Abstract
The miR17~92 cluster plays important roles in haematopoiesis. However, it is not clear at what stage of differentiation and through which targets miR17~92 exerts this function. Therefore, we generated miR17~92fl/fl; RosaCreERT2 mice for inducible deletion of miR17~92 in haematopoietic cells. Bone marrow reconstitution experiments revealed that miR17~92-deleted cells were not capable to contribute to mature haematopoietic lineages, which was due to defects in haematopoietic stem/progenitor cells (HSPCs). To identify the critical factor targeted by miR17~92 we performed gene expression analysis in HSPCs, demonstrating that mRNA levels of pro-apoptotic Bim inversely correlated with the expression of the miR17~92 cluster. Strikingly, loss of pro-apoptotic BIM completely prevented the loss of HSPCs caused by deletion of miR17~92. The BIM/miR17~92 interaction is conserved in human CD34+ HSPCs, as miR17~92 inhibition or blockade of its binding to the BIM 3′UTR reduced the survival and growth of these cells. Despite the prediction that miR17~92 functions by impacting a plethora of different targets, the absence of BIM alone is sufficient to prevent all defects caused by deletion of miR17~92 in haematopoietic cells.
Similar content being viewed by others
Introduction
MicroRNAs (miRNAs) regulate many biological processes by binding and post-transcriptionally regulating multiple different gene products [1, 2]. However, currently the regulation of gene products by a specific miRNA is mostly based on prediction and overexpression assays but often lacks the study of physiological functions within a whole organism.
The miR17~92 cluster consists of miR17, miR18a, miR19a, miR20a, miR19b1 and miR92-1, which belong to four different seed families [3]. Studies of mice constitutively deficient for miR17~92 revealed that this microRNA cluster is essential for the normal development of several organs, including the heart, lung and skeleton, as well as for the production of B and T lymphocytes [4,5,6,7,8]. Mice deficient for miR17~92 (miR17~92del/del) die soon after birth owing to severe lung and ventricular septal defects [5]. Conditional deletion using tissue-specific CRE transgenes revealed that the miR17~92 cluster also plays a critical role in the development of several haematopoietic cell populations [9,10,11,12]. Accordingly, haematopoietic stem/progenitor cells (HSPCs) from the foetal livers of miR17~92del/del embryos (E14.5) produced substantially fewer pro-B/pre-B cells compared with wild-type HSPCs when transplanted into lethally irradiated recipient mice [5, 13]. Moreover, haematopoietic cell-specific loss of miR17~92 in miR17~92fl/fl; vav-Cre mice impaired T cell development both at the level of T cell progenitors in the thymus and at later stages of differentiation [14].
Of note, aberrant expression of the miR17~92 cluster can contribute to lymphoma development. Overexpression of miR-17-92 resulted in a lymphoproliferative disorder [6] similar to that observed in mice with loss of pro-apoptotic BIM [15] or overexpression of anti-apoptotic BCL-2 [16, 17]. Moreover, miR17~92 is overexpressed in certain B cell lymphomas and enforced expression of miR17~92 accelerated MYC-driven lymphomagenesis in mice [18,19,20,21]. In vitro studies indicated that MYC increases the transcription of the miR17~92 cluster [22], and this is thought to promote tumorigenesis through repression of several genes that promote apoptosis (Bim), proliferation and differentiation (Pten) [5]. However, the precise functional connections between miR17~92 and its targets and their relevance for lymphoma development remain unresolved.
Importantly, the studies published thus far do not discriminate whether the loss of the different haematopoietic cell types caused by the deletion of miR17~92 is due to a direct impact of the loss of this microRNA cluster in these cells or a consequence of its loss in a more primitive stem/progenitor population. To address this question, we generated mice in which miR17~92 could be inducibly deleted only in haematopoietic cells by using the RosaCreERT2 transgene and haematopoietic reconstitution studies. Deletion of the miR17~92 cluster substantially reduced the competitiveness of HSPCs in mixed bone marrow reconstitution experiments. Interestingly, the absence of the pro-apoptotic BH3-only protein BIM, one of many predicted targets of miR17~92, completely prevented the defects in the survival of HSPCs that is caused by the deletion of miR17~92. These findings demonstrate that miR17~92 safeguards the survival of HSPCs by restraining the levels of the pro-apoptotic BH3-only protein BIM.
Material and methods
Mice
Experiments with mice (8–12 weeks, male and female) were approved by and conducted according to the guidelines of The Walter and Eliza Hall Institute Animal Ethics Committee. Expert animal technicians were fully blinded to the genotypes and treatments of mice in the experiments. No animals were excluded from any experiment unless technical issues were present. The generation of miR17~92fl/fl, RosaCreERT2+/Ki, Bim−/− and Puma−/− mice, all maintained on a C57BL/6-Ly5.2 background, have been described previously [5, 15, 23, 24]. To activate CreERT2, mice were given 60 mg/kg tamoxifen (Sigma-Aldrich, Rowville, VIC, Australia) in peanut oil/10% ethanol each day for 3 days by oral gavage 8–15 weeks post transplantation with bone marrow or foetal liver cells [25].
Haematopoietic reconstitution experiments
Bone marrow or foetal liver cells of the test and competitor populations (GFP) were mixed 1:1 in PBS/10% FCS. A total of 6 × 106 bone marrow cells or 2–5 × 105 foetal liver cells were injected i.v. into each lethally irradiated (2 × 5.5 Gy, 3 h between doses) female C57BL/6-Ly5.1 recipient (8–10 weeks, randomly chosen) 2 h after the second dose of γ-irradiation. Retro-orbital bleeds were taken 8 weeks after transplantation to confirm successful haematopoietic reconstitution by determining the frequency of Ly5.2-positive cells by FACS analysis.
