Abstract
Background
Youth populations with overweight/obesity (OW/OB) exhibit heterogeneity in cardiometabolic health phenotypes. The underlying mechanisms for those differences are still unclear. This study aimed to analyze the whole-blood transcriptome profile (RNA-seq) of children with metabolic healthy overweight/obesity (MHO) and metabolic unhealthy overweight/obesity (MUO) phenotypes.
Methods
Twenty-seven children with OW/OB (10.1 ± 1.3 years, 59% boys) from the ActiveBrains project were included. MHO was defined as having none of the following criteria for metabolic syndrome: elevated fasting glucose, high serum triglycerides, low high-density lipoprotein-cholesterol, and high systolic or diastolic blood pressure, while MUO was defined as presenting one or more of these criteria. Inflammatory markers were additionally determined. Total blood RNA was analyzed by 5’-end RNA-sequencing.
Results
Whole-blood transcriptome analysis revealed a distinct pattern of gene expression in children with MHO compared to MUO children. Thirty-two genes differentially expressed were linked to metabolism, mitochondrial, and immune functions.
Conclusions
The identified gene expression patterns related to metabolism, mitochondrial, and immune functions contribute to a better understanding of why a subset of the population remains metabolically healthy despite having overweight/obesity.
Impact
-
A distinct pattern of whole-blood transcriptome profile (RNA-seq) was identified in children with metabolic healthy overweight/obesity (MHO) compared to metabolic unhealthy overweight/obesity (MUO) phenotype.
-
The most relevant genes in understanding the molecular basis underlying the MHO/MUO phenotypes in children could be: RREB1, FAM83E, SLC44A1, NRG1, TMC5, CYP3A5, TRIM11, and ADAMTSL2.
-
The identified whole-blood transcriptome profile related to metabolism, mitochondrial, and immune functions contribute to a better understanding of why a subset of the population remains metabolically healthy despite having overweight/obesity.
Similar content being viewed by others
Introduction
Childhood obesity is strongly associated with increased cardiometabolic risk factors (e.g., dyslipidemia and hypertension) and type 2 diabetes.1,2 However, there is a subset of children and adolescents with overweight/obesity (OW/OB) (6–22%, depending on the classification criteria,3,4 who present a normal/healthy metabolic profile that might reflect a better cardiovascular disease prognosis.3,5 Metabolically “healthy” obesity (MHO) is defined as a subset of individuals with obesity (OB) according to the standard definition of body mass index (BMI) who lack metabolic syndrome symptoms. In contrast, MUO (i.e., metabolically unhealthy overweight/obesity) children and adolescents have 1–4 metabolic syndrome classification criteria5,6: fasting glucose (≥100.9 mg/dl in boys and girls), high serum triglycerides (≥127.4 mg/dl in boys and ≥141.6 mg/dl in girls), low high-density lipoprotein (HDL)-cholesterol (≤43.7 mg/dl boys and ≤48.3 mg/dl in girls), and systolic or diastolic blood pressure (systolic ≥121 mmHg in boys and girls, diastolic ≥76 mmHg in boys and ≥80 mmHg in girls). In pediatric populations, MHO has mostly included children with both OW and OB (for simplicity, the abbreviation MHO will be hereinafter used instead of MHO-O or others) to maximize sample size and power.7,8
The underlying mechanisms for the normal/healthy metabolic profile of individuals with OW/OB are poorly understood.9 Naukkarinen et al. reported lower expression of inflammatory genes and higher expression of mitochondrial genes in subcutaneous adipose tissue of young adults with MHO compared to MUO.10 Das et al. also evaluated the transcriptome profile in the subcutaneous adipose tissue of middle-aged adults with MHO compared to MUO11 and found differences in genes involved in cardiovascular disease-related processes and immune/inflammatory responses. However, a study by Gómez-Ambrosi et al.12 analyzed candidate genes involved in inflammation and tissue remodeling and detected no differences between adults with MHO and MUO in the liver and visceral adipose tissue. The inconsistent results in the different studies may be explained by different ways to classify individuals with MHO and MUO5,13,14 and/or different tissues used to analyze transcriptome profiles (e.g., visceral adipose tissue, liver, subcutaneous adipose tissue, and peripheral blood mononuclear cells).10,11,12,15
Nevertheless, to the best of our knowledge, no previous studies have characterized the transcriptome profiles of children with MHO and MUO. Due to ethical considerations, tissue biopsies are not permitted in pediatric populations, so that the main tissue to be studied in this population is whole blood, which can reflect the systemic inflammation in the body. Obesity is characterized by a low-grade systemic inflammation, which is involved in the pathophysiology of type 2 diabetes mellitus and arteriosclerosis.16,17 Studies have pointed to the tissue macrophages as a source, which may contribute to increases in inflammation in adipose tissue, resulting in the dysregulation of the endocrine function inducing low-grade systemic inflammation in persons with obesity.18 In this regard, one of the main factors that could contribute to an MHO phenotype is the presence of a low pro-inflammatory profile.19 However, why some individuals would have a lower pro-inflammatory profile than others in the presence of an excessive weight/adiposity is still poorly understood and the study of differences in gene expression levels between MHO vs. MUO can shed light on this.
