Abstract
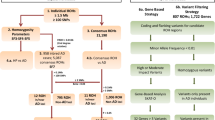
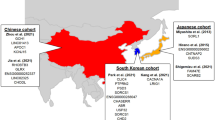
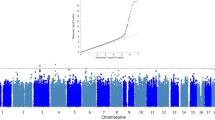
Late-onset Alzheimer’s disease (LOAD) is significantly more frequent in Hispanics than in non-Hispanic Whites. Ancestry may explain these differences across ethnic groups. To this end, we studied a large cohort of Caribbean Hispanics (CH, N = 8813) and tested the association between Local Ancestry (LA) and LOAD (“admixture mapping”) to identify LOAD-associated ancestral blocks, separately for ancestral components (European [EUR], African [AFR], Native American[NA]) and jointly (AFR + NA). Ancestral blocks significant after permutation were fine-mapped employing multi-ethnic whole-exome sequencing (WES) to identify rare variants associated with LOAD (SKAT-O) and replicated in the UK Biobank WES dataset. Candidate genes were validated studying (A) protein expression in human LOAD and control brains; (B) two animal AD models, Drosophila and Zebrafish. In the joint AFR + NA model, we identified four significant ancestral blocks located on chromosomes 1 (p value = 8.94E-05), 6 (p value = 8.63E-05), 21 (p value = 4.64E-05) and 22 (p value = 1.77E-05). Fine-mapping prioritized the GCAT gene on chromosome 22 (SKAT-O p value = 3.45E-05) and replicated in the UK Biobank (SKAT-O p value = 0.05). In LOAD brains, a decrease of 28% in GCAT protein expression was observed (p value = 0.038), and GCAT knockdown in Amyloid-β42 Drosophila exacerbated rough eye phenotype (68% increase, p value = 4.84E-09). In zebrafish, gcat expression increased after acute amyloidosis (34%, p value = 0.0049), and decreased upon anti-inflammatory Interleukin-4 (39%, p value = 2.3E-05). Admixture mapping uncovered genomic regions harboring new LOAD-associated loci that might explain the observed different frequency of LOAD across ethnic groups. Our results suggest that the inflammation-related activity of GCAT is a response to amyloid toxicity, and reduced GCAT expression exacerbates AD pathology.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Tang MX, Cross P, Andrews H, Jacobs DM, Small S, Bell K, et al. Incidence of AD in African-Americans, Caribbean Hispanics, and Caucasians in northern Manhattan. Neurology. 2001;56:49–56. https://doi.org/10.1212/wnl.56.1.49. PubMed PMID: 11148235.
Vardarajan BN, Faber KM, Bird TD, Bennett DA, Rosenberg R, Boeve BF, et al. Age-specific incidence rates for dementia and Alzheimer disease in NIA-LOAD/NCRAD and EFIGA families: National Institute on Aging Genetics Initiative for Late-Onset Alzheimer Disease/National Cell Repository for Alzheimer Disease (NIA-LOAD/NCRAD) and Estudio Familiar de Influencia Genetica en Alzheimer (EFIGA). JAMA Neurol. 2014;71:315–23. https://doi.org/10.1001/jamaneurol.2013.5570. PubMed PMID: 24425039; PubMed Central PMCID: PMCPMC4000602.
Tang MX, Stern Y, Marder K, Bell K, Gurland B, Lantigua R, et al. The APOE-epsilon4 allele and the risk of Alzheimer disease among African Americans, whites, and Hispanics. JAMA. 1998;279:751–5. https://doi.org/10.1001/jama.279.10.751. PubMed PMID: 9508150.
Tosto G, Reitz C. Genome-wide association studies in Alzheimer’s disease: a review. Curr Neurol Neurosci Rep. 2013;13:381 https://doi.org/10.1007/s11910-013-0381-0. Epub 2013/08/21PubMed PMID: 23954969; PubMed Central PMCID: PMCPMC3809844.
Sariya S, Felsky D, Reyes-Dumeyer D, Lali R, Lantigua RA, Vardarajan B, et al. Polygenic risk score for Alzheimer’s Disease in Caribbean Hispanics. Ann Neurol. 2021. Epub 2021/05/27. https://doi.org/10.1002/ana.26131. PubMed PMID: 34038570.
McKhann GM, Knopman DS, Chertkow H, Hyman BT, Jack CR Jr., Kawas CH, et al. The diagnosis of dementia due to Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011;7:263–9. https://doi.org/10.1016/j.jalz.2011.03.005. PubMed PMID: 21514250; PubMed Central PMCID: PMCPMC3312024.