Blood and flow cytometric analysis
Haemogram analysis was performed using the ADVIA system. FACS analysis was performed as previously described [26]. HSPCs were analysed by staining with fluorochrome-conjugated monoclonal antibodies against the following cell surface markers: CD150-BV421 (clone TC15-12F12.2, Biolegend), CD127-APC (IL-7 receptor, clone A7R34, eBioscience), CD48-PECy7 (clone HM48-1, eBioscience), CD105-PE (clone MJ718, eBioscience), CD34-A647 (clone RAM34, BD), CD117-BV711 (clone 2B8, BD), CD16/32-PerCPCy5.5 (clone 2.4G2, BD), CD135-PE (eBioscience), CD41-PECy7 (clone MWReg30, BD), SCA-1-A595, CD127-APCCy7 (eBioscience), CD9-A647, CD16_32-FITC, CD41-PECy7, CD2-A700, CD4-A700, CD8-A700, GR-1-A700, F4/80-A700, CD19-A700, B220-A700, Ly6G-A700, TER-119-A700 and NK1.1-A700 [27].
Cell culture
Total bone marrow cells were isolated from the tibia and femur of mice with the indicated genotypes. Bone marrow cells were cultured on OP9 stromal cells in Iscove’s Modified Dulbecco’s medium (IMDM) containing 10% newborn calf serum (Sigma), 1% BSA, 2 mM glutamine, 50 mM beta-mercaptoethanol and supplemented with 20 ng/mL IL-6. To obtain T-cell blasts, 2 × 106 spleen cells were stimulated with recombinant mIL-2 and 2 µg/mL concanavalin A (ConA) (Sigma) in Dulbecco’s modified Eagle’s medium supplemented with 10% FCS, 50 mM beta-mercaptoethanol and 100 mM asparagin for 3 days. Subsequently, cells were washed five times to remove ConA and then cultured in the presence of recombinant mIL-2. Cells were cultured in a fully humidified atmosphere of 5% CO2 in air in the presence or absence of 10 µM 4-Hydroxy-tamoxifen (Sigma) and 25 nM QVD-OPh (JOMAR LIFE RESEARCH).
RNA extraction and quantitative RT-PCR
RNA was extracted using the standard phenol–chloroform extraction protocol using TRIzolTM (Ambion Inc). Isolated RNA was reverse transcribed using oligo-dT primers and the TaqManTM Reverse Transcription Kit (Applied Biosystems) and cDNA synthesis was carried out according to the manufacturer’s instructions. Quantitative RT-PCR reactions were performed in triplicate using the TaqManTM gene expression assays (Applied Biosystems) for Bcl2l11 mRNA (Cat#Mm00437796_m1). Data were collected using the Viia7 PCR System (Thermo Fischer Scientific) using the Viia7TM software package (Thermo Fischer Scientific). Probing for HMBS (Cat#Mm01143545_m1) served as a house-keeping gene. Data were calculated using the 2−ΔΔCT method.
Cell lysis and western blotting
Cell lysates were generated using CHAPS lysis buffer (10 mM HEPES, pH 7.4; 150 mM NaCl; 1% CHAPS; complete protease inhibitor cocktail). Protein concentration was determined by Bradford assay using the Protein Assay Dye Reagent Concentrate (Bio-Rad). Proteins were size fractionated by gel electrophoresis on NuPAGE 4–12% Bis-Tris 1.5 mm gels (Life Technologies) in MES buffer and then transferred onto nitrocellulose membranes (Life Technologies) using the iBlot membrane transfer system. The following primary antibodies were diluted in 3% BSA in PBS/0.1%Tween buffer: rabbit anti pAKT (Cell Signalling), AKT (Cell Signalling), BIM (ENZO) and ACTIN (Sigma). Secondary anti-rat/mouse/rabbit IgG antibodies conjugated to HRP (Southern BioTech) were applied, followed by Luminata Forte Western HRP substrate (Millipore) for band visualisation. Membranes were imaged using the ChemiDoc XRS+ machine with ImageLab software (Bio-Rad).
Microarray analyses
Gene expression data from mature haematopoietic cells were taken from the ImmGen dataset (www.immgen.org) [28]. Statistical significance was tested by calculation of Pearson correlation.
Gene deletion analysis using genotyping PCR
Genomic DNA was isolated using tail lysis buffer (Invitrogen). The genotyping PCR reaction detecting miRfl (floxed allele) and miRdel (deleted allele) was performed using forward primer (5′ TCGAGTATCTGACAATGTGG) and specific reverse primers detecting the floxed (fl) (5′ TAGCCAGAAGTTCCAAATTGG) or deleted (del) (5′ ATAGCCTGAAACCAACTGTGC) allele. Deletion efficiency was calculated by densitometric analysis of deleted vs. non-deleted PCR products using the Image Lab software (BioRad).
