Abstract
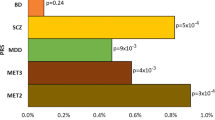
Lithium is a first-line medication for bipolar disorder (BD), but only one in three patients respond optimally to the drug. Since evidence shows a strong clinical and genetic overlap between depression and bipolar disorder, we investigated whether a polygenic susceptibility to major depression is associated with response to lithium treatment in patients with BD. Weighted polygenic scores (PGSs) were computed for major depression (MD) at different GWAS p value thresholds using genetic data obtained from 2586 bipolar patients who received lithium treatment and took part in the Consortium on Lithium Genetics (ConLi+Gen) study. Summary statistics from genome-wide association studies in MD (135,458 cases and 344,901 controls) from the Psychiatric Genomics Consortium (PGC) were used for PGS weighting. Response to lithium treatment was defined by continuous scores and categorical outcome (responders versus non-responders) using measurements on the Alda scale. Associations between PGSs of MD and lithium treatment response were assessed using a linear and binary logistic regression modeling for the continuous and categorical outcomes, respectively. The analysis was performed for the entire cohort, and for European and Asian sub-samples. The PGSs for MD were significantly associated with lithium treatment response in multi-ethnic, European or Asian populations, at various p value thresholds. Bipolar patients with a low polygenic load for MD were more likely to respond well to lithium, compared to those patients with high polygenic load [lowest vs highest PGS quartiles, multi-ethnic sample: OR = 1.54 (95% CI: 1.18–2.01) and European sample: OR = 1.75 (95% CI: 1.30–2.36)]. While our analysis in the Asian sample found equivalent effect size in the same direction: OR = 1.71 (95% CI: 0.61–4.90), this was not statistically significant. Using PGS decile comparison, we found a similar trend of association between a high genetic loading for MD and lower response to lithium. Our findings underscore the genetic contribution to lithium response in BD and support the emerging concept of a lithium-responsive biotype in BD.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Ferrari AJ, Stockings E, Khoo JP, Erskine HE, Degenhardt L, Vos T, et al. The prevalence and burden of bipolar disorder: findings from the Global Burden of Disease Study 2013. Bipolar Disord. 2016;18:440–50.
Chesney E, Goodwin GM, Fazel S. Risks of all-cause and suicide mortality in mental disorders: a meta-review. World Psychiatry. 2014;13:153–60.
Schulze TG, Alda M, Adli M, Akula N, Ardau R, Bui ET, et al. The International Consortium on Lithium Genetics (ConLiGen): an initiative by the NIMH and IGSLI to study the genetic basis of response to lithium treatment. Neuropsychobiology. 2010;62:72–8.
Malhi GS, Bassett D, Boyce P, Bryant R, Fitzgerald PB, Fritz K, et al. Royal Australian and New Zealand College of Psychiatrists clinical practice guidelines for mood disorders. Aust N Z J Psychiatry. 2015;49:1087–206.
Hou L, Heilbronner U, Degenhardt F, Adli M, Akiyama K, Akula N, et al. Genetic variants associated with response to lithium treatment in bipolar disorder: a genome-wide association study. The Lancet. 2016;387:1085–93.
Grof P, Duffy A, Cavazzoni P, Grof E, Garnham J, MacDougall M, et al. Is response to prophylactic lithium a familial trait? J Clin Psychiatry. 2002;63:942–7.
Garnham J, Munro A, Slaney C, Macdougall M, Passmore M, Duffy A, et al. Prophylactic treatment response in bipolar disorder: results of a naturalistic observation study. J Affect Disord. 2007;104:185–90.
Lahteenvuo M, Tanskanen A, Taipale H, Hoti F, Vattulainen P, Vieta E, et al. Real-world effectiveness of pharmacologic treatments for the prevention of rehospitalization in a finnish nationwide cohort of patients with bipolar disorder. JAMA Psychiatry. 2018;75:347–55.
Grande I, Berk M, Birmaher B, Vieta E. Bipolar disorder. Lancet (London, England). 2016;387:1561–72.
Bauer M, Gitlin M. Practical management of lithium. In: The essential guide to lithium treatment. Springer International Publishing: Cham, 2016, pp 113–28.
