Abstract
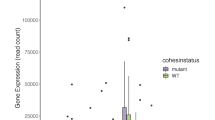
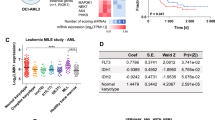
Acute myeloid leukemia (AML) is driven by mutations that occur in numerous combinations. A better understanding of how mutations interact with one another to cause disease is critical to developing targeted therapies. Approximately 50% of patients that harbor a common mutation in NPM1 (NPM1cA) also have a mutation in the cohesin complex. As cohesin and Npm1 are known to regulate gene expression, we sought to determine how cohesin mutation alters the transcriptome in the context of NPM1cA. We utilized inducible Npm1cAflox/+ and core cohesin subunit Smc3flox/+ mice to examine AML development. While Npm1cA/+;Smc3Δ/+ mice developed AML with a similar latency and penetrance as Npm1cA/+ mice, RNA-seq suggests that the Npm1cA/+; Smc3Δ/+ mutational combination uniquely alters the transcriptome. We found that the Rac1/2 nucleotide exchange factor Dock1 was specifically upregulated in Npm1cA/+;Smc3Δ/+ HSPCs. Knockdown of Dock1 resulted in decreased growth and adhesion and increased apoptosis only in Npm1cA/+;Smc3Δ/+ AML. Higher Rac activity was also observed in Npm1cA/+;Smc3Δ/+ vs. Npm1cA/+ AMLs. Importantly, the Dock1/Rac pathway is targetable in Npm1cA/+;Smc3Δ/+ AMLs. Our results suggest that Dock1/Rac represents a potential target for the treatment of patients harboring NPM1cA and cohesin mutations and supports the use of combinatorial genetics to identify novel precision oncology targets.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Heimbruch KE, Meyer AE, Agrawal P, Viny AD, Rao S. A cohesive look at leukemogenesis: The cohesin complex and other driving mutations in AML. Neoplasia (US). 2021;23:337–47.
Ley TJ, Miller C, Ding L, Raphael BJ, Mungall AJ, Robertson A, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med 2013;368:2059–74.
Papaemmanuil E, Gerstung M, Bullinger L, Gaidzik VI, Paschka P, Roberts ND, et al. Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med 2016;374:2209–21.
Thol F, Bollin R, Gehlhaar M, Walter C, Dugas M, Suchanek KJ, et al. Mutations in the cohesin complex in acute myeloid leukemia: Clinical and prognostic implications. Blood. 2014;123:914–20.
Thota S, Viny AD, Makishima H, Spitzer B, Radivoyevitch T, Przychodzen B, et al. Genetic alterations of the cohesin complex genes in myeloid malignancies. Blood. 2014;124:1790–8.
Kong X, Ball AR, Pham HX, Zeng W, Chen H-Y, Schmiesing JA, et al. Distinct functions of human Cohesin-SA1 and Cohesin-SA2 in double-strand break repair. Mol Cell Biol. 2014;34:685–98.
Heimbruch KE, Fisher JB, Stelloh CT, Phillips E, Reimer MH, Wargolet AJ, et al. DOT1L inhibitors block abnormal self-renewal induced by cohesin loss. Sci Rep. 2021;11:7288.
Fisher JB, Peterson J, Reimer M, Stelloh C, Pulakanti K, Gerbec ZJ, et al. The cohesin subunit Rad21 is a negative regulator of hematopoietic self-renewal through epigenetic repression of Hoxa7 and Hoxa9. Leukemia. 2017;31:712–9.
Viny AD, Ott CJ, Spitzer B, Rivas M, Meydan C, Papalexi E, et al. Dose-dependent role of the cohesin complex in normal and malignant hematopoiesis. J Exp Med. 2015;212:1819–32.
Mullenders J, Aranda-Orgilles B, Lhoumaud P, Keller M, Pae J, Wang K, et al. Cohesin loss alters adult hematopoietic stem cell homeostasis, leading to myeloproliferative neoplasms. J Exp Med. 2015;212:1833–50.
Mazumdar C, Shen Y, Xavy S, Zhao F, Reinisch A, Li R, et al. Leukemia-associated cohesin mutants dominantly enforce stem cell programs and impair human hematopoietic progenitor differentiation. Cell Stem Cell. 2015;17:675–88.
Galeev R, Baudet A, Kumar P, Nilsson AR, Nilsson B, Torngren T, et al. Genome-wide RNAi Screen Identifies Cohesin Genes as Modifiers of Renewal and Differentiation in Human HSCs. Cell Rep. 2016;14:2988–3000.
Kon A, Shih LY, Minamino M, Sanada M, Shiraishi Y, Nagata Y, et al. Recurrent mutations in multiple components of the cohesin complex in myeloid neoplasms. Nat Genet. 2013;45:1232–7.