The present study aimed to characterize the whole-blood transcriptome profile of children with MHO and MUO to promote a better understanding of why a subset of the population remains metabolically healthy despite having OW/OB.
Methods
Participants
In total, 27 children with OW and OB, ages 10.1 ± 1.3 years, 59% boys, from the ActiveBrains project (www.profith.ugr.es/activebrains, Clinical Trial: NCT02295072) were included in this analysis. Detailed design, methods, and inclusion/exclusion criteria have been described elsewhere.20 The study was approved by the Committee for Research Involving Human Subjects at the University of Granada (Reference: 848, February 2014). All parents were informed about the study objective and written informed consent following the Declaration of Helsinki.
Body composition measurements and maturational status
Body weight and height were measured with an electronic scale and a stadiometer (Seca Instruments, Germany, Ltd). BMI was calculated as kg/m2 and participants were then classified as children with OW or OB according to the sex- and age-specific World Obesity Federation cut-off points.21 Waist circumference (WC) was evaluated as using the International Society for the Advancement of Kinanthropometry procedures.22 Body weight, height, and WC were collected twice consecutively by the same trained evaluator, and the average for each metric was used. Body fat percentage, fat-free mass, and visceral adipose tissue were measured by dual energy X-ray absorptiometry (DXA; Discovery densitometer from Hologic). The positioning of the participants and the analyses of the results were undertaken following recommendations from the International Society of Clinical Densitometry.23 All DXA analyses were performed by the same evaluator using the GE encore software (version 4.0.2).
The maturational status of the participants was reflected by peak height velocity (PHV) using age and height in validated algorithms24: in boys: −8.1 + (0.0070346 × (age × sitting height); and in girls: −7.7 + (0.0042232 × (age × height). Maturity offset was calculated by subtracting the PHV age from the chronological age.
Definition of MHO and MUO in youth
Cardiometabolic risk factors used in our study to define the MHO and MUO (i.e., fasting glucose, serum triglycerides, and HDL-cholesterol) were measured from fasting blood samples. Blood pressure (systolic and diastolic) was measured from the left arm with an automatic sphygmomanometer (Omron M6, the Netherlands) on two different days, and the minimum value was registered for the analyses.
Our participants were classified using the harmonization definition for MHO and MUO in youth using the Jolliffe and Janssen metabolic syndrome study,25 based on a comprehensive review of different criteria available (for more details, see Ortega et al.5,6). In this study, age- and sex-specific cut-off points for each marker of metabolic syndrome were developed for adolescent populations from 12 to 18 years.25 Cut-off points for boys and girls aged 12 years25 were used in our study, since it was the closest to the age range of our sample, i.e., 9–11 years. The strength of these cut-off points are equivalent to those proposed for adults by the International Diabetes Federation and Adults Treatment Panel and the adaptation by age- and sex-specific based on growth curves in youth.5 The metabolic syndrome is defined as having fasting glucose (≥100.9 mg/dl in boys and girls), high serum triglycerides (≥127.4 mg/dl in boys and ≥141.6 mg/dl in girls), low HDL-cholesterol (≤43.7 mg/dl boys and ≤48.3 mg/dl in girls), and systolic or diastolic blood pressure (systolic ≥ 121 mmHg in boys and girls, diastolic ≥76 mmHg in boys and ≥80 mmHg in girls). Children lacking all the metabolic syndrome criteria (excluding high WC) were classified as MHO. Children with one or more of the previous criteria were classified as MUO. Both OW and OB were included in the analysis (n = 27; 6 OW and 21 OB) to maximize sample size and in line with previous studies in pediatric populations.7,8 Homeostasis model assessment for insulin resistance as an index of insulin resistance were provided.
Blood sampling and analysis
Participants arrived at the laboratory between 8 and 9 a.m. after an overnight fasting of at least 12 h. Venous blood collected in tubes with EDTA was spun immediately at 1000 × g for 10 min. Plasma was isolated and stored at −80 °C until analyses. In addition, 500 μl blood were collected into tubes containing 1.3 ml RNAlater solution (Ambion Inc.; Austin, TX, USA) for transcriptome analysis and was stored at −80 °C until further processing.