Oedegaard KJ, Alda M, Anand A, Andreassen OA, Balaraman Y, Berrettini WH, et al. The Pharmacogenomics of Bipolar Disorder study (PGBD): identification of genes for lithium response in a prospective sample. BMC Psychiatry. 2016;16:129.
Severus E, Taylor MJ, Sauer C, Pfennig A, Ritter P, Bauer M, et al. Lithium for prevention of mood episodes in bipolar disorders: systematic review and meta-analysis. Int J Bipolar Disord. 2014;2:15.
Joas E, Karanti A, Song J, Goodwin GM, Lichtenstein P, Landen M. Pharmacological treatment and risk of psychiatric hospital admission in bipolar disorder. Br J Psychiatry. 2017;210:197–202.
Chen CH, Lee CS, Lee MT, Ouyang WC, Chen CC, Chong MY, et al. Variant GADL1 and response to lithium therapy in bipolar I disorder. N Engl J Med. 2014;370:119–28.
Song J, Bergen SE, Di Florio A, Karlsson R, Charney A, Ruderfer DM, et al. Genome-wide association study identifies SESTD1 as a novel risk gene for lithium-responsive bipolar disorder. Molecular Psychiatry. 2016;21:1290–7.
Amare AT, Schubert KO, Hou L, Clark SR, Papiol S, Heilbronner U, et al. Association of polygenic score for schizophrenia and HLA antigen and inflammation genes with response to lithium in bipolar affective disorder: a genome-wide association study. JAMA Psychiatry. 2018;75:65–74.
Goldberg JF, Garno JL, Leon AC, Kocsis JH, Portera L. A history of substance abuse complicates remission from acute mania in bipolar disorder. J Clin Psychiatry. 1999;60:733–40.
Calkin C, van de Velde C, Ruzickova M, Slaney C, Garnham J, Hajek T, et al. Can body mass index help predict outcome in patients with bipolar disorder? Bipolar Disorders. 2009;11:650–6.
Calkin CV, Ruzickova M, Uher R, Hajek T, Slaney CM, Garnham JS, et al. Insulin resistance and outcome in bipolar disorder. Brit J Psychiatry. 2015;206:52–7.
Mitchell PB, Frankland A, Hadzi-Pavlovic D, Roberts G, Corry J, Wright A, et al. Comparison of depressive episodes in bipolar disorder and in major depressive disorder within bipolar disorder pedigrees. Brit J Psychiatry. 2011;199:303–9.
Lee SH, Ripke S, Neale BM, Faraone SV, Purcell SM. Cross-Disorder Group of the Psychiatric Genomics C et al. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet. 2013;45:984–94.
Cross-Disorder Group of the Psychiatric Genomics C. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. The Lancet. 2013;381:1371–9.
Amare AT, Schubert KO, Klingler-Hoffmann M, Cohen-Woods S, Baune BT. The genetic overlap between mood disorders and cardiometabolic diseases: a systematic review of genome wide and candidate gene studies. Transl Psychiatry. 2017;7:e1007.
Amare AT, Schubert KO, Baune BT. Pharmacogenomics in the treatment of mood disorders: strategies and opportunities for personalized psychiatry. EPMA J. 2017;8:211–27.
Abou-Saleh MT, Muller-Oerlinghausen B, Coppen AJ. Lithium in the episode and suicide prophylaxis and in augmenting strategies in patients with unipolar depression. Int J Bipolar Disord. 2017;5:11.
Zhou X, Ravindran AV, Qin B, Del Giovane C, Li Q, Bauer M, et al. Comparative efficacy, acceptability, and tolerability of augmentation agents in treatment-resistant depression: systematic review and network meta-analysis. J Clin Psychiatry. 2015;76:e487–98.
Tiihonen J, Tanskanen A, Hoti F, Vattulainen P, Taipale H, Mehtala J, et al. Pharmacological treatments and risk of readmission to hospital for unipolar depression in Finland: a nationwide cohort study. Lancet Psychiatry. 2017;4:547–53.
Popovic D, Reinares M, Goikolea JM, Bonnin CM, Gonzalez-Pinto A, Vieta E. Polarity index of pharmacological agents used for maintenance treatment of bipolar disorder. Eur Neuropsychopharmacol. 2012;22:339–46.