Bolli N, Nicoletti I, De Marco MF, Bigerna B, Pucciarini A, Mannucci R, et al. Born to be exported: COOH-terminal nuclear export signals of different strength ensure cytoplasmic accumulation of nucleophosmin leukemic mutants. Cancer Res. 2007;67:6230–7.
Falini B, Nicoletti I, Bolli N, Martelli MP, Liso A, Gorello P, et al. Translocations and mutations involving the nucleophosmin (NPM1) gene in lymphomas and leukemias. Haematologica. 2007;92:519–32.
Alpermann T, Schnittger S, Eder C, Dicker F, Meggendorfer M, Kern W, et al. Molecular subtypes of npm1 mutations have different clinical profiles, specific patterns of accompanying molecular mutations and varying outcomes in intermediate risk acute myeloid leukemia. Haematologica. 2016;101:e55–e58.
Vassiliou GS, Cooper JL, Rad R, Li J, Rice S, Uren A, et al. Mutant nucleophosmin and cooperating pathways drive leukemia initiation and progression in mice. Nat Genet 2011;43:470–5.
Woolthuis CM, Han L, Verkaik-Schakel RN, van Gosliga D, Kluin PM, Vellenga E, et al. Downregulation of MEIS1 impairs long-term expansion of CD34 + NPM1-mutated acute myeloid leukemia cells. Leukemia. 2012;26:848–53.
Loberg MA, Bell RK, Goodwin LO, Eudy E, Miles LA, SanMiguel JM, et al. Sequentially inducible mouse models reveal that Npm1 mutation causes malignant transformation of Dnmt3a-mutant clonal hematopoiesis. Leukemia. 2019;33:1635–49.
Brunetti L, Gundry MC, Sorcini D, Guzman AG, Huang YH, Ramabadran R, et al. Mutant NPM1 maintains the leukemic state through HOX expression. Cancer Cell. 2018;34:499–512.
Dovey OM, Cooper JL, Mupo A, Grove CS, Lynn C, Conte N, et al. Molecular synergy underlies the co-occurrence patterns and phenotype of NPM1-mutant acute myeloid leukemia. Blood. 2017;130:1911–22.
Uckelmann HJ, Kim SM, Antonissen NJC, Krivtsov AV, Hatton C, McGeehan GM, et al. MLL-menin inhibition reverses pre-leukemic progenitor self-renewal induced By NPM1 mutations and prevents AML development. Blood. 2018;132:546–546.
Kühn MWM, Song E, Feng Z, Sinha A, Chen C-W, Deshpande AJ, et al. Targeting chromatin regulators inhibits leukemogenic gene expression in NPM1 mutant leukemia. Cancer Disco. 2016;6:1166–81.
Brugnera E, Haney L, Grimsley C, Lu M, Walk SF, Tosello-Trampont AC, et al. Unconventional Rac-GEF activity is mediated through the Dock180-ELMO complex. Nat Cell Biol. 2002;4:574–82.
Lu M, Ravichandran KS. Dock180-ELMO cooperation in Rac activation. Methods Enzymol. 2006;406:388–402.
Gu Y, Filippi MD, Cancelas JA, Siefring JE, Williams EP, Jasti AC, et al. Hematopoietic cell regulation by Rac1 and Rac2 guanosine triphosphatases. Science. 302:445–9.
Müller LUW, Schore RJ, Zheng Y, Thomas EK, Kim M-O, Cancelas JA, et al. Rac guanosine triphosphatases represent a potential target in AML. Leukemia. 2008;22:1803–6.
Rozenveld-Geugien M, Baas IO, van Gosliga D, Vellenga E, Schuringa JJ. Expansion of normal and leukemic human hematopoietic stem/progenitor cells requires Rac-mediated interaction with stromal cells. Exp Hematol. 2007;35:782–92.
Capala ME, Vallenga E, Schuringa JJ. ELMO1 is upregulated in AML CD34+ stem/progenitor cells, mediates chemotaxis and predicts poor prognosis in normal karyotype AML. PLoS One. 2014;9:e111568.
Lee S-H, Chiu Y-C, Li Y-H, Lin C-C, Hou H-A, Chou W-C, et al. High expression of dedicator of cytokinesis 1 (DOCK1) confers poor prognosis in acute myeloid leukemia. Oncotarget. 2017;8:72250–9.
Sha K, Lu Y, Zhang P, Pei R, Shi X, Fan Z, et al. Identifying a novel 5-gene signature predicting clinical outcomes in acute myeloid leukemia. Clin Transl Oncol. 2021;23:648–56.