Human HSPC isolation and colony formation assays
Human cord blood-derived CD34+ cells (human HSPCs) were obtained from the Bone Marrow Donor Institute/Cord Blood Research Unit (Murdoch Children’s Research Institute, Parkville, VIC, Australia) under approval from The Walter and Eliza Hall Institute of Medical Research Human Research Ethics Committee. CD34+ cells were isolated using immunomagnetic positive selection (Stemcell Technologies #17896) according to the manufacturer’s instructions and expanded in complete StemMACS HSC Expansion medium (Miltentyi Biotech 130-100-473 & 130-100-843). 5000 cells were cultured in 1 mL volumes of 0.3% agar in IMDM containing 20% newborn calf serum, 1% BSA, 2 mM glutamine, 0.1 mM 2-mercaptoethanol and supplemented with 50 ng/mL stem cell factor, 20 ng/mL GM-CSF, 20 ng/mL G-CSF, 20 ng/mL IL-3, 20 ng/mL IL-6 and 3 U/mL erythropoietin treated with 20 µM of MIR17PTi [29], scrambled control or no treatment in a fully humidified atmosphere of 5% CO2 in air. Colonies were fixed, dried onto glass slides, and stained for acetylcholinesterase as well as with Luxol fast blue and haematoxylin, and the numbers of colonies were determined by microscopic examination. The mean of two technical replicates was determined for each treatment, and the proportion of colonies from cells treated with MiR17PTi and scrambled control are shown relative to no drug treatment with mean and standard error of the mean from the analysis of two independent human cord blood samples.
Target protector transfection
Target protectors specific for the four predicted seed regions in the human BIM 3′UTR for mature miRNAs of the miR17~92 cluster were designed using the online tool from Qiagen. Sequences: miR17 seed: CCAGTCTCCTGACTAGAGCACTTTACTCTGTTTCCTCAGC, miR19 seed: CCCTGGCTTACTTGTGTTTTGCACTGATGAATTTTGACAG, miR92.1 seed: CTGAGCCAAATGTCTGTGTGCAATTGTGTTTCCTTTACCT, miR92.2 seed: TAATTGCCACTTTACTTGTGCAATACTGCTGTAAATAACT.
Cells were transfected with 2 µM target protector or scrambled oligos using the Lonza nucleofection kit (P3 primary cell media with supplement 1, DK-100). Cells were co-transfected with pEX-GFP to allow identification of transfected cells by flow cytometric analysis. At 48 h after transfection, the viability of the transfected cells was assessed by flow cytometric analysis with viable transfected cells defined as GFP+AnnexinV−DAPI−.
Results
Deletion of miR17~92 causes a competitive disadvantage in diverse haematopoietic cell types in vivo that can be rescued by deletion of BIM
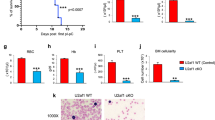
To assess the impact of miR17~92 deletion in adult animals, we generated miR17~92fl/fl; RosaCreERT2+/Ki mice. Upon induction of miR17~92 deletion by treatment with tamoxifen, we observed no obvious differences in appearance and behavior between these mice and wild-type controls (data not shown). However, we found a significant reduction of various haematopoietic cell populations in the spleen, blood and bone marrow (BM) in miR17~92-deleted mice (Supplementary Fig. S1A–C), which is in line with previous reports [5, 6, 13, 14, 30]. The reduction in the various haematopoietic cell types we observed is probably an underestimation of the effects exerted by the loss of miR17~92, given that although miR17~92 deletion in spleen and BM cells was initially almost complete, this was found to be <50% after 1 month (Fig. 1a).
Induced deletion of miR17~92 in adult mice leads to BIM dependent loss of diverse haematopoietic cell subsets. a Genotyping to detect miR17~92 wt, floxed and del bands of bone marrow (top left) and spleen cells (bottom left) from adult miR17~92fl/fl; RosaCreERT2+/Ki mice at the indicated time points after CreERT2 activation by treatment with tamoxifen (left panel). The efficiency of miR17~92 deletion was calculated by densitometric analysis of the PCR products of the deleted vs. the non-deleted alleles (right panel). b Correlation analysis of mir17hg (miR17~92), Bcl2l11 (Bim) and Pten RNA expression in the indicated cell types was determined by examination of microarray data provided by immgen.org [28]. Statistical significance of Pearson correlation was assessed for mir17hg (miR17~92) and Bcl2l11 (Bim) (−0.2305488, p-value: 0.0006382) as well as mir17hg (miR17~92) and Pten (0.16, p-value > 0.05). c Total bone marrow cells were isolated from miR17~92fl/fl; RosaCreERT2+/Ki and RosaCreERT2+/Ki mice and cultured for 24 h on OP9 stromal cells in the presence or absence of QVD-OPh (25 nM) and tamoxifen (10 µM) to induce CreERT2 activation and miR17~92 deletion. qPCR analysis using Taq-man probes detecting Bcl2L11 (Bim) relative to HMBS was performed using the ABI700 machine. Data are presented as mean 2^-ddCT ± SEM pooled from two independent experiments with each performed in triplicates. *p < 0.05 (Student's t test). d Genotyping to detect miR17~92 wt, floxed and del bands in cells from the bone marrow, spleen, blood and thymus of adult miR17~92fl/fl; RosaCreERT2+/Ki (n = 3) and miR17~92fl/fl; RosaCreERT2+/Ki; Bim−/− mice (n = 3) 3 months after CreERT2 activation by treatment with tamoxifen (left panel). The efficiency of miR17~92 deletion was calculated by densitometric analysis of the PCR products of the deleted vs. the non-deleted alleles (right panel) *p < 0.05, **p < 0.01 (Student's t test)
These findings demonstrate an important role of miR17~92 in haematopoietic cells, but how this microRNA cluster exerts its function in the haematopoietic compartment is not understood. In order to identify the most critical factor targeted by miR17~92 we performed gene expression analysis using data provided in the ImmGen microarray database (https://www.immgen.org) [28]. This revealed a negative correlation of the expression of Bim (BCL2L11) mRNA and the miR17~92 cluster host gene (miR17hg) in all immature and mature lymphoid cell subsets, several haematopoietic progenitor populations and mature cell types of myeloid origin, including macrophages, monocytes and granulocytes (Fig. 1b, Pearson correlation: −0.23, p-value < 0.001). Remarkably, this correlation was associated with high expression of miR17hg in progenitors, which decreases in lineage-committed cells, with the opposite pattern observed for Bim mRNA. A similar pattern was observed upon analysis of murine RNA-Seq data sets that are available in the Haemopedia database (data not shown) [31, 32]. In contrast, no significant correlation was observed in gene expression analysis of miR17~92 and Pten, another recognized target of the miR17~92 cluster [5, 6, 21], in hematopoietic progenitor populations.