Vieta E, Berk M, Schulze TG, Carvalho AF, Suppes T, Calabrese JR, et al. Bipolar disorders. Nat Rev Dis Primers. 2018;4:18008.
Colom F, Vieta E, Daban C, Pacchiarotti I, Sanchez-Moreno J. Clinical and therapeutic implications of predominant polarity in bipolar disorder. J Affect Disord. 2006;93:13–7.
Kessing LV, Hellmund G, Andersen PK. Predictors of excellent response to lithium: results from a nationwide register-based study. Int Clin Psychopharmacol. 2011;26:323–8.
Kleindienst N, Engel R, Greil W. Which clinical factors predict response to prophylactic lithium? A systematic review for bipolar disorders. Bipolar Disorders. 2005;7:404–17.
Pfennig A, Schlattmann P, Alda M, Grof P, Glenn T, Muller-Oerlinghausen B, et al. Influence of atypical features on the quality of prophylactic effectiveness of long-term lithium treatment in bipolar disorders. Bipolar Disorders. 2010;12:390–6.
Fountoulakis KN, Kontis D, Gonda X, Siamouli M, Yatham LN. Treatment of mixed bipolar states. Int J Neuropsychopharmacol. 2012;15:1015–26.
Sportiche S, Geoffroy PA, Brichant-Petitjean C, Gard S, Khan JP, Azorin JM, et al. Clinical factors associated with lithium response in bipolar disorders. Aust N Z J Psychiatry. 2017;51:524–30.
Wray NR, Ripke S, Mattheisen M, Trzaskowski M, Byrne EM, Abdellaoui A et al. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nature Genetics 2018;50:668–81.
Duffy A, Alda M, Milin R, Grof P. A consecutive series of treated affected offspring of parents with bipolar disorder: is response associated with the clinical profile? Can J Psychiatry. 2007;52:369–76.
Manchia M, Adli M, Akula N, Ardau R, Aubry JM, Backlund L, et al. Assessment of response to lithium maintenance treatment in bipolar disorder: a consortium on lithium genetics (ConLiGen) report. PloS ONE. 2013;8:e65636.
Scott J, Etain B, Manchia M, Brichant-Petitjean C, Geoffroy PA, Schulze T et al. An examination of the quality and performance of the Alda scale for classifying lithium response phenotypes. Bipolar Disorders. 2019. https://doi.org/10.1111/bdi.12829.
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75.
Das S, Forer L, Schonherr S, Sidore C, Locke AE, Kwong A, et al. Next-generation genotype imputation service and methods. Nat Genet. 2016;48:1284–7.
Purcell SM, Wray NR, Stone JL, Visscher PM, O’Donovan MC, Sullivan PF, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–52.
Stahl EA, Breen G, Forstner AJ, McQuillin A, Ripke S, Trubetskoy V, et al. Genome-wide association study identifies 30 loci associated with bipolar disorder. Nat Genet. 2019;51:793–803.
Zheng HF, Forgetta V, Hsu YH, Estrada K, Rosello-Diez A, Leo PJ, et al. Whole-genome sequencing identifies EN1 as a determinant of bone density and fracture. Nature. 2015;526:112–7.
Amare AT, Vaez A, Hsu YH, Direk N, Kamali Z, Howard DM et al. Bivariate genome-wide association analyses of the broad depression phenotype combined with major depressive disorder, bipolar disorder or schizophrenia reveal eight novel genetic loci for depression. Mol Psychiatry. 2019. https://doi.org/10.1038/s41380-018-0336-6.
Choi SW, O’Reilly PF. PRSice-2: polygenic risk score software for biobank-scale data. GigaScience. 2019;8:giz082.
Bland JM, Altman DG. Multiple significance tests: the Bonferroni method. BMJ. 1995;310:170.
Malhi GS, Tanious M, Das P, Berk M. The science and practice of lithium therapy. Aust N Z J Psychiatry. 2012;46:192–211.
Grof E, Haag M, Grof P, Haag H. Lithium response and the sequence of episode polarities: preliminary report on a Hamilton sample. Prog Neuropsychopharmacol Biol Psychiatry. 1987;11:199–203.