Inflammatory markers
Interleukin-6 (IL-6), IL-1β, and tumor necrosis factor-α (TNF-α) cytokines were measured in plasma using multiple analyte profiling technology (MILLIPLEX® MAP Human High Sensitivity T Cell Magnetic Bead Panel, EMD Millipore Corporation, MO, USA) with a kit plex (HCYIL6-MAG Anti-Human IL-6 Beads set, HCYIL1B-MAG Anti-Human IL-1β Bead, and HCYTNFA-MAG Anti-Human TNFα Bead set).26 The intra- and inter-assay coefficients of variation (CVs) for IL-6 were ≤5% and ≤20%, respectively, and sensitivity was 0.11 pg/ml. For both IL-1β and TNF-α, the intra- and inter-assay CVs were ≤5% and ≤15%, respectively, with a sensitivity of 0.14 pg/ml for IL-1β, and of 0.16 pg/ml for TNF-α.26 Vascular endothelial growth factor A (VEGF-A) and epidermal growth factor (EGF) were quantified by multiple analyte profiling technology (MILLIPLEX® MAP Human Angiogenesis/Growth Factor Magnetic Bead Panel 1, EMD Millipore Corporation, MO, USA) using a kit plex (HVEGF-MAG Anti-Human VEGF-A Bead and HAGEGF-MAG Anti-Human EGF Bead).27 The intra- and inter-assay CVs for VEGF-A were ≤3.5% and ≤10%, respectively, and sensitivity was 8.1 pg/ml. For EGF, the intra- and inter-assay CVs were ≤3.2% and ≤6.8%, respectively, with a sensitivity of 1 pg/ml.27
RNA extraction and sequencing
Total RNA was isolated from the blood samples that contained RNAlater using the RiboPureTM-Blood Kit (Thermo Fisher Scientific; Waltham, MA, USA). Subsequently, GlobinLock molecular mechanism was applied to block the high globin mRNA content of erythrocytes, which is abundant in the blood and could hamper the whole-blood RNA analyses.28 Full transcriptome analysis was performed following the modified version of single-cell tagged reverse transcription (STRT) protocol as described in detail before,29 where 10 ng of high-quality input RNA was converted into cDNA and amplified to form an Illumina-compatible library. The STRTprep pipeline, available at https://github.com/shka/STRTprep/tree/v3dev, was used for processing raw sequencing reads, aligning to the hg19 genome, and quantitating the expression levels. RNA-seq data are available at www.ncbi.nlm.nih.gov/geo under accession number GSE146869.
Statistical analyses
The differences in sample characteristics between children with MHO and MUO were tested using the non-parametric Mann–Whitney U and chi-squared test for continuous and categorical variables, respectively. Winsorizing the data was performed to limit the influence of extreme values. Briefly, the winsorization method replaces extreme high/low values for the closest (highest/lowest) valid values.30 The analyses were performed using SPSS version 21.0 (IBM Corporation, NY, USA), and the statistical significance was defined at the level of p < 0.05. Differential expression analysis between children with MHO and MUO was performed with Limma R/Bioconductor software package.31 Prior to performing the analyses, quantile normalization was performed on gene expression data of RNA-sequencing (RNA-seq). These analyses were adjusted by sex and maturational status. Statistically differentially regulated genes were defined by a false discovery rate (FDR) < 5% (Benjamini and Hochberg correction on multiple testing). These genes were characterized by functional enrichment analysis using g:Profiler.32 Biological process and pathways with an FDR < 0.05 were considered significantly enriched.
Additionally, we performed a weighted gene coexpression network analysis (WGCNA) following the guidelines for these analyses33,34,35 in order to explore possible gene networks. Two different networks of genes coexpressed based on terms of correlations were created, i.e., one network in children with MHO and another network of genes coexpressed in the group of children with MUO. The subsequent step was to define and characterize the non-preserved modules or subnetworks (i.e., clusters of genes named by colors) and their function in the network constructed in children with MHO respect to the network computed in children with MUO. The “modulePreservation” WGCNA function was used to compute a range of preservation across the two networks. Module preservation statistics were calculated to evaluate whether a given color module defined in one data set (reference network, in our study the reference network was computed in children with MHO) can also be found in another data set (test network, in our study the test network was computed in children with MUO). Module preservation among children with MHO and MUO was reported as a composite Zsummary measurement of connectivity and density. Zsummary scores >10 indicate high network preservation (i.e., a module color or group of genes is similarly clustered in children with MHO and MUO), scores between 2 and 10 are indicative of moderate network preservation, and Zsummary measures <2 provide no evidence of preservation (i.e., a color module or group of genes is clustered in children with MHO but not clustered in MUO). Hub genes (i.e., highly connected intramodular genes, referring to bigger role within the network) were identified in color modules with low preservation based on the module membership value (MM). The MM has been calculated as the correlation between the module eigengene (i.e., first principal component) of a module and the expression profile of a gene. Genes whose MM was >0.8 were considered as hub genes in each module.36
The Venn diagram37 was used to perform the overlapping between genes differentially expressed (i.e., Limma analyses) and the hub genes in color modules with low preservation (i.e., WGCNA analyses) in children with MHO compared to MUO. A molecular interaction network was created using GeneMania app in CYTOSCAPE platform,38 and the most relevant genes in the molecular network were defined based on the highest value of centrality degree, which is defined as the number of interactions in which a node (i.e., gene) is involved.