Zhang W, Zheng X, Xie S, Zhang S, Mao J, Cai Y, et al. TBOPP enhances the anticancer effect of cisplatin by inhibiting DOCK1 in renal cell carcinoma. Mol Med Rep. 2020;22:1187–94.
Yang X, Wang Y, Pang S, Li X, Wang P, Ma R, et al. LINC00665 promotes the progression of acute myeloid leukemia by regulating the miR-4458/DOCK1 pathway. Sci Rep. 2021;11:5009.
Bagci H, Laurin M, Huber J, Muller WJ, Côté JF. Impaired cell death and mammary gland involution in the absence of Dock1 and Rac1 signaling. Cell Death Dis. 2014;5:e1374.
Schäker K, Bartsch S, Patry C, Stoll SJ, Hillebrands J-L, Wieland T, et al. The bipartite Rac1 guanine nucleotide exchange factor engulfment and cell motility 1/dedicator of cytokinesis 180 (Elmo1/Dock180) protects endothelial cells from apoptosis in blood vessel development. J Biol Chem. 2015;290:6408–18.
Yan A, Li G, Zhang X, Zhu B, Linghu H. Pro-survival effect of Dock180 overexpression on rat-derived H9C2 cardiomyocytes. Med Sci Monit Basic Res 2013;19:12–19.
Akakura S, Singh S, Spataro M, Akakura R, Kim J-I, Albert ML, et al. The opsonin MFG-E8 is a ligand for the αvβ5 integrin and triggers DOCK180-dependent Rac1 activation for the phagocytosis of apoptotic cells. Exp Cell Res. 2004;292:403–16.
Mulloy JC, Cancelas JA, Filippi M-D, Kalfa TA, Guo F, Zheng Y. Rho GTPases in hematopoiesis and hemopathies. Blood. 2010;115:936–47.
Durand-Onaylı V, Haslauer T, Harzschel A, Hartmann TN. Rac GTPases in Hematological Malignancies. Int J Mol Sci. 2018;19:4041.
Nishikimi A, Uruno T, Duan X, Cao Q, Okamura Y, Saitoh T, et al. Blockade of inflammatory responses by a small-molecule inhibitor of the Rac activator DOCK2. Chem Biol. 2012;19:488–97.
Shutes A, Onesto C, Picard V, Leblond B, Schweighoffer F, Der CJ. Specificity and mechanism of action of EHT 1864, a novel small molecule inhibitor of Rac family small GTPases. J Biol Chem. 2007;282:35666–78.
Fisher JB, McNulty M, Burke MJ, Crispino JD, Rao S. Cohesin mutations in myeloid malignancies. Trends Cancer. 2017;3:282–93.
Darracq A, Pak H, Bourgoin V, Zmiri F, Dellaire G, Affar EB, et al. NPM and NPM-MLF1 interact with chromatin remodeling complexes and influence their recruitment to specific genes. PLoS Genet. 2019;15:e1008463.
Tajiri H, Uruno T, Shirai T, Takaya D, Matsunaga S, Setoyama D, et al. Targeting Ras-driven cancer cell survival and invasion through selective inhibition of DOCK1. Cell Rep. 2017;19:969–80.
Watanabe M, Terasawa M, Miyano K, Yanagihara T, Uruno T, Sanematsu F, et al. DOCK2 and DOCK5 Act Additively in Neutrophils To Regulate Chemotaxis, Superoxide Production, and Extracellular Trap Formation. J Immunol. 2014;193:5660–7.
Acknowledgements
The authors would like to acknowledge Scott Armstrong and Michael Kühn for providing the OCI-AML3 Cas9 cell line and Benedetta Bonacci for aid with flow cytometry experiments.
Funding
This work was funded by: NCI R01 CA204231 and the Midwest Athletes against Childhood Cancer to SR. AEM is supported by a generous gift from Ms. Nan Gardetto.
Author information
Authors and Affiliations
Contributions
AEM designed research studies, conducted experiments, acquired and analyzed data, and wrote the manuscript. CS conducted experiments and acquired data. KP, RB, QF, and JB analyzed data. ST provided statistical advice and analyzed data. JBF designed research studies, conducted experiments, acquired data, and analyzed data. KEH conducted experiments and acquired data. YZ conducted experiments. ADV and GSV created and provided mouse models for the experiments and provided comments and edits to the manuscript. SR designed research studies and aided in writing the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Meyer, A.E., Stelloh, C., Pulakanti, K. et al. Combinatorial genetics reveals the Dock1-Rac2 axis as a potential target for the treatment of NPM1;Cohesin mutated AML. Leukemia 36, 2032–2041 (2022). https://doi.org/10.1038/s41375-022-01632-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41375-022-01632-y