Hui TP, Kandola A, Shen L, Lewis G, Osborn DPJ, Geddes JR, et al. A systematic review and meta-analysis of clinical predictors of lithium response in bipolar disorder. Acta Psychiatr Scand. 2019;140:94–115.
Grof P, Alda M, Grof E, Zvolsky P, Walsh M. Lithium response and genetics of affective disorders. J Affect Disord. 1994;32:85–95.
Bschor T. Lithium in the treatment of major depressive disorder. Drugs. 2014;74:855–62.
Alevizos B, Alevizos E, Leonardou A, Zervas I. Low dosage lithium augmentation in venlafaxine resistant depression: an open-label study. Psychiatriki. 2012;23:143–8.
Bauer M, Döpfmer S. Lithium augmentation in treatment-resistant depression: meta-analysis of placebo-controlled studies. Prim Care Companion J Clin Psych. 2000;2:31.
Bauer M, Adli M, Ricken R, Severus E, Pilhatsch M. Role of lithium augmentation in the management of major depressive disorder. CNS Drugs. 2014;28:331–42.
Bauer M, Bschor T, Kunz D, Berghofer A, Strohle A, Muller-Oerlinghausen B. Double-blind, placebo-controlled trial of the use of lithium to augment antidepressant medication in continuation treatment of unipolar major depression. Am J Psychiatry. 2000;157:1429–35.
Bauer M, Adli M, Baethge C, Berghöfer A, Sasse J, Heinz A, et al. Lithium augmentation therapy in refractory depression: clinical evidence and neurobiological mechanisms. Can J Psychiatry. 2003;48:440–8.
Bschor T, Bauer M. Efficacy and mechanisms of action of Lithium augmentation in refractory major depression. Curr Pharm Des. 2006;12:2985–92.
Kan C, Pedersen NL, Christensen K, Bornstein SR, Licinio J, MacCabe JH, et al. Genetic overlap between type 2 diabetes and depression in Swedish and Danish twin registries. Mol Psychiatry. 2016;21:903.
Bigdeli TB, Ripke S, Peterson RE, Trzaskowski M, Bacanu SA, Abdellaoui A, et al. Genetic effects influencing risk for major depressive disorder in China and Europe. Transl Psychiatry. 2017;7:e1074.
Shan GW, Makmor-Bakry M, Omar MS. Long term use of lithium and factors associated with treatment response among patients with bipolar disorder. Psychiatr Danub. 2016;28:146–53.
Wray NR, Lee SH, Mehta D, Vinkhuyzen AA, Dudbridge F, Middeldorp CM. Research review: polygenic methods and their application to psychiatric traits. J Child Psychol Psychiatry Allied Discip. 2014;55:1068–87.
Acknowledgements
The authors are grateful to all patients who participated in the study and we appreciate the contributions of clinicians, scientists, research assistants and study staffs who have helped with patient recruitment, data collection and sample preparation for the studies. We are also indebted to the members of the ConLi+Gen Scientific Advisory Board (http://www.conligen.org/) for critical input over the course of the project. The analysis of this study was carried out using the high-performance computational capabilities of the University of Adelaide, Phoenix supercomputer https://www.adelaide.edu.au/phoenix/.
Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium
Naomi R. Wray77,78, Stephan Ripke79,80,81, Manuel Mattheisen82,83,84,85, Maciej Trzaskowski77, Enda M. Byrne77, Abdel Abdellaoui86, Mark J. Adams87, Esben Agerbo85,88,89, Tracy M. Air90, Till F. M. Andlauer91,92, Silviu-Alin Bacanu93, Marie Bækvad-Hansen85,94, Aartjan T. F. Beekman95, Tim B. Bigdeli93,96, Elisabeth B. Binder91,97, Douglas H. R. Blackwood87, Julien Bryois98, Henriette N. Buttenschøn84,85,99, Jonas Bybjerg-Grauholm85,94, Na Cai100,101, Enrique Castelao102, Jane varregaard Christensen83,84,85, Toni-Kim Clarke87, Jonathan R. I. Coleman103, Lucía Colodro-Conde104, Baptiste Couvy-Duchesne105,106, Nick Craddock107, Gregory E. Crawford108,109, Gail Davies110, Ian J. Deary110, Franziska Degenhardt111,112, Eske M. Derks104, Nese Direk113,114, Conor V. Dolan86, Erin C. Dunn115,116,117, Thalia C. Eley103, Valentina Escott-Price118, Farnush Farhadi Hassan Kiadeh119, Hilary K. Finucane120,121, Andreas J. Forstner111,112,122,123, Josef Frank124, Héléna A. Gaspar103, Michael Gill125, Fernando S. Goes126, Scott D. Gordon127, Jakob Grove83,84,85,128, Lynsey S. Hall87,129, Christine Søholm Hansen85,94, Thomas F. Hansen130,131,132, Stefan Herms111,112,123, Ian B. Hickie133, Per Hoffmann111,112,123, Georg Homuth134, Carsten Horn135, Jouke-Jan Hottenga86, David M. Hougaard85,94, Marcus Ising136, Rick Jansen95, Eric Jorgenson137, James A. Knowles138, Isaac S. Kohane139,140,141, Julia Kraft80, Warren W. Kretzschmar142, Jesper Krogh143, Zoltán Kutalik144,145, Yihan Li142, Penelope A. Lind104, Donald J. MacIntyre146,147, Dean F. MacKinnon126, Robert M. Maier78, Wolfgang Maier148, Jonathan Marchini149, Hamdi Mbarek86, Patrick McGrath150, Peter McGuffin103, Sarah E. Medland104, Divya Mehta78,151, Christel M. Middeldorp86,152,153, Evelin Mihailov154, Yuri Milaneschi95, Lili Milani154, Francis M. Mondimore126, Grant W. Montgomery77, Sara Mostafavi155,156, Niamh Mullins103, Matthias Nauck157,158, Bernard Ng156, Michel G. Nivard86, Dale R. Nyholt159, Paul F. O’Reilly103, Hogni Oskarsson160, Michael J. Owen161, Jodie N. Painter104, Carsten Bøcker Pedersen85,88,89, Marianne Giørtz Pedersen85,88,89, Roseann E. Peterson93,162, Erik Pettersson98, Wouter J. Peyrot95, Giorgio Pistis102, Danielle Posthuma163,164, Jorge A. Quiroz165, Per Qvist77,84,85, John P. Rice166, Brien P. Riley93, Margarita Rivera103,167, Saira Saeed Mirza113, Robert Schoevers168, Eva C. Schulte169,170, Ling Shen137, Jianxin Shi171, Stanley I. Shyn172, Engilbert Sigurdsson173, Grant C. B. Sinnamon174, Johannes H. Smit95, Daniel J. Smith175, Hreinn Stefansson176, Stacy Steinberg176, Fabian Streit124, Jana Strohmaier124, Katherine E. Tansey177, Henning Teismann178, Alexander Teumer179, Wesley Thompson85,131,180,181, Pippa A. Thomson182, Thorgeir E. Thorgeirsson176, Matthew Traylor183, Jens Treutlein124, Vassily Trubetskoy80, André G. Uitterlinden184, Daniel Umbricht185, Sandra Van der Auwera186, Albert M. van Hemert187, Alexander Viktorin98, Peter M. Visscher77,78, Yunpeng Wang85,131,181, Bradley T. Webb188, Shantel Marie Weinsheimer85,131, Jürgen Wellmann178, Gonneke Willemsen86, Stephanie H. Witt124, Yang Wu77, Hualin S. Xi189, Jian Yang78,190, Futao Zhang77, Volker Arolt191, Bernhard T. Baune90, Klaus Berger178, Dorret I. Boomsma86, Sven Cichon111,123,192,193, Udo Dannlowski191, E. J. C. de Geus86,194, J. Raymond DePaulo126, Enrico Domenici195, Katharina Domschke196, Tõnu Esko81,154, Hans J. Grabe186, Steven P. Hamilton197, Caroline Hayward198, Andrew C. Heath166, Kenneth S. Kendler93, Stefan Kloiber136,199,200, Glyn Lewis201, Qingqin S. Li202, Susanne Lucae136, Pamela A. F. Madden166, Patrik K. Magnusson98, Nicholas G. Martin127, Andrew M. McIntosh87,110, Andres Metspalu154,203, Ole