In silico validation analysis was performed using two publicly available (PHENOPEDIA database) lists of OB and metabolism genes. We performed the overlapping between the most important genes found in our study (i.e., genes overlapped between Limma and WGCNA analyses) and the two lists. These relevant genes in OB and metabolism were identified following a semi-automatic workflow for the identification of the most relevant genes in a pathology.39 For the interested reader wishing to retrace our analyses, all files are available from the Open Science Framework (https://osf.io/xt8v4/).
Results
Anthropometric characteristics and cardiometabolic and inflammatory markers of participants are presented in Table 1. Ten boys and three girls were classified as MHO and six boys and eight girls were classified as MUO. MUO had abnormal values of triglycerides and HDL-cholesterol (p < 0.05), and higher values of pro-inflammatory cytokines (e.g., TNF-α and IL-6) compared to MHO (p < 0.05).
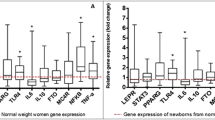
Differential gene expression analyses, using Limma R/Bioconductor software package, showed 40 genes differentially expressed (19 upregulated and 21 downregulated, FDR < 0.05) in children with MHO compared to MUO (Fig. 1). These 40 genes were mainly related to metabolism, mitochondrial, and immune functions (Supplemental Table S1 [online]). Genes differentially expressed were enriched in pathways related to cardiovascular growth via hypertrophy (FDR < 0.05; Table 2). Additionally, WGCNA analysis identified 23 gene coexpression modules with low preservation (Zsummary < 2) (i.e., genes clustered in children with MHO but not clustered in MUO), 3 with moderate preservation (2 > Zsummary < 10), and 2 with high preservation (Zsummary > 10) (i.e., genes similarly clustered in children with MHO and MUO) in children with MHO compared to MUO (Fig. 2). The gene coexpression modules size ranged from 65 to 1217 genes (Supplemental Table S2 [online]). The hub genes (i.e., highly connected intramodular genes) identified in the 23 modules with low preservation ranged from 35 to 826 genes (Supplemental Table S2 [online]).
Fold change (FC) of genes differentially expressed in children with metabolic healthy overweight/obesity (MHO) compared to metabolic unhealthy overweight/obesity (MUO) according to Limma analyses. In total, 19 genes were upregulated (orange) and 21 were downregulated (blue), false discovery rate (FDR) < 0.05.
Gene coexpression module preservation between children with metabolic healthy overweight/obesity (MHO) and children with metabolic unhealthy overweight/obesity (MUO) as compound Zsummary (y-axis). In total, 23 gene coexpression modules were low preserved (Zsummary < 2, y-axis) in children with MHO compared with MUO (i.e., genes clustered in children with MHO but not clustered in MUO). Saddlebrown: 1, White: 2, Skyblue: 3, Orange: 4, Darkorange: 5, Darkgrey: 6, Darkgreen: 7, Darkturquoise: 8, Darkred: 9, Royalblue: 10, Lightyellow: 11, Grey60: 12, Midnightblue: 13, Lightcyan: 14, Lightgreen: 15, Cyan: 16, Salmon: 17, Tan: 18, Purple: 19, Pink: 20, Red: 21, Yellow: 22, Brown: 23, Greenyellow: 24, Black: 25, Green: 26, Turquoise: 27, Blue: 28.
From 40 genes differentially expressed in the Limma analyses, 32 genes were found among the hub genes (i.e., highly connected intramodular genes) identified within WGCNA modules with low preservation clustered in the Tan and midnightblue color. These genes were ZNF559, MPRIP, MYO3A, ADAMTSL2, LOC388780, FAM83E, TOMM20L, DSC1, TMC5, ZNF560, CMIP, UPF1, SLC44A1, KPTN, NXN, SPATA5L1, TMEM253, NRG1, SEMA7A, CCDC180, FBXW4, FAM53A, CYP3A5, ARL17A, IRF2BP2, HIPK1, MOV10L1, COL26A1, GPRC5D, CCL17, RREB1, and ST14. From the molecular network composed of these 32 genes (Fig. 3), we selected the top 5 based on the highest values in centrality degree (i.e., the number of interactions in which a gene is involved). These genes, RREB1, FAM83E, SLC44A1, NRG1, and TMC5, are involved in type 2 diabetes, macrophage phospholipid metabolism, inflammation, and cardiovascular growth through exercise (Table 3 and Supplemental Table S3 [online]).
In silico corroboration analysis using PHENOPEDIA database provided a list of 2263 genes with the term “obesity” and 5324 genes with the term “metabolic diseases”. Next, GeneMania app in CYTOSCAPE platform was applied for the identification of the most significant genes in both lists, creating 2 molecular interaction networks composed of 2263 and 5324 genes, respectively. When testing our common list of 32 genes (Fig. 3), 3 genes were found in the in silico lists of top genes involved in obesity and metabolism, based on the highest centrality degree (i.e., the number of interactions in which a gene is involved) threshold by their means (Supplemental Tables S4 and S5 [online]): RREB1, NRG1, and CYP3A5. Genes RREB1 and NRG1 were identified in the top five genes based on the highest values in centrality degree (Table 3 and Fig. 3).