In line with these observations, we noticed a marked upregulation of Bim mRNA and BIM protein levels when we deleted miR17~92 in bone marrow cells in vitro (Fig. 1c and Supplementary Fig. S1D). A similar increase of BIM protein levels (BIMEL and BIML) were detected upon deletion of miR17~92 in cultured T cell blasts (Supplementary Fig. S1E). Of note, we did not observe changes in PI3K/AKT signaling upon deletion of miR17~92, consistent with the notion that the increase of BIM protein is a direct consequence of the loss of Bim regulation by miR17~92 and not due to an increase in PTEN with a consequent inhibition of PI3K/AKT signaling (Supplementary Fig. S1D).
This prompted us to test whether the deletion of BIM could rescue miR17~92-deleted haematopoietic cells by generating miR17~92fl/fl; RosaCreERT2+/Ki; Bim−/− mice. Firstly, we analysed the deletion efficiency of the miR17~92 cluster in the haematopoietic organs of these mice and control animals after treatment with tamoxifen. While the efficiency of miR17~92 deletion in haematopoietic cells was significantly <70% at 3 months post tamoxifen-treatment in the miR17~92fl/fl; RosaCreERT2+/Ki mice, almost complete deletion was observed in cells from the bone marrow, spleen, blood and thymus of the miR17~92fl/fl; RosaCreERT2+/Ki;Bim−/− mice (Fig. 1d). These findings indicate that loss of BIM may be sufficient to rescue the defects in haematopoietic cells that are caused by the deletion of the miR17~92 cluster.
Loss of pro-apoptotic BIM prevents the reduction in lymphoid, myeloid and erythroid cells that is caused by the induced deletion of miR17~92
The findings described above suggest that BIM may be critical for the loss of the haematopoietic cell populations that occurs upon the induced deletion of the miR17~92 cluster. To examine the functional interaction of miR17~92 and BIM-mediated apoptosis, we performed competitive bone marrow reconstitution experiments. Lethally irradiated wild-type (wt; C57BL/6-Ly5.1) mice were reconstituted with a 1:1 mixture of GFP-expressing wt (GFP-C57BL/6-Ly5.2) competitor bone marrow cells and test bone marrow cells of the genotypes of interest, i.e. miR17~92fl/fl; RosaCreERT2+/Ki or miR17~92fl/fl; RosaCreERT2+/Ki; Bim−/− (all on a C57BL/6-Ly5.2 background). After confirming efficient haematopoietic reconstitution of the competitor and the test cells by FACS analysis ~8–10 weeks post-transplantation, the recipient mice were treated with tamoxifen to delete the miR17~92 cluster in the test cells. These animals were analysed after another 60 days (experimental design depicted in Supplementary Fig. S2A). As predicted, we observed significant reductions in several haematopoietic cell populations, including immature as well as mature B and T cells, myeloid cells and erythroid cells, that had lost the miR17~92 cluster, whereas wt as well as RosaCreERT2+/Ki cells were not outcompeted by the GFP-expressing wt cells (Fig. 2). Remarkably, deletion of miR17~92 on a BIM-deficient background did not decrease cellular competitiveness, as demonstrated by an equal contribution of miR17~92fl/fl; RosaCreERT2+/Ki; Bim−/− cells and GFP competitor cells to the lymphoid (Fig. 2a–c), myeloid (Fig. 2d) and erythroid compartments (Fig. 2e and Supplementary Fig. S2F). In fact, the contributions of miR17~92fl/fl; RosaCreERT2+/Ki; Bim−/− cells to the B lymphoid compartment in the bone marrow as well as the mature B and T cells in the spleen were significantly >50%. This is consistent with the accumulation of these cell types seen in BIM-deficient mice [15].