Mors85,204, Preben Bo Mortensen84,85,88,89, Bertram Müller-Myhsok91,92,205, Merete Nordentoft85,206, Markus M. Nöthen111,112, Michael C. O’Donovan161, Sara A. Paciga207, Nancy L. Pedersen98, Brenda W. J. H. Penninx95, Roy H. Perlis115,208, David J. Porteous182, James B. Potash209, Martin Preisig102, Marcella Rietschel124, Catherine Schaefer137, Thomas G. Schulze124,170,210,211,212, Jordan W. Smoller115,116,117, Kari Stefansson176,213, Henning Tiemeier113,214,215, Rudolf Uher216, Henry Völzke179, Myrna M. Weissman150,217, Thomas Werge85,131,218, Cathryn M. Lewis103,219, Douglas F. Levinson220, Gerome Breen103,221, Anders D. Børglum83,84,85, Patrick F. Sullivan98,222,223
Funding
ATA received a Postgraduate Research Scholarship support from the University of Adelaide through the Adelaide Scholarship International (ASI) program. We thank 23andMe, Inc. staffs for giving us the permission to utilize GWAS summary data for depression which was included as part of the Psychiatric Genomics Consortium (PGC) study. The primary sources of funding were the Deutsche Forschungsgemeinschaft (DFG; grant no. RI 908/7–1; grant FOR2107, RI 908/11–1 to MR, NO 246/10–1 to MMN, WI3429/3–1 to SHW) and the Intramural Research Program of the National Institute of Mental Health (ZIA-MH00284311; ClinicalTrials.gov identifier: NCT00001174). The genotyping was in part funded by the German Federal Ministry of Education and Research (BMBF) through the Integrated Network IntegraMent (Integrated Understanding of Causes and Mechanisms in Mental Disorders), under the auspices of the e:Med Programme (grants awarded to TGS, MR, and MMN). Some data and biomaterials were collected as part of eleven projects (Study 40) that participated in the National Institute of Mental Health (NIMH) Bipolar Disorder Genetics Initiative. From 2003–2007, the Principal Investigators and Co-Investigators were: Indiana University, Indianapolis, IN, R01 MH59545, John Nurnberger, M.D., Ph.D., Marvin J. Miller, M.D., Elizabeth S. Bowman, M.D., N. Leela Rau, M.D., P. Ryan Moe, M.D., Nalini Samavedy, M.D., Rif El-Mallakh, M.D. (at University of Louisville), Husseini Manji, M.D. (at Johnson and Johnson), Debra A.Glitz, M.D. (at Wayne State University), Eric T. Meyer, Ph.D., M.S. (at Oxford University, UK), Carrie Smiley, R.N., Tatiana Foroud, Ph.D., Leah Flury, M.S., Danielle M. Dick, Ph.D. (at Virginia Commonwealth University), Howard Edenberg, Ph.D.; Washington University, St. Louis, MO, R01 MH059534, John Rice, Ph.D., Theodore Reich, M.D., Allison Goate, Ph.D., Laura Bierut, M.D.K02 DA21237; Johns Hopkins University, Baltimore, M.D., R01 MH59533, Melvin McInnis, M.D., JRD, M.D., Dean F. MacKinnon, M.D., FMM, M.D., JBP, M.D., PPZ, Ph.D., Dimitrios Avramopoulos, and Jennifer Payne; University of Pennsylvania, PA, R01 MH59553, Wade Berrettini, M.D., Ph.D.; University of California at San Francisco, CA, R01 MH60068, William Byerley, M.D., and Sophia Vinogradov, M.D.; University of Iowa, IA, R01 MH059548, William Coryell, M.D., and Raymond Crowe, M.D.; University of Chicago, IL, R01 MH59535, Elliot Gershon, M.D., Judith Badner, Ph.D., Francis McMahon, M.D., Chunyu Liu, Ph.D., Alan Sanders, M.D., Maria Caserta, Steven Dinwiddie, M.D., Tu Nguyen, Donna Harakal; University of California at San Diego, CA, R01 MH59567, JK, M.D., Rebecca McKinney, B.A.; Rush University, IL, R01 MH059556, William Scheftner, M.D., Howard M. Kravitz, D.O., M.P.H., Diana Marta, B.S., Annette Vaughn-Brown, M.S.N., R.N., and Laurie Bederow, M.A.; NIMH Intramural Research Program, Bethesda, MD, 1Z01MH002810-01, FJM, M.D., LK, Psy.D., Sevilla Detera-Wadleigh, Ph.D., Lisa Austin, Ph.D., Dennis L. Murphy, M.D.; Howard University, William B. Lawson, M.D., Ph.D., Evarista