Discussion
We identified 32 genes differentially expressed in children with MHO compared to MUO, which were related to metabolism, mitochondrial, and immune functions. These findings provide the first step toward the understanding the underlying mechanisms that differentiate MHO from MUO in young ages. Concerning the most upregulated and downregulated genes in our study (i.e., MHO vs. MUO), we found the highest expression of TRIM11 gene and the lowest expression of the ADAMTSL2 gene in children with MHO compared to MUO. TRIM11 is involved in the degradation of AIM2 inflammasome.40 The AIM2 inflammasome cytosolic signaling complex lead to the maturation of pro-inflammatory cytokines.40 Indeed, children with MHO showed TRIM11 gene upregulation and lower levels of pro-inflammatory cytokines compared to MUO. In fact, the MHO phenotype has been characterized by a favorable inflammatory profile compared to MUO.9
Body mass index and adiposity correlate positively with circulating levels of transforming growth factor beta (TGF-β1) protein.41 This protein is involved in the regulation of inflammation, immune function, and glucose homeostasis, among other biological processes.41,42,43 Interestingly, mutations in the ADAMTSL2 gene may lead to dysregulation of TGF-β signaling.44 Furthermore, previous evidence has shown that the blockade of TGF-β signaling protects mice from obesity, diabetes, and hepatic steatosis.41 Likewise, Snelling et al. reported upregulation of ADAMTSL2 gene in human damaged cartilage compared with undamaged cartilage.45 In our study, inflammatory markers TNF-α and IL-6 were significantly higher in MUO compared to MHO children. We hypothesize that a higher ADAMTSL2 together with lower TRIM11 gene expression levels in children with MUO could promote higher inflammatory profile in those children compared to children with MHO.
Telle-Hansen et al. reported higher peripheral blood mononuclear cell expression of some candidate genes involved in lipid metabolism (i.e., UCP2, HSL, and PPARG) in young and middle-aged adults with MHO compared to MUO.15 Similarly, we found higher whole-blood expression of some genes involved in fatty acids synthesis and cholesterol metabolism (e.g., CYP3A5 and IRF2BP2) in children with MHO compared to MUO. The increase in peripheral blood mononuclear CYP3A4 and CYP3A5 gene expression has been associated with a greater low-density lipoprotein-cholesterol reduction in adults with hypercholesterolemia after treatment with atorvastatin.46 On the contrary, to our knowledge, the relation of the higher expression of CYP3A5 gene with HDL-cholesterol metabolism has not been reported. Furthermore, the IRF2BP2 protein is a novel regulator of lipid metabolism and inflammation in macrophages.47 This protein is necessary for the expression of the anti-inflammatory transcription factor KLF2 in macrophages, and deficiency in IRF2BP2 protein leads to an increase in lipid accumulation.47 Likewise, IRF2BP2 protein deficiency has been associated with an increase in atherosclerosis and coronary artery disease in mice and humans, respectively.47 Interesting, IRF2BP2 gene was upregulated in children with MHO compared to MUO and could contribute partially to a better cardiometabolic and inflammatory profile. Interestingly, Sánchez et al. identified some genes (e.g., ZNF418, NPPA, POLR1C, ADRB3, P2RX2) that might discriminate children aged 4.7–8 years with OW presenting high or low triglycerides levels.48 These genes were not detected in our study; some differences between studies could partially explain the lack of gene overlapping. In this regard, different high-throughput technologies (microarrays vs. RNA-seq) were used, different weight status (children with OW vs. children with OW/OB), and different age range of the participants (4.7–8 years vs. 9–11 years) among others.
On the other hand, Zhang et al. showed association between metabolic abnormalities (i.e., MUO and metabolic unhealthy non-obesity) and left ventricular hypertrophy in young adults, while not in the MHO group.49 In our study, key genes involved in cardiovascular growth via hypertrophy that are regulated by physical exercise, i.e., HIPK1 and NRG1 genes,50,51 were found upregulated and downregulated in children with MHO compared to MUO. Our results suggest a “balance” on the expression of genes and pathways involved in cardiac growth via hypertrophy between children with MHO and MUO. As we did not perform echocardiography measurements, it is not possible to elucidate the clinical implications of these results in cardiovascular health via cardiac hypertrophy in children with weight disturbances.
Our constructed molecular network composed of 32 genes could highlight the most relevant genes in understanding the molecular basis underlying the MHO phenotype in children. RREB1 and NRG1 genes could be of special interest, as we found them among top five genes of our network of 32 genes and that were also confirmed as top genes in the in silico analyses on two different publicly available data sets. RREB1 gene (i.e., the gene with the highest value of centrality degree of our network of 32 genes) is considered a novel candidate gene for type 2 diabetes in humans.52 Furthermore, genome-wide association studies identified loci in RREB1 gene associated with fasting glucose and fat distribution.53,54 Further, the RREB1 gene might modulate different clinical phenotypes (e.g., hypertension, fat distribution, fasting glucose, non-diabetic end-stage kidney disease).52 RREB1 gene could serve as a promising candidate for prognosis of cardiometabolic health in pediatric population.
