Abstract
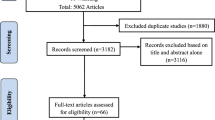
The gene encoding adhesion G protein-coupled receptor L3 (ADGRL3, also referred to as latrophilin 3 or LPHN3) has been associated with ADHD susceptibility in independent ADHD samples. We conducted a systematic review and a comprehensive meta-analysis to summarize the associations between the most studied ADGRL3 polymorphisms (rs6551665, rs1947274, rs1947275, and rs2345039) and both childhood and adulthood ADHD. Eight association studies (seven published and one unpublished) fulfilled criteria for inclusion in our meta-analysis. We also incorporated GWAS data for ADGRL3. In order to avoid overlapping samples, we started with summary statistics from GWAS samples and then added data from gene association studies. The results of our meta-analysis suggest an effect of ADGRL3 variants on ADHD susceptibility in children (n = 8724/14,644 cases/controls and 1893 families): rs6551665 A allele (Z score = −2.701; p = 0.0069); rs1947274 A allele (Z score = −2.033; p = 0.0421); rs1947275 T allele (Z score = 2.339; p = 0.0978); and rs2345039 C allele (Z score = 3.806; p = 0.0026). Heterogeneity was found in analyses for three SNPs (rs6551665, rs1947274, and rs2345039). In adults, results were not significant (n = 6532 cases/15,874 controls): rs6551665 A allele (Z score = 2.005; p = 0.0450); rs1947274 A allele (Z score = 2.179; p = 0.0293); rs1947275 T allele (Z score = −0.822; p = 0.4109); and rs2345039 C allele (Z score = −1.544; p = 0.1226). Heterogeneity was found just for rs6551665. In addition, funnel plots did not suggest publication biases. Consistent with ADGRL3’s role in early neurodevelopment, our findings suggest that the gene is predominantly associated with childhood ADHD.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
American Psychiatric Association. Diagnostic and statistical manual of mental disorders. 5th ed. Arlington, VA: American Psychiatric Publishing; 2013.
Polanczyk GV, Salum GA, Sugaya LS, Caye A, Rohde LA. Annual research review: a meta-analysis of the worldwide prevalence of mental disorders in children and adolescents. J Child Psychol Psychiatry. 2015;56:345–65.
Simon V, Czobor P, Bálint S, Mészáros A, Bitter I. Prevalence and correlates of adult attention-deficit hyperactivity disorder: meta-analysis. Br J Psychiatry. 2009;194:204–11.
Asherson P, Trzaskowski M. Attention-deficit/hyperactivity disorder is the extreme and impairing tail of a continuum. J Am Acad Child Adolesc Psychiatry. 2015;54:249–50.
Demontis D, Walters RK, Martin J, Mattheisen M, Als TD, Agerbo E, et al. Discovery of the first genome-wide significant risk loci for ADHD. Nat Genet. 2019;51:63–75.
Faraone SV, Larsson H. Genetics of attention deficit hyperactivity disorder. Mol Psychiatry. 2019;24:562–75.
Akutagava-Martins GC, Rohde LA, Hutz MH. Genetics of attention-deficit/hyperactivity disorder: an update. Expert Rev Neurother. 2016;16:145–56.
Arcos-Burgos M, Jain M, Acosta MT, Shively S, Stanescu H, Wallis D, et al. A common variant of the latrophilin 3 gene, LPHN3, confers susceptibility to ADHD and predicts effectiveness of stimulant medication. Mol Psychiatry. 2010;15:1053–66.
Hwang IW, Lim MH, Kwon HJ, Jin HJ. Association of LPHN3 rs6551665 A/G polymorphism with attention deficit and hyperactivity disorder in Korean children. Gene. 2015;566:68–73.
Ribasés M, Ramos-Quiroga JA, Sánchez-Mora C, Bosch R, Richarte V, Palomar G, et al. Contribution of LPHN3 to the genetic susceptibility to ADHD in adulthood: a replication study. Genes, Brain Behav. 2011;10:149–57.
Bruxel EM, Salatino-Oliveira A, Akutagava-Martins GC, Tovo-Rodrigues L, Genro JP, Zeni CP, et al. LPHN3 and attention-deficit/hyperactivity disorder: a susceptibility and pharmacogenetic study. Genes, Brain Behav. 2015;14:419–27.