Prince M, Ferri CP, Acosta D, Albanese E, Arizaga R, Dewey M, et al. The protocols for the 10/66 dementia research group population-based research programme. BMC Public Health. 2007;7:165 https://doi.org/10.1186/1471-2458-7-165. PubMed PMID: 17659078; PubMed Central PMCID: PMCPMC1965476.
Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience.2015;4:7 https://doi.org/10.1186/s13742-015-0047-8. PubMed PMID: 25722852; PubMed Central PMCID: PMCPMC4342193.
Das S, Forer L, Schonherr S, Sidore C, Locke AE, Kwong A, et al. Next-generation genotype imputation service and methods. Nat Genet. 2016;48:1284–7. https://doi.org/10.1038/ng.3656. PubMed PMID: 27571263; PubMed Central PMCID: PMCPMC5157836.
Taliun D, Harris DN, Kessler MD, Carlson J, Szpiech ZA, Torres R, et al. Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. Nature.2021;590:290–9. https://doi.org/10.1038/s41586-021-03205-y. PubMed PMID: 33568819; PubMed Central PMCID: PMCPMC7875770.
Loh PR, Palamara PF, Price AL. Fast and accurate long-range phasing in a UK Biobank cohort. Nat Genet. 2016;48:811–6. https://doi.org/10.1038/ng.3571. PubMed PMID: 27270109; PubMed Central PMCID: PMCPMC4925291.
Fuchsberger C, Abecasis GR, Hinds DA. minimac2: faster genotype imputation. Bioinformatics. 2015;31:782–4. https://doi.org/10.1093/bioinformatics/btu704. PubMed PMID: 25338720; PubMed Central PMCID: PMCPMC4341061.
Sariya S, Lee JH, Mayeux R, Vardarajan BN, Reyes-Dumeyer D, Manly JJ, et al. Rare variants imputation in admixed populations: comparison across reference panels and bioinformatics tools. Front Genet. 2019;10:239. https://doi.org/10.3389/fgene.2019.00239. Epub 2019/04/20PubMed PMID: 31001313; PubMed Central PMCID: PMCPMC6456789.
Alexander DH, Lange K. Enhancements to the ADMIXTURE algorithm for individual ancestry estimation. BMC Bioinforma. 2011;12:246 https://doi.org/10.1186/1471-2105-12-246. PubMed PMID: 21682921; PubMed Central PMCID: PMCPMC3146885.
Guan Y. Detecting structure of haplotypes and local ancestry. Genetics .2014;196:625–42. https://doi.org/10.1534/genetics.113.160697. Epub 2014/01/07PubMed PMID: 24388880; PubMed Central PMCID: PMCPMC3948796.
Zhou X, Stephens M. Genome-wide efficient mixed-model analysis for association studies. Nat Genet. 2012;44:821–4. https://doi.org/10.1038/ng.2310. PubMed PMID: 22706312; PubMed Central PMCID: PMCPMC3386377.
Gogarten SM, Sofer T, Chen H, Yu C, Brody JA, Thornton TA, et al. Genetic association testing using the GENESIS R/Bioconductor package. Bioinformatics. 2019;35:5346–8. https://doi.org/10.1093/bioinformatics/btz567. PubMed PMID: 31329242.
Shriner D, Adeyemo A, Rotimi CN. Joint ancestry and association testing in admixed individuals. PLoS Comput Biol. 2011;7:e1002325 https://doi.org/10.1371/journal.pcbi.1002325. PubMed PMID: 22216000; PubMed Central PMCID: PMCPMC3245293.
Joo JW, Hormozdiari F, Han B, Eskin E. Multiple testing correction in linear mixed models. Genome Biol. 2016;17:62 https://doi.org/10.1186/s13059-016-0903-6. PubMed PMID: 27039378; PubMed Central PMCID: PMCPMC4818520.
Liu Z, Shriner D, Hansen NF, Rotimi CN, Mullikin JC, Program NCS. Admixture mapping identifies genetic regions associated with blood pressure phenotypes in African Americans. PLoS ONE. 2020;15:e0232048 https://doi.org/10.1371/journal.pone.0232048. PubMed PMID: 32315356; PubMed Central PMCID: PMCPMC7173845.