Several limitations in our study need to be highlighted. First, we analyzed whole-blood transcriptome profile. Different subsets of leukocytes have different roles in immune function and metabolism. However, the whole-blood RNA-sequencing has been considered promising for the identification and tracking of biomarkers and useful as a diagnostic tool for rare diseases.55 Second, our sample size was relatively small and most of children in the MHO group were boys (10 of 13), which could have influenced WGCNA analyses. Nevertheless, we adjusted the Limma analyses by sex (in addition to maturation, i.e., PHV), and the selected genes overlapped with those from the WGCNA analyses, aiming to minimize the potential confounding effect of sex. Likewise, we cannot assume a causal relationship due to our cross-sectional study design. Lastly, the cut-off points used in our study were specific for boys and girls aged 12 years, as it was the closest to the age range of 9–11 years. Despite these limitations, to the best of our knowledge, this is the first study that reports differences on whole-blood transcriptome profile in children with MHO and MUO. Further, blood samples for transcriptome analysis were obtained in a unified manner (fasting blood in first hour in the morning), we applied GlobinLock as a novel robust globin mRNA reduction tool to preserve RNA quality,28 and two different bioinformatics approaches together with in silico data mining were applied to corroborate the results.
Conclusion
The distinct gene expression pattern includes genes related to metabolism, mitochondrial, and immune functions. The most relevant genes in understanding the molecular basis underlying the MHO phenotype in children could be: RREB1, FAM83E, SLC44A1, NRG1, TMC5, CYP3A5, TRIM11, and ADAMTSL2. The results contribute to the development of hypothesis directed to a better understanding of why a subset of the population remains metabolically healthy despite having OW/OB. Future studies should confirm our findings in larger cohorts of children and in other populations.
References
Weiss, R. & Kaufman, F. R. Metabolic complications of childhood obesity: identifying and mitigating the risk. Diabetes Care 31, S310–S316 (2008).
Franks, P. W. et al. Childhood obesity, other cardiovascular risk factors, and premature death. N. Engl. J. Med. 362, 485–493 (2010).
Bervoets, L. & Massa, G. Classification and clinical characterization of metabolically “healthy” obese children and adolescents. J. Pediatr. Endocrinol. Metab. 29, 553–560 (2016).
Prince, R. L., Kuk, J. L., Ambler, K. A., Dhaliwal, J. & Ball, G. D. C. Predictors of metabolically healthy obesity in children. Diabetes Care 37, 1462–1468 (2014).
Ortega, F. B., Lavie, C. J. & Blair, S. N. Obesity and cardiovascular disease. Circ. Res. 118, 1752–1770 (2016).
Lavie, C. J. et al. Healthy weight and obesity prevention: JACC Health Promotion Series. J. Am. Coll. Cardiol. 72, 1506–1531 (2018).
Ubago-Guisado, E. et al. Differences in areal bone mineral density between metabolically healthy and unhealthy overweight/obese children: the role of physical activity and cardiorespiratory fitness. Pediatr. Res. 87, 1219–1225 (2019).
Sénéchal, M. et al. Cardiorespiratory fitness and adiposity in metabolically healthy overweight and obese youth. Pediatrics 132, 85–92 (2013).
Primeau, V. et al. Characterizing the profile of obese patients who are metabolically healthy. Int. J. Obes. 35, 971–981 (2011).
Naukkarinen, J. et al. Characterising metabolically healthy obesity in weight-discordant monozygotic twins. Diabetologia 57, 167–176 (2014).
Das, S. K., Ma, L. & Sharma, N. K. Adipose tissue gene expression and metabolic health of obese adults. Int. J. Obes. 39, 869–873 (2015).
Gómez-Ambrosi, J. et al. Increased cardiometabolic risk factors and inflammation in adipose tissue in obese subjects classified as metabolically healthy. Diabetes Care 37, 2813–2821 (2014).
Muñoz-Garach, A., Cornejo-Pareja, I. & Tinahones, F. J. Does metabolically healthy obesity exist? Nutrients 8, 1–10 (2016).
Beh, S. Is metabolically healthy obesity a useful concept? Diabet. Med. 36, 539–545 (2019).
Telle-Hansen, V. H. et al. Altered expression of genes involved in lipid metabolism in obese subjects with unfavourable phenotype. Genes Nutr. 8, 425–434 (2013).
Duncan, B. B. et al. Low-grade systemic inflammation and the development of type 2 diabetes. Diabetes 52, 1799–1805 (2003).
Libby, P., Ridker, P. M. & Maseri, A. Inflammation and atherosclerosis. Circulation 105, 1135–1143 (2002).