Loss of BIM rescues haematopoietic cells with deleted miR17~92. a–d C57BL/6 wt mice (Ly5.1+) were lethally irradiated and reconstituted with a 1:1 mixture of bone marrow cells from UBC-GFP mice (competitor cells) and bone marrow cells from mice of the indicated genotypes (test cells). Reconstituted mice were treated with tamoxifen (three doses oral gavage, 60 mg/kg/day) 8–15 weeks post-transplantation to activate the CreERT2 recombinase. After an additional 8–10 weeks, the immature as well as mature lymphoid and myeloid cell populations were analysed by flow cytometry. a Mature B and T cells in the spleen and peripheral blood were identified by staining with antibodies against B220 and TCRβ. b Immature and mature thymic T cell populations were identified by staining with antibodies against CD4 and CD8. DN = double negative CD4−CD8−; DP = double positive CD4+CD8+ cells. c Immature and mature B cell populations of the bone marrow were identified by staining with antibodies against B220 and IgM: pro-B/pre-B (B220+IgM−), immature B (B220lowIgM+, transitional B (B220+IgMhigh) and mature B cells (B220highIgM+). d Monocytes/macrophages (MAC-1+GR-1low) and granulocytes (MAC-1+GR-1high) were identified in the bone marrow and spleen by staining with antibodies against MAC-1 and GR-1. e Immature and mature erythroid cell populations were identified in the bone marrow by staining with antibodies against TER119 and CD71 as well as by size discrimination using forward light scatter (FSC-A). Ery = erythroid. a–e Representative FACS plots for the gating strategies are provided for cells from wt mice. Percentages of GFP-negative cells (test cells) were determined for each cell subset. Data represent mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 (Student's t test miR17~92fl/fl; RosaCreERT2+/Ki vs. wt, RosaCreERT2+/Ki, miR17~92fl/fl; RosaCreERT2+/Ki; Bim−/−, Bim−/−, miR17~92fl/fl; RosaCreERT2+/Ki; Puma−/− and Puma−/− mice). n = number of donor/number of recipient mice as indicated
We next wanted to validate that the rescue of miR17~92-deleted cells by the loss of BIM is not due to a general rescue from apoptosis but due to direct derepression of BIM function. Therefore, we assessed the impact of the absence of PUMA another pro-apoptotic BH3-only protein that also plays critical roles in the haematopoietic system [24, 33, 34], on the reduction of haematopoietic cells caused by the induced deletion of miR17~92. In contrast to the Bim3′UTR, the Puma3′UTR does not harbor putative binding sites for any of the mature members of the miR17~92 cluster (targetscan.org [35]). While the loss of BIM could rescue most mature and immature haematopoietic cell populations from the consequences of miR17~92 deletion, loss of PUMA had significant impact on only a subset of these cell populations (Fig. 2). Most effects of PUMA loss were seen in immature and mature B cells, although their rescue from miR17~92 deletion was still less pronounced than that observed by the absence of BIM (Fig. 2a, c). Collectively, these data demonstrate that the loss of competitiveness of haematopoietic cells caused by the deletion of the miR17~92 cluster is driven by apoptotic cell death triggered by derepression of the BH3-only protein BIM.
miR17~92 regulates the survival of haematopoietic stem and progenitor cells by restraining BIM-induced apoptosis
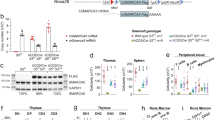
The reduction of all haematopoietic cell subsets observed after deletion of miR17~92 could be due to the independent loss of both immature and mature cell types. Alternatively, it could be a consequence of the loss of common HSPCs causing subsequent reductions in their differentiated progeny. To examine this, we first analysed the fate of HSPCs in competitive haematopoietic reconstitution assays using foetal liver cells from E14.5 miR17~92del/del, miR17~92+/del (del = gene deleted in all cells of the embryo) or wt embryos as test cells and foetal liver cells from E14.5 GFP embryos as competitor cells. A significant competitive disadvantage to reconstitute the host haematopoietic system was observed for E14.5 foetal liver cells from miR17~92del/del embryos (Fig. 3). Of note, there was almost no contribution observed of the GFP-negative miR17~92del/del cells to the lineage−c-KIT+SCA-1+ (LSK), haematopoietic stem/progenitor (Fig. 3a) as well as lineage-committed pluripotent progenitor cells, including common lymphoid progenitors (CLPs), common myeloid progenitors (CMPs), megakaryocyte erythroid progenitors (MEPs) and granulocyte monocyte progenitors (GMPs) (Fig. 3b). In contrast the control donor cells (wt, miR17~92+/del) contributed at least 50% to these HSPC compartments (Fig. 3a, b). A more detailed analysis of the HSPC populations, using either the stem cell markers of the FLK (foetal liver kinase) series [36] (Fig. 3c) or the SLAM (signalling lymphocytic activation molecule) series [37] (Fig. 3d), revealed that deletion of miR17~92 greatly reduced the competitiveness of all HSPC subsets.
Induced deletion of the miR17~92 cluster causes a loss of haematopoietic stem and progenitor cells (HSPCs). C57BL/6 wt mice (Ly5.1+) were lethally irradiated and reconstituted with a 1:1 mixture of E14.5 foetal liver cells from UBC-GFP embryos (competitor cells) and E14.5 foetal liver cells from miR17~92deldel, miR17~92+/del or wt embryos (test cells). At 8–10 weeks after transplantation, the fraction of reconstituted test cells was determined by FACS analysis gating on Ly5.1+GFP− cells (donor test cells). Representative FACS plots for the gating strategies are provided for wt mice. Percentages of GFP-negative cells (test cells) were determined for the indicated cell populations. Data represent mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 (Student's t test miR17~92deldel, miR17~92+/del and wt). a LSK (lineage−c-KIT+SCA-1+) cells and progenitor cells (lineage−c-KIT+SCA-1−) were identified in the lineage marker negative compartment (TER119−, CD4−, CD8a−, Ly6G−, B220−, CD2−, CD3−, CD19−, F4/80−, NK1.1−, GR-1−) by staining with antibodies against c-KIT and SCA-1. b Oligopotent progenitors, including common myeloid progenitors (CMP), granulocyte/macrophage progenitors (GMP) and megakaryocyte/erythroid progenitors, were identified by cell surface staining for the SLAM markers (CD16/32, CD150, CD34) in the progenitor cell population (lineage−c-KIT+SCA-1−). Common lymphoid progenitors (CLP) were identified as c-KITmedSCA-1medIL-7 receptor+ within the lineage marker negative cell populations. c Long-term haematopoietic stem cells (LT-HSC), short-term haematopoietic stem cells (ST-HSC) as well as multipotent progenitor cells (MPP) were identified using the FLK series markers (CD34, CD135) within the LSK cell population. d Haematopoietic stem cells (HSC), multipotent progenitor cells (MPP) and haematopoietic progenitor cells (HPC-1/2) were identified using cell surface staining for the SLAM markers (CD48, CD150) within the LSK cell population. n = number of donor/number of recipient mice as indicated
A significant decrease in the numbers of LSK, HSC and most progenitor cells, including CLPs, CMPs, MEPs and GMPs was also evident in foetal livers from E14.5 miR17~92del/del embryos when compared with foetal livers from wt controls (Supplementary Fig. S3). This demonstrates that the loss of miR17~92 impairs the survival of all HSPC populations in both a transplantation as well as a steady state setting.