Tosto G, Vardarajan B, Sariya S, Brickman AM, Andrews H, Manly JJ, et al. Association of Variants in PINX1 and TREM2 With Late-Onset Alzheimer Disease. JAMA Neurol. 2019. https://doi.org/10.1001/jamaneurol.2019.1066. PubMed PMID: 31058951; PubMed Central PMCID: PMCPMC6503572.
Karczewski K, Solomonson M, Chao KR, Goodrich JK, Tiao G, Lu W, et al. Systematic single-variant and gene-based association testing of 3,700 phenotypes in 281,850 UK Biobank exomes. medRxiv. 2021:2021.06.19.21259117. https://doi.org/10.1101/2021.06.19.21259117.
Casas-Tinto S, Zhang Y, Sanchez-Garcia J, Gomez-Velazquez M, Rincon-Limas DE, Fernandez-Funez P. The ER stress factor XBP1s prevents amyloid-beta neurotoxicity. Hum Mol Genet. 2011;20:2144–60. https://doi.org/10.1093/hmg/ddr100. Epub 2011/03/11PubMed PMID: 21389082; PubMed Central PMCID: PMCPMC3090193.
Reid DW, Muyskens JB, Neal JT, Gaddini GW, Cho LY, Wandler AM, et al. Identification of genetic modifiers of CagA-induced epithelial disruption in Drosophila. Front Cell Infect Microbiol. 2012;2:24 https://doi.org/10.3389/fcimb.2012.00024. PubMed PMID: 22919616; PubMed Central PMCID: PMCPMC3417398.
Santa-Maria I, Alaniz ME, Renwick N, Cela C, Fulga TA, Van Vactor D, et al. Dysregulation of microRNA-219 promotes neurodegeneration through post-transcriptional regulation of tau. J Clin Invest. 2015;125:681–6. https://doi.org/10.1172/JCI78421. PubMed PMID: 25574843; PubMed Central PMCID: PMCPMC4319412.
Bhattarai P, Cosacak MI, Mashkaryan V, Demir S, Popova S, Govindarajan N, et al. Neuron-glia interaction through Serotonin-BDNF-NGFR axis enables regenerative neurogenesis in Alzheimer’s model of adult zebrafish brain. PLoS Biol. 2020;18:e3000585 https://doi.org/10.1371/journal.pbio.3000585.
Cosacak MI, Bhattarai P, Reinhardt S, Petzold A, Dahl A, Zhang Y, et al. Single-Cell transcriptomics analyses of neural stem cell heterogeneity and contextual plasticity in a Zebrafish Brain Model of Amyloid Toxicity. Cell Rep. 2019;27:1307–18 e3. https://doi.org/10.1016/j.celrep.2019.03.090. PubMed PMID: 31018142.
Bhattarai P, Thomas AK, Zhang Y, Kizil C. The effects of aging on Amyloid-β42-induced neurodegeneration and regeneration in adult zebrafish brain. Neurogenesis. 2017;4:e1322666 https://doi.org/10.1080/23262133.2017.1322666. Epub 2 May 2017.
Bhattarai P, Thomas AK, Papadimitriou C, Cosacak MI, Mashkaryan V, Froc C, et al. IL4/STAT6 signaling activates neural stem cell proliferation and neurogenesis upon Amyloid-β42 aggregation in adult zebrafish brain. Cell Rep. 2016;17:941–8. https://doi.org/10.1016/j.celrep.2016.09.075.
Meyer D, VR CA, Bitarello BD, DY CB, Nunes K. A genomic perspective on HLA evolution. Immunogenetics. 2018;70:5–27. https://doi.org/10.1007/s00251-017-1017-3. PubMed PMID: 28687858; PubMed Central PMCID: PMCPMC5748415.
Moreno-Estrada A, Gravel S, Zakharia F, McCauley JL, Byrnes JK, Gignoux CR, et al. Reconstructing the population genetic history of the Caribbean. PLoS Genet. 2013;9:e1003925. https://doi.org/10.1371/journal.pgen.1003925. PubMed PMID: 24244192; PubMed Central PMCID: PMCPMC3828151 Ancestry.com, 23andMe’s “Roots into the Future” project, and Personalis, Inc. He is on the medical advisory board of Invitae and Med-tek. None of these entities played any role in the project or research results reported here.