Kappel DB, Schuch JB, Rovaris DL, da Silva BS, Cupertino RB, Winkler C, et al. Further replication of the synergistic interaction between LPHN3 and the NTAD gene cluster on ADHD and its clinical course throughout adulthood. Prog Neuro-Psychopharmacol Biol Psychiatry. 2017;79:120–7.
Huang X, Zhang Q, Gu X, Hou Y, Wang M, Chen X, et al. LPHN3 gene variations and susceptibility to ADHD in Chinese Han population: a two-stage case-control association study and gene-environment interactions. Eur Child Adolesc Psychiatry. 2019;28:861–73.
Lange M, Norton W, Coolen M, Chaminade M, Merker S, Proft F, et al. The ADHD-susceptibility gene lphn3.1 modulates dopaminergic neuron formation and locomotor activity during zebrafish development. Mol Psychiatry. 2012;17:946–54.
Wallis D, Hill DS, Mendez IA, Abbott LC, Finnell RH, Wellman PJ, et al. Initial characterization of mice null for Lphn3, a gene implicated in ADHD and addiction. Brain Res. 2012;1463:85–92.
O’Sullivan ML, Martini F, von Daake S, Comoletti D, Ghosh A. LPHN3, a presynaptic adhesion-GPCR implicated in ADHD, regulates the strength of neocortical layer 2/3 synaptic input to layer 5. Neural Dev. 2014;9:1–11.
Orsini CA, Setlow B, DeJesus M, Galaviz S, Loesch K, Ioerger T, et al. Behavioral and transcriptomic profiling of mice null for Lphn3, a gene implicated in ADHD and addiction. Mol Genet Genom Med. 2016;4:322–43.
Martinez AF, Abe Y, Hong S, Molyneux K, Yarnell D, Löhr H, et al. An ultraconserved brain-specific enhancer within ADGRL3 (LPHN3) underpins attention-deficit/hyperactivity disorder susceptibility. Biol Psychiatry. 2016;80:943–54.
Rovira P, Demontis D, Sanchez-Mora C, Zayats T, Klein M, Roth Mota N, et al. Shared genetic background between children and adults with attention deficit/hyperactivity disorder. 2019. https://doi.org/10.1101/589614.
Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–1.
Higgins JPT, Green S (editors). Cochrane handbook for systematic reviews of interventions version 5.1.0 [updated March 2011]. The Cochrane Collaboration; 2011. www.handbook.cochrane.org.
Acosta MT, Swanson J, Stehli A, Molina BSG, Martinez AF, Arcos-Burgos M, et al. ADGRL3 (LPHN3) variants are associated with a refined phenotype of ADHD in the MTA study. Mol Genet Genom Med. 2016;4:540–7.
Dark C, Homman-Ludiye J, Bryson-Richardson RJ. The role of ADHD associated genes in neurodevelopment. Dev Biol. 2018;438:69–83.
The Brainstorm Consortium, Anttila V, Bulik-Sullivan B, Finucane HK, Walters R, Bras J, et al. Analysis of shared heritability in common disorders of the brain. Science. 2018;360:eaap8757. https://doi.org/10.1126/science.aap8757.
Cross-Disorder Group of the Psychiatric Genomics C. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet. 2013;45:984–94.
Visscher PM, Wray NR, Zhang Q, Sklar P, Mccarthy MI, Brown MA, et al. 10 years of GWAS discovery: biology, function, and translation. Am J Hum Genet. 2017;101:5–22.
Choudhry Z, Sengupta SM, Grizenko N, Fortier ME, Thakur GA, Bellingham J, et al. LPHN3 and attention-deficit/hyperactivity disorder: Interaction with maternal stress during pregnancy. J Child Psychol Psychiatry Allied Discip. 2012;53:892–902.
Jain M, Vélez JI, Acosta MT, Palacio LG, Balog J, Roessler E, et al. A cooperative interaction between LPHN3 and 11q doubles the risk for ADHD. Mol Psychiatry. 2012;17:741–7.
Acosta MT, Vélez JI, Bustamante ML, Balog JZ, Arcos-Burgos M, Muenke M. A two-locus genetic interaction between LPHN3 and 11q predicts ADHD severity and long-term outcome. Transl Psychiatry. 2011;1:1–8.
Boyle EA, Li YI, Pritchard JK. An expanded view of complex traits: from polygenic to omnigenic. Cell. 2017;169:1177–86.