Maurizi, G., Della Guardia, L., Maurizi, A. & Poloni, A. Adipocytes properties and crosstalk with immune system in obesity-related inflammation. J. Cell. Physiol. 233, 88–97 (2018).
Blüher, S. & Schwarz, P. Metabolically healthy obesity from childhood to adulthood - does weight status alone matter? Metabolism 63, 1084–1092 (2014).
Cadenas-Sánchez, C. et al. An exercise-based randomized controlled trial on brain, cognition, physical health and mental health in overweight/obese children (ActiveBrains project): rationale, design and methods. Contemp. Clin. Trials 47, 315–324 (2016).
Cole, T. J. & Lobstein, T. Extended international (IOTF) body mass index cut-offs for thinness, overweight and obesity. Pediatr. Obes. 7, 284–294 (2012).
Marfell-Jones, M., Olds, T., Stewart, A., & Carter, L. International Standards for Anthropometric Assessment (International Society for the Advancement of Kinanthropometry, 2011).
Crabtree, N. J. et al. Dual-energy X-ray absorptiometry interpretation and reporting in children and adolescents: the Revised 2013 ISCD Pediatric Official Positions. J. Clin. Densitom. 17, 225–242 (2014).
Moore, S. A. et al. Enhancing a somatic maturity prediction model. Med Sci. Sports Exerc. 47, 1755–1764 (2015).
Jolliffe, C. J. & Janssen, I. Development of age-specific adolescent metabolic syndrome criteria that are linked to the adult treatment panel iii and international diabetes federation criteria. J. Am. Coll. Cardiol. 49, 891–898 (2007).
Adelantado-Renau, M. et al. Inflammatory biomarkers and brain health indicators in children with overweight and obesity: the ActiveBrains Project. Brain Behav. Immun. 81, 588–597 (2019).
Gil-Cosano, J. J. et al. Inflammatory markers and bone mass in children with overweight/obesity: the role of muscular fitness. Pediatr. Res. 87, 42–47 (2020).
Krjutškov, K. et al. Globin mRNA reduction for whole-blood transcriptome sequencing. Sci. Rep. 6, 1–7 (2016).
Krjutškov, K. et al. Single-cell transcriptome analysis of endometrial tissue. Hum. Reprod. 31, 844–853 (2016).
Sink, K. M. et al. Effect of a 24-month physical activity intervention vs health education on cognitive outcomes in sedentary older adults: the LIFE randomized trial. JAMA 314, 781–790 (2015).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, 1–13 (2015).
Raudvere, U. et al. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 47, W191–W198 (2019).
Langfelder, P. & Horvath, S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9, 559 (2008).
Zhao, W. et al. Weighted gene coexpression network analysis: state of the art. J. Biopharm. Stat. 20, 281–300 (2010).
Langfelder, P., Luo, R., Oldham, M. C. & Horvath, S. Is my network module preserved and reproducible? PLoS Comput. Biol. 7, 1–29 (2011).
Cox, B. et al. A co-expression analysis of the placental transcriptome in association with maternal pre-pregnancy BMI and newborn birth weight. Front. Genet. 10, 1–13 (2019).
VIB/UGent, Bioinformatics & Evolutionary Genomics. Draw Venn diagram. http://bioinformatics.psb.ugent.be/webtools/Venn/.
Montojo, J. et al. GeneMANIA cytoscape plugin: fast gene function predictions on the desktop. Bioinformatics 26, 2927–2928 (2010).
Marín, M., Esteban, F. J., Ramírez-Rodrigo, H., Ros, E. & Sáez-Lara, M. J. An integrative methodology based on protein-protein interaction networks for identification and functional annotation of disease-relevant genes applied to channelopathies. BMC Bioinformatics 20, 565 (2019).
Liu, T. et al. TRIM11 suppresses AIM2 inflammasome by degrading AIM2 via p62-dependent selective autophagy. Cell Rep. 16, 1988–2002 (2016).
Yadav, H. et al. Protection from obesity and diabetes by blockade of TGF-β/Smad3 signaling. Cell Metab. 14, 67–79 (2011).
Romano, M. et al. Association of inflammation markers with impaired insulin sensitivity and coagulative activation in obese healthy women. J. Clin. Endocrinol. Metab. 88, 5321–5326 (2003).
Varga, J. & Pasche, B. Transforming growth factor β as a therapeutic target in systemic sclerosis. Nat. Rev. Rheumatol. 5, 200–206 (2009).
Le Goff, C. et al. ADAMTSL2 mutations in geleophysic dysplasia demonstrate a role for ADAMTS-like proteins in TGF-β bioavailability regulation. Nat. Genet. 40, 1119–1123 (2008).
Snelling, S. et al. A gene expression study of normal and damaged cartilage in anteromedial gonarthrosis, a phenotype of osteoarthritis. Osteoarthr. Cartil. 22, 334–343 (2014).