Bonnemaijer PWM, Cook C, Nag A, Hammond CJ, van Duijn CM, Lemij HG, et al. Genetic African Ancestry Is Associated With Central Corneal Thickness and Intraocular Pressure in Primary Open-Angle Glaucoma. Invest Ophthalmol Vis Sci. 2017;58:3172–80. https://doi.org/10.1167/iovs.17-21716. Epub 2017/06/28PubMed PMID: 28654982.
Hohman TJ, Cooke-Bailey JN, Reitz C, Jun G, Naj A, Beecham GW, et al. Global and local ancestry in African-Americans: Implications for Alzheimer’s disease risk. Alzheimers Dement. 2016;12:233–43. https://doi.org/10.1016/j.jalz.2015.02.012. Epub 2015/06/21PubMed PMID: 26092349; PubMed Central PMCID: PMCPMC4681680.
Tosto G, Vardarajan B, Sariya S, Brickman AM, Andrews H, Manly JJ, et al. Association of Variants in PINX1 and TREM2 With Late-Onset Alzheimer Disease. JAMA Neurol. 2019;76:942–8. https://doi.org/10.1001/jamaneurol.2019.1066. Epub 2019/05/07PubMed PMID: 31058951; PubMed Central PMCID: PMCPMC6503572.
Ravichandran M, Priebe S, Grigolon G, Rozanov L, Groth M, Laube B, et al. Impairing L-Threonine Catabolism Promotes Healthspan through Methylglyoxal-Mediated Proteohormesis. Cell Metab. 2018;27:914–25 e5. https://doi.org/10.1016/j.cmet.2018.02.004. PubMed PMID: 29551589.
Montine TJ, Phelps CH, Beach TG, Bigio EH, Cairns NJ, Dickson DW, et al. National Institute on Aging-Alzheimer’s Association guidelines for the neuropathologic assessment of Alzheimer’s disease: a practical approach. Acta Neuropathol. 2012;123:1–11. https://doi.org/10.1007/s00401-011-0910-3. PubMed PMID: 22101365; PubMed Central PMCID: PMCPMC3268003.
Haass C, Selkoe DJ. Soluble protein oligomers in neurodegeneration: lessons from the Alzheimer’s amyloid beta-peptide. Nat Rev Mol Cell Biol. 2007;8:101–12. https://doi.org/10.1038/nrm2101. PubMed PMID: 17245412.
Krautwald M, Munch G. Advanced glycation end products as biomarkers and gerontotoxins - A basis to explore methylglyoxal-lowering agents for Alzheimer’s disease? Exp Gerontol. 2010;45:744–51. https://doi.org/10.1016/j.exger.2010.03.001. PubMed PMID: 20211718.
Angeloni C, Zambonin L, Hrelia S. Role of methylglyoxal in Alzheimer’s disease. Biomed Res Int. 2014;2014:238485. https://doi.org/10.1155/2014/238485. PubMed PMID: 24734229; PubMed Central PMCID: PMCPMC3966409.
Pandey UB, Nichols CD. Human disease models in Drosophila melanogaster and the role of the fly in therapeutic drug discovery. Pharm Rev. 2011;63:411–36. https://doi.org/10.1124/pr.110.003293. PubMed PMID: 21415126; PubMed Central PMCID: PMCPMC3082451.
Prussing K, Voigt A, Schulz JB. Drosophila melanogaster as a model organism for Alzheimer’s disease. Mol Neurodegener. 2013;8:35 https://doi.org/10.1186/1750-1326-8-35. PubMed PMID: 24267573; PubMed Central PMCID: PMCPMC4222597.
Kizil C, Bhattarai P. Is Alzheimer’s also a stem cell disease? - The Zebrafish Perspective. Front Cell Dev Biol. 2018;6:159 https://doi.org/10.3389/fcell.2018.00159. PubMed PMID: 30533414; PubMed Central PMCID: PMCPMC6265475.
Kizil C. Mechanisms of Pathology-Induced Neural Stem Cell Plasticity and Neural Regeneration in Adult Zebrafish Brain. Curr Pathobiol Rep. 2018;6:71–7. https://doi.org/10.1007/s40139-018-0158-x. PubMed PMID: 29938129; PubMed Central PMCID: PMCPMC5978899.
Kizil C, Kyritsis N, Brand M. Effects of inflammation on stem cells: together they strive? EMBO Rep. 2015;16:416–26. https://doi.org/10.15252/embr.201439702. PubMed PMID: 25739812.