To expand on these findings, we again performed competitive bone marrow reconstitution assays and treated reconstituted mice with tamoxifen to delete miR17~92 in the test cells (strategy depicted in Supplementary Fig. S2A). We found that 2 months after miR17~92 deletion only a few LSK stem cell-enriched or early progenitors (lineage−c-KIT+SCA-1–) with deleted miR17~92 cluster were present in the competitive setting (Fig. 4a). A more detailed analysis of the HSPC populations, using either the stem cell markers of the SLAM series [37] (Fig. 4b) or the FLK series markers [36] (Fig. 4c), revealed that deletion of miR17~92 did not substantially affect the long-term (LT)-HSCs but greatly diminished the competitiveness of the early HSPCs with self-renewal capacity, such as short-term (ST)-HSCs and multipotent progenitors (MPPs). In all of these tests, oligopotent progenitors (Fig. 5a) as well as early lineage-committed progenitors of the myeloid (pre-GMs and GMs), erythroid (pre-CFU-E, CFU-E) and megakaryocyte lineages (MegE, MK) were significantly disadvantaged upon deletion of miR17~92 (Fig. 5a–c). In striking contrast, miR17~92fl/fl; RosaCreERT2+/Ki; Bim−/− donor cells were able to contribute efficiently to all HSPC subsets (Figs. 4 and 5). In fact, the miR17~92fl/fl; RosaCreERT2+/Ki; Bim−/− donor cells contributed significantly >50% to several of these compartments. Importantly, miR17~92fl/fl; RosaCreERT2+/Ki; Puma−/− donor cells did not contribute more than the miR17~92fl/fl; RosaCreERT2+/Ki donor cells to the LSK and progenitor compartments (Fig. 4a). This reveals that BIM, but not PUMA, is the critical initiator of apoptosis targeted by the miR17~92 cluster in HSPCs. Interestingly, in multipotent progenitor cells the loss of PUMA could alleviate the lack of competitiveness caused by the deletion of the miR17~92 cluster. However, this protection was still less pronounced than that observed by the loss of BIM (Fig. 4c). These findings further demonstrate that HSPCs rely for their survival on miR17~92-mediated suppression of apoptotic death mainly via direct suppression of the pro-apoptotic BH3-only protein BIM.
The reduction of haematopoietic stem and progenitor cells (HSPCs) caused by the induced deletion of miR17~92 can be fully prevented by the loss of pro-apoptotic BIM. a–c C57BL/6 wt mice (Ly5.1+) were lethally irradiated and reconstituted with a 1:1 mixture of bone marrow cells from UBC-GFP mice (competitor cells) and bone marrow cells from mice of the indicated genotypes (test cells). Reconstituted mice were treated with tamoxifen 8–15 weeks post-transplantation to activate the CreERT2 recombinase. After an additional 8–10 weeks the haematopoietic stem/progenitor cell populations (HSPC) indicated were identified by staining with antibodies against cell type specific surface markers. Representative FACS plots for the gating strategies are provided for wt mice in Fig. 3. Percentages of GFP-negative cells (test cells) were determined for each cell subset. Data represent mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 (Student's t test miR17~92fl/fl; RosaCreERT2+/Ki vs. wt, RosaCreERT2+/Ki, miR17~92fl/fl; RosaCreERT2+/Ki; Bim−/−, Bim−/−, miR17~92fl/fl; RosaCreERT2+/Ki; Puma−/− and Puma−/− mice). n = number of donor/number of recipient mice as indicated
The induced deletion of miR17~92 causes a substantial reduction in lineage-committed haematopoietic progenitor cells and this can be completely prevented by the concomitant loss of BIM. C57BL/6 wt mice (Ly5.1+) were lethally irradiated and reconstituted with a 1:1 mixture of bone marrow cells from UBC-GFP mice (competitor cells) and bone marrow cells from mice of the indicated genotypes (test cells). Reconstituted mice were treated with tamoxifen 8–10 weeks post-transplantation to activate the CreERT2 recombinase. After an additional 8–10 weeks, the haematopoietic progenitor cell populations were identified by staining with antibodies against cell type specific surface markers as indicated in the representative FACS plots. The percentages of GFP-negative cells (test cells) were determined for the indicated cell populations. Data represent mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 (Student's t test miR17~92fl/fl; RosaCreERT2+/Ki vs. wt, RosaCreERT2+/Ki, miR17~92fl/fl; RosaCreERT2+/Ki; Bim−/− and Bim−/− mice). Representative FACS plots indicating the gating strategy and staining for the cell surface markers used are provided for wt mice. a Lineage-committed progenitor cells, including CLP (common lymphoid progenitor), CMP (common myeloid progenitor), GMP (granulocyte/macrophage progenitor) and MEP (megakaryocyte/erythroid progenitor) populations, b granulocyte progenitors, including the pre-GM and GM cell populations, and c pre-megakaryocyte/erythroid progenitors (Pre-MegE), megakaryocyte (MK progenitors) and the erythroid progenitors, pre-CFU-E (colony forming unit erythroid) and CFU-E were examined in the competitive haematopoietic reconstitution assay using staining for the indicated markers
miR17~92 is critical for the survival of human CD34+ haematopoietic stem/progenitor cells by restraining BIM-induced apoptosis
Our study identified the importance of miR17~92-mediated suppression of BIM-induced apoptosis for the survival of murine HSPCs. To extend our studies into the human system, we tested whether miR17~92-mediated suppression of BIM expression also plays a critical role in human HSPCs. To this end we assessed the colony forming capacity of human CD34+ cells from cord blood in the absence or presence of MIR17PTi, a recently described specific inhibitor of pri-miR17~92 [29]. Remarkably, the colony forming capacity of human CD34+ cells derived from two individual donors was reduced in the presence of the MIR17PTi, whereas no impact was observed when colony formation was assessed in the presence of a non-targeting (scrambled) oligo (Fig. 6a).