Willrich, M. A. V. et al. Effects of atorvastatin on CYP3A4 and CYP3A5 mRNA expression in mononuclear cells and CYP3A activity in hypercholeresterolemic patients. Clin. Chim. Acta 421, 157–163 (2013).
Zhang, H. & Reilly, M. P. IRF2BP2: a new player at the crossroads of inflammation and lipid metabolism. Circ. Res. 117, 656–658 (2015).
Sánchez, J. et al. Transcriptome analysis in blood cells from children reveals potential early biomarkers of metabolic alterations. Int. J. Obes. 41, 1481–1488 (2017).
Zhang, N. et al. Metabolic abnormalities, but not metabolically healthy obesity, are associated with left ventricular hypertrophy. Hear Lung Circ. 26, 251–257 (2017).
Liu, X. et al. MiR-222 is necessary for exercise-induced cardiac growth and protects against pathological cardiac remodeling. Cell Metab. 21, 584–595 (2015).
Tao, L., Bei, Y., Zhang, H., Xiao, J. & Li, X. Exercise for the heart: signaling pathways. Oncotarget 6, 20773–20784 (2015).
Bonomo, J. A. et al. The ras responsive transcription factor RREB1 is a novel candidate gene for type 2 diabetes associated end-stage kidney disease. Hum. Mol. Genet. 23, 6441–6447 (2014).
Scott, R. A. et al. Large-scale association analyses identify new loci influencing glycemic traits and provide insight into the underlying biological pathways. Nat. Genet. 44, 991–1005 (2012).
Liu, C. T. et al. Genome-wide association of body fat distribution in african ancestry populations suggests new loci. PLoS Genet. 9, 1–16 (2013).
Fresard, L. et al. Identification of rare-disease genes in diverse undiagnosed cases using whole blood transcriptome sequencing and large control cohorts. Nat. Med. 25, 911–919 (2018).
Acknowledgements
The authors would like to thank all the participants who participated in this investigation. This manuscript is part of a PhD thesis conducted in the Biomedicine Doctoral Studies of the University of Granada, Spain. The project was funded by the Spanish Ministry of Economy and Competitiveness (Reference DEP2013-47540, DEP2016-79512-R, and DEP2017-91544-EXP), the Spanish Ministry of Economy, Industry and Competitiveness (MINECO) and European Regional Development Fund (FEDER): grants RYC-2016- 21199 and ENDORE SAF2017-87526-R, and Programa Operativo FEDER Andalucía (BCTS-500-UGR18). F.J.E. is supported by the Junta de Andalucía [BIO-302; US-1254251] and the University of Jaén [PAIUJA-EI_CTS02]. C.C.-S. is supported by a grant from the Spanish Ministry of Science and Innovation (FJC2018-037925-I). A.P.-F. is supported by the Spanish Ministry of Education, Culture and Sport (FPU 16/02760). S.R.-A. is supported by NIH UO1 TR002004 and PERC Systems Biology Fund. Additional support was obtained from EXERNET Research Network on Exercise and Health in Special Populations (DEP2005-00046/ACTI). This study has been partially funded by the University of Granada, Plan Propio de Investigación 2016, Excellence actions: Units of Excellence; Unit of Excellence on Exercise and Health (UCEES); and by the Junta de Andalucía, Consejería de Conocimiento, Investigación y Universidades and European Regional Development Fund (ERDF), ref. SOMM17/6107/UGR. Additional funding was obtained from the Andalusian Operational Programme supported with European Regional Development Funds (ERDF in English, FEDER in Spanish, project ref: B-CTS-355-UGR18).
Author information
Authors and Affiliations
Contributions
A.P.-F. was involved in the conception and design of the paper, analysis, interpretation of data, and drafting the manuscript. S.A. and F.B.O. were involved in the conception and design of the paper/project, interpretation of data, investigation (experiments), and critical revision. F.J.E. was involved in the formal analysis and interpretation of data. C.C.-S. was involved in the conception and design of the project and critical revision. C.M.A. was involved in critical revision. E.E., S.K., K.K., and J.K. were involved in the experiments and critical revision. S.R.-A. and F.Z. were involved in critical revision. All authors have approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Patient consent
The study was approved by the Committee for Research Involving Human Subjects at the University of Granada (Reference: 848, February 2014). All parents were informed about the study objective and written informed consent following the Declaration of Helsinki.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Plaza-Florido, A., Altmäe, S., Esteban, F.J. et al. Distinct whole-blood transcriptome profile of children with metabolic healthy overweight/obesity compared to metabolic unhealthy overweight/obesity. Pediatr Res 89, 1687–1694 (2021). https://doi.org/10.1038/s41390-020-01276-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41390-020-01276-7
This article is cited by
-
Eight-year longitudinal study of whole blood gene expression profiles in individuals undergoing long-term medical follow-up
Scientific Reports (2021)
-
Visceral adiposity index performed better than traditional adiposity indicators in predicting unhealthy metabolic phenotype among Chinese children and adolescents
Scientific Reports (2021)