Moffitt TE, Houts R, Asherson P, Belsky DW, Corcoran DL, Hammerle M, et al. Is adult ADHD a childhood-onset neurodevelopmental disorder? Evidence from a four-decade longitudinal cohort study. Am J Psychiatry. 2015;172:967–77.
Riglin L, Collishaw S, Thapar AK, Dalsgaard S, Langley K, Smith GD, et al. Association of genetic risk variants with attention-deficit/hyperactivity disorder trajectories in the general population. JAMA Psychiatry. 2016;73:1285.
Acknowledgements
The authors thank Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, Brazil) and Fundo de Incentivo à Pesquisa e Eventos—Hospital de Clínicas de Porto Alegre (FIPE/HCPA, Brazil) for financial support. The authors also thank Han Jun Jin from the Department of Nanobiomedical Science, Dankook University in South Korea for the provided clarifications.
Author information
Authors and Affiliations
Contributions
EMB, GCAM, CRMM, LAR, and MHH conceived and designed the study. MK, BF, MR, DBK, NRM, EHG, CHDB, and MAB contributed with data. EMB, GCAM, CRMM, PR, and TPQ conducted statistical analyses and wrote the first draft of the manuscript. PR provided SNP-based and gene-based results for ADGRL3 on ADHD in children and on persistent ADHD in adults from their GWAS-MA [19]. LAR and MHH provided funding for the project. All coauthors provided critical feedback on the manuscript, suggested additional analyses, and critical revisions, and edited the manuscript for clarity and precision. All authors contributed to and have approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
CRMM receives financial research support from the government agency: Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq). CRMM received fees for the development of educational materials for Libbs, Novartis, and Pfizer. He has served as a consultant and served on the speakers’ bureau of Shire. Libbs, Novartis, and Shire make products for the treatment of ADHD. CRMM received travel, accommodation, and registration support to the fourth and fifth World Congress on ADHD from the World Federation of ADHD. EHG was on the speaker’s bureau for Novartis and Shire, and Shire’s national and international advisory boards for the last 3 years. He also received travel awards (air tickets and hotel accommodations) for participating in two psychiatric meetings from Shire and Novartis. LAR has received grant or research support from, served as a consultant to, and served on the speakers’ bureau of Eli Lilly and Co., Janssen, Medice, Novartis and Shire. The ADHD and Juvenile Bipolar Disorder Outpatient Programs chaired by LAR have received unrestricted educational and research support from the following pharmaceutical companies: Eli Lilly and Co., Janssen, Novartis, and Shire. LAR has received authorship royalties from Oxford Press and ArtMed and travel grants from Shire to take part in the 2018 APA and from Novartis to take part of the 2015 WFADHD congresses. MHH receives financial research support from the Brazilian governmental agency: CNPq. EMB and GCAM have received financial research support from CNPq. CHDB and DBK have received financial support from the following Brazilian agencies: CNPq, Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) and HCPA (FIPE-HCPA). BF has received educational speaking fees from Shire and Medice. She also is supported by the personal Vici grant from the Netherlands Organisation for Scientific Research (NWO). PR is a recipient of a predoctoral fellowship from the Agència de Gestió d’Ajuts Universitaris i de Recerca, Generalitat de Catalunya, Spain (2016FI_B 00899). TPQ, MK, MR, NRM, and MAB have no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Bruxel, E.M., Moreira-Maia, C.R., Akutagava-Martins, G.C. et al. Meta-analysis and systematic review of ADGRL3 (LPHN3) polymorphisms in ADHD susceptibility. Mol Psychiatry 26, 2277–2285 (2021). https://doi.org/10.1038/s41380-020-0673-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41380-020-0673-0
This article is cited by
-
Novel non-stimulants rescue hyperactive phenotype in an adgrl3.1 mutant zebrafish model of ADHD
Neuropsychopharmacology (2023)
-
Relation between latrophilin 3 (LPHN3) gene polymorphism (rs2345039) and attention deficit hyperactivity disorder in children
Egyptian Pediatric Association Gazette (2022)
-
ADGRL3 genomic variation implicated in neurogenesis and ADHD links functional effects to the incretin polypeptide GIP
Scientific Reports (2022)
-
Brain structural and functional substrates of ADGRL3 (latrophilin 3) haplotype in attention-deficit/hyperactivity disorder
Scientific Reports (2021)
-
Adhesion G protein-coupled receptor L3 gene variants: Statistically significant association observed in the male Indo-caucasoid Attention deficit hyperactivity disorder probands
Molecular Biology Reports (2021)