nlm.nih.gov/pubmed/18707582.
El-Amraoui A, Petit C. Cadherins as targets for genetic diseases. Cold Spring Harb Perspect Biol. 2010;2:a003095. https://doi.org/10.1101/cshperspect.a003095.
Jamora C, Fuchs E. Intercellular adhesion, signalling and the cytoskeleton. Nat Cell Biol. 2002;4:E101–8. http://www.nature.com/articles/ncb0402-e101.
Fujimori T, Takeichi M. Disruption of epithelial cell-cell adhesion by exogenous expression of a mutated nonfunctional N-cadherin. Mol Biol Cell. 1993;4:37–47. http://www.ncbi.nlm.nih.gov/pubmed/8443408.
Pontén FK, Schwenk JM, Asplund A, Edqvist PHD. The Human Protein Atlas as a proteomic resource for biomarker discovery. J Intern Med. 2011;270:428–46.
Yin L, Coelho SG, Ebsen D, Smuda C, Mahns A, Miller SA, et al. Epidermal gene expression and ethnic pigmentation variations among individuals of Asian, European and African ancestry. Exp Dermatol. 2014;23:731–5. https://doi.org/10.1111/exd.12518.
Del Bino S, Duval C, Bernerd F. Clinical and biological characterization of skin pigmentation diversity and its consequences on UV impact. Int J Mol Sci. 2018;19:2668. https://doi.org/10.3390/ijms19092668
Acknowledgements
We would like to warmly acknowledge the patients and their families for their collaboration and all clinicians involved the care of the patients.
Funding
This work was supported by the Tunisian Ministry of Public Health, the Ministry of Higher Education and Scientific Research (LR16IPT05), and RARE-MED project (A* MIDEX Initiative d’excellence). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Charfeddine, C., Dallali, H., Abdessalem, G. et al. Identification of a CDH12 potential candidate genetic variant for an autosomal dominant form of transgrediens and progrediens palmoplantar keratoderma in a Tunisian family. J Hum Genet 65, 397–410 (2020). https://doi.org/10.1038/s10038-019-0711-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s10038-019-0711-4