Kizil C, Dudczig S, Kyritsis N, Machate A, Blaesche J, Kroehne V, et al. The chemokine receptor cxcr5 regulates the regenerative neurogenesis response in the adult zebrafish brain. Neural Dev. 2012;7:27. Epub 2012/07/25. 1749-8104-7-27 [pii] 10.1186/1749-8104-7-27. PubMed PMID: 22824261.
Kizil C, Kyritsis N, Dudczig S, Kroehne V, Freudenreich D, Kaslin J, et al. Regenerative neurogenesis from neural progenitor cells requires injury-induced expression of Gata3. Dev Cell. 2012;23:1230–7. Epub 2012/11/22. doi: S1534-5807(12)00477-7 [pii] 10.1016/j.devcel.2012.10.014. PubMed PMID: 23168169.
Kyritsis N, Kizil C, Zocher S, Kroehne V, Kaslin J, Freudenreich D, et al. Acute inflammation initiates the regenerative response in the adult zebrafish brain. Science. 2012;338:1353–6. Epub 2012/11/10. doi: science.1228773 [pii] 10.1126/science.1228773. PubMed PMID: 23138980.
Jurisch-Yaksi N, Yaksi E, Kizil C. Radial glia in the zebrafish brain: Functional, structural, and physiological comparison with the mammalian glia. Glia. 2020. https://doi.org/10.1002/glia.23849. PubMed PMID: 32476207.
Acknowledgements
The National Institutes of Health (NIH), National Institute on Aging (NIH-NIA) and National Institute of Neurological Disorders and Stroke supported this work through the following grants: R56AG069118, R56AG066889, R56AG059756, R01AG056531, and R01NS095922. For WHICAP: Data collection and sharing for this project was supported by the Washington Heights-Inwood Columbia Aging Project (WHICAP, R01AG037212, RF1AG054023, RF1AG066107) funded by the NIH-NIA and by the National Center for Advancing Translational Sciences, NIH, through Grant Number UL1TR001873. This manuscript has been reviewed by WHICAP investigators for scientific content and consistency of data interpretation with previous WHICAP Study publications. We acknowledge the WHICAP study participants and the WHICAP research and support staff for their contributions to this study. For EFIGA: Data collection for this project was supported by the Genetic Studies of Alzheimer’s disease in Caribbean Hispanics (EFIGA) funded by the NIH-NIA and by the NIH (5R37AG015473, RF1AG015473, R56AG051876, R01AG067501, R56AG063908, RF1AG015473. We acknowledge the EFIGA study participants and the EFIGA research and support staff for their contributions to this study. For ADI 10/66 PR Alzheimer’s Disease International Epidemiological Study: Data collection for this project was supported by a recurrent PR Legislature grant, Pfizer Co. Grant # GA9001NE, and for PR Apo-E labs: Human Genetics Core Award from Columbia University Irving Institute for Clinical and Translational Research. Zebrafish work was supported by grants from Helmholtz Association (VH-NG-1021) and Schaefer Research Scholars Award. We thank Drs. Mehmet Ilyas Cosacak and Prabesh Bhattarai for gene expression analyses in zebrafish.
Author information
Authors and Affiliations
Contributions
GT, ISM, CK contributed to the conception and design of the study. GT, SS, RM, BV, IJV, AM, YA, DRD, JH, FR, EM contributed to the acquisition and analysis of data. GT, SS, ISM, CK contributed to drafting the text or preparing the figures.
Corresponding author
Ethics declarations
Competing interests
CK has executive role in Neuron-D GmbH, which had no academic or financial relationship to the design and execution of this project. Supplementary Information is available at MP’s website.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Kizil, C., Sariya, S., Kim, Y.A. et al. Admixture Mapping of Alzheimer’s disease in Caribbean Hispanics identifies a new locus on 22q13.1. Mol Psychiatry 27, 2813–2820 (2022). https://doi.org/10.1038/s41380-022-01526-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41380-022-01526-6
This article is cited by
-
Rare genetic variation in fibronectin 1 (FN1) protects against APOEε4 in Alzheimer’s disease
Acta Neuropathologica (2024)
-
A global view of the genetic basis of Alzheimer disease
Nature Reviews Neurology (2023)
-
Nerve growth factor receptor (Ngfr) induces neurogenic plasticity by suppressing reactive astroglial Lcn2/Slc22a17 signaling in Alzheimer’s disease
npj Regenerative Medicine (2023)
-
FMNL2 regulates gliovascular interactions and is associated with vascular risk factors and cerebrovascular pathology in Alzheimer’s disease
Acta Neuropathologica (2022)