miR17~92 is critical for the survival of human CD34+ haematopoietic stem/progenitor cells (HSPCs) by restraining BIM-induced apoptosis. a Human CD34+ cord blood cells where cultured with 20 µM MIR17PTi [29] or scrambled control (scr) and the total numbers of colonies were counted and calculated as relative to no drug treated samples. Data are presented as mean ± SEM from two human CD34+ cord blood cell samples, each tested in duplicates. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. b human CD34+ cord blood cells (n = 5) were transfected in triplicates with the indicated mirScript target protectors (Qiagen) together with a reporter plasmid encoding for GFP. At 48 h after transfection the cell viability of the transfected (GFP+) cells was assessed by flow cytometric analysis upon staining with AnnexinV-647 and DAPI with viable cells defined as AnnexinV negative/DAPI negative. Data (normalised to scrambled (scr) control samples) are presented as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 (Student's t test comparing individual target protectors to the scr control)
To investigate the direct interaction of individual mature miRNAs of the miR17~92 cluster with the human BIM mRNA, we exploited miR17~92 target protectors, which specifically block the binding of miR17~92 family members to specific sites in the 3′UTR of the BIM mRNA. The BIM 3′UTR harbors at least four potential binding sites, including miR17, miR19 and miR92 seed family sequences [5]. Human CD34+ cells were transfected with four individual target protectors covering the four highly conserved predicted binding sites for mature miRNAs of the miR17~92 cluster. Remarkably, CD34+ cells from four of the five human donors underwent apoptotic cell death upon transfection with the aforementioned target protectors, with the most prominent effect observed in response to the miR19 or miR92 target protectors (Fig. 6b). These results demonstrate that miR17~92-mediated suppression of BIM-induced apoptosis is also crucial for the survival of human HSPCs.
Discussion
The miR17~92 cluster has been shown to play a critical role in the survival of immature as well as mature B lymphoid cells [7]. Our present study reveals a previously unknown pro-survival function of the miR17~92 cluster in HSPCs. Remarkably, amongst the many predicted targets reported to be bound and suppressed by the miR17~92 cluster, Bim appears to be the most important, given that the mere loss of this pro-apoptotic BH3-only protein fully prevented all defects caused by the deletion of miR17~92.
While the conditional deletion of miR17~92 in adult mice did not impact the LT-HSCs, all subsequent stages, including the ST-HSC, MPP and lineage-committed progenitors, were found to depend on miR17~92-mediated suppression of BIM-induced apoptosis for their survival. Notably, loss of PUMA, another highly potent pro-apoptotic BH3-only protein [38, 39], could rescue the survival defect caused by the deletion of miR17~92 only in a subset of haematopoietic cell populations, including MPPs and B cells, and to a considerably lesser extent compared with the loss of BIM. This partial rescue afforded by the absence of PUMA in some haematopoietic cell subsets is consistent with the observation that BIM and PUMA have overlapping functions in developmentally programmed as well as cytotoxic stress-induced death of lymphoid cells [40]. Why does loss of PUMA show any protection when miR17~92 is deleted? The most likely explanation is that the absence of PUMA frees up pro-survival BCL-2 family members in the cells that are then capable of neutralising BIM. Hence, deletion of miR17~92 causes an increase in the levels of BIM, which can be more efficiently neutralised by BCL-2, MCL-1 and their relatives when PUMA is deleted as more such free pro-survival proteins are available. Remarkably, even though Puma−/− cells had a survival advantage over wt cells (without deletion of miR17~92) in almost all haematopoietic cell subsets tested, loss of PUMA was not sufficient to rescue the survival defects induced by the deletion of the miR17~92 cluster in most of these cell populations. Of note loss of BIM on a wt background resulted in a survival advantage of several mature haematopoietic cell subsets but not HSPCs or immature cells of the lymphoid and erythroid lineage in the bone marrow (Figs. 2, 4). This is in line with the notion that miR17~92 is highly expressed in HSPCs (Fig. 1b), thereby capable of repressing Bim mRNA effectively in these cell populations. These high levels of miR17~92 result in low expression of BIM, which is similar to the loss of BIM by genetic deletion. Therefore, it would be expected that the genetic loss of Bim in miR17~92 high-expressing cells has no or only little impact on their survival, because this microRNA cluster already efficiently restrains the expression of this initiator of apoptosis.