Nwulia, M.D., and Maria Hipolito, M.D. This work was supported by the NIH grants P50CA89392 from the National Cancer Institute and 5K02DA021237 from the National Institute of Drug Abuse. The Canadian part of the study was supported by the Canadian Institutes of Health Research to MA grant #64410 to MA. Collection and phenotyping of the Australian UNSW sample, by PBM, PRS, JMF and AW, was funded by an Australian NHMRC Program Grant (No.1037196)), with personnel supported by NHMRC project grants (Nos. 1063960, 1066177) and the Janette Mary O’Neil Research Fellowship to JMF. The collection of the Barcelona sample was supported by the Centro de Investigación en Red de Salud Mental (CIBERSAM), IDIBAPS, the CERCA Programme / Generalitat de Catalunya, Miguel Servet II and Instituto de Salud Carlos III (grant numbers PI080247, PI1200906, PI12/00018, 2017 SGR 1577 and 2017 SGR 1365). Dr FC was funded by unrestricted funding from CIBERSAM and “Secretaria d′Universitats i Recerca del Departament d′Economia i Coneixement (2017 SGR 134), Generalitat de Catalunya (Government of Catalonia). The Swedish Research Council, the Stockholm County Council, Karolinska Institutet and the Söderström-Königska Foundation supported this research through grants awarded to LB, LF, CL and MS. The collection of the Geneva sample was supported by the Swiss National Foundation (grants Synapsy 51NF40-158776 and 32003B-125469). The work by the French group was supported by INSERM (Institut National de la Santé et de la Recherche Médicale); AP-HP (Assistance Publique des Hôpitaux de Paris); the Fondation FondaMental (RTRS Santé Mentale) and the labex Bio-PSY (Investissements d’Avenir program managed by the ANR under reference ANR-11-IDEX-0004-02). The German Research Foundation (DFG, grant FOR2107 DA1151/5-1 and DA1151/5–2 to UD; SFB-TRR58, Project C09 and Z02 to UD), and the Interdisciplinary Center for Clinical Research (IZKF) of the Medical Faculty of the University of Münster (grant Dan3/012/17 to UD) supported this work. The collection of the Romanian sample was supported by U.E.F.I.S.C.D.I., Romania, grant awarded to MG-S. The collection of the Czech sample was supported by the project No. LO1611 with a financial support from the MEYS under the NPU I program and by the Czech Science Foundation, grant No. 17-07070S.
Author information
Authors and Affiliations
Consortia
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Members of the Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium are listed below acknowldgments.
Supplementary information
Rights and permissions
About this article
Cite this article
Amare, A.T., Schubert, K.O., Hou, L. et al. Association of polygenic score for major depression with response to lithium in patients with bipolar disorder. Mol Psychiatry 26, 2457–2470 (2021). https://doi.org/10.1038/s41380-020-0689-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41380-020-0689-5
This article is cited by
-
Effectiveness of ultra-long-term lithium treatment: relevant factors and case series
International Journal of Bipolar Disorders (2024)
-
Key subphenotypes of bipolar disorder are differentially associated with polygenic liabilities for bipolar disorder, schizophrenia, and major depressive disorder
Molecular Psychiatry (2024)
-
Pharmacogenomic overlap between antidepressant treatment response in major depression & antidepressant associated treatment emergent mania in bipolar disorder
Translational Psychiatry (2024)
-
Polygenic risk scores of lithium response and treatment resistance in major depressive disorder
Translational Psychiatry (2023)
-
Focal adhesion is associated with lithium response in bipolar disorder: evidence from a network-based multi-omics analysis
Molecular Psychiatry (2023)