Our studies with human CD34+ HSPCs treated with a specific inhibitor of pri-miR17~92 (MIR17PTi) [29] confirmed the critical role of miR17~92-mediated repression of BIM for their survival given that this reagent substantially reduced the haematopoietic colony forming potential of these cells. Accordingly, the expression of microRNA target protectors (mirScript) designed to bind to four specific regions in the BIM 3′UTR [5] in human CD34+ cells induced marked apoptotic cell death. Interestingly, we observed apoptotic cell death upon targeting of several miR17~92 seed-match sequences in the human BIM 3′UTR. However, one out of five human CD34+ HSPC samples did not respond to any of the target protectors. It is tempting to speculate, that polymorphisms in the miR17~92 seed regions of the BIM 3′UTR in this donor may be responsible for the observed resistance. So how could BIM be kept in check in such a scenario? One could imagine that positive regulators of gene expression that act on the BIM 3′UTR are downregulated to allow less dependence on miR17~92-mediated repression of BIM, but further studies are required to confirm this hypothesis.
While several in silico and in vitro findings provided evidence for the regulation of BIM expression through its 3′UTR by the miR17~92 cluster [5, 6, 22, 29, 41, 42], functional demonstration that BIM is regulated in physiological settings in vivo by such a process has never been reported before. This makes the post-transcriptional regulation of Bim by miR17~92 thus far the only in vivo validated regulatory mechanism of BIM expression, given that the importance of previously in vitro reported transcriptional (FOXO transcription factors) and post-translational (ERK-mediated phosphorylation) processes for regulating BIM expression could not be confirmed in physiological settings within the context of a whole organism [43, 44].
In conclusion, our studies reveal that miR17~92 plays a critical role in the survival of both mouse and human HSPCs and committed progenitors by restraining the expression of pro-apoptotic BIM. Moreover, our results uncover for the first time a regulatory mechanism of BIM that is important in the context of haematopoietic cell survival and development within the whole organism. The observation that the mere absence of BIM is capable to completely prevent all defects caused by the deletion of the miR17~92 cluster is a remarkable finding as many targets bound and suppressed by this microRNA cluster have been reported with their repression proposed to be critical for normal physiology. Based on our findings, it is tempting to speculate that microRNA functions in the context of a whole organism might be less complex than previously assumed and this could be uncovered by more in vivo experimentation through comparing the impact of the absence of a microRNA cluster with the phenotype of the combined absence of this microRNA cluster and one of its targets at the same time.
References
Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97.
Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–98.
Tanzer A, Stadler PF. Molecular evolution of a microRNA cluster. J Mol Biol. 2004;339:327–35.
Lu Y, Thomson JM, Wong HY, Hammond SM, Hogan BL. Transgenic over-expression of the microRNA miR-17-92 cluster promotes proliferation and inhibits differentiation of lung epithelial progenitor cells. Dev Biol. 2007;310:442–53.
Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–86.
Xiao C, Srinivasan L, Calado DP, Patterson HC, Zhang B, Wang J, et al. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol. 2008;9:405–14.
Jiang S, Li C, Olive V, Lykken E, Feng F, Sevilla J, et al. Molecular dissection of the miR-17-92 cluster’s critical dual roles in promoting Th1 responses and preventing inducible Treg differentiation. Blood. 2011;118:5487–97.
Shan SW, Lee DY, Deng Z, Shatseva T, Jeyapalan Z, Du WW, et al. MicroRNA MiR-17 retards tissue growth and represses fibronectin expression. Nat Cell Biol. 2009;11:1031–8.
Chamorro-Jorganes A, Lee MY, Araldi E, Landskroner-Eiger S, Fernandez-Fuertes M, Sahraei M, et al. VEGF-induced expression of miR-17-92 cluster in endothelial cells is mediated by ERK/ELK1 activation and regulates angiogenesis. Circ Res. 2016;118:38–47.
Landskroner-Eiger S, Qiu C, Perrotta P, Siragusa M, Lee MY, Ulrich V, et al. Endothelial miR-17 approximately 92 cluster negatively regulates arteriogenesis via miRNA-19 repression of WNT signaling. Proc Natl Acad Sci USA. 2015;112:12812–7.
Lau KW, Stiffel VM, Rundle CH, Amoui M, Tapia J, White TD, et al. Conditional disruption of miR17~92 in osteoclasts led to activation of osteoclasts and loss of trabecular bone in part through suppression of the miR17-mediated downregulation of protein-tyrosine phosphatase-oc in mice. JBMR. 2017;1:73–85.
Bian S, Hong J, Li Q, Schebelle L, Pollock A, Knauss JL, et al. MicroRNA cluster miR-17-92 regulates neural stem cell expansion and transition to intermediate progenitors in the developing mouse neocortex. Cell Rep. 2013;3:1398–406.
de Pontual L, Yao E, Callier P, Faivre L, Drouin V, Cariou S, et al. Germline deletion of the miR-17 approximately 92 cluster causes skeletal and growth defects in humans. Nat Genet. 2011;43:1026–30.
Regelin M, Blume J, Pommerencke J, Vakilzadeh R, Witzlau K, Lyszkiewicz M, et al. Responsiveness of developing T cells to IL-7 signals is sustained by miR-17 approximately 92. J Immunol. 2015;195:4832–40.
Bouillet P, Metcalf D, Huang DC, Tarlinton DM, Kay TW, Kontgen F, et al. Proapoptotic Bcl-2 relative Bim required for certain apoptotic responses, leukocyte homeostasis, and to preclude autoimmunity. Science. 1999;286:1735–8.
Strasser A, Whittingham S, Vaux DL, Bath ML, Adams JM, Cory S, et al. Enforced BCL2 expression in B-lymphoid cells prolongs antibody responses and elicits autoimmune disease. Proc Natl Acad Sci USA. 1991;88:8661–5.
McDonnell TJ, Deane N, Platt FM, Nunez G, Jaeger U, McKearn JP, et al. bcl-2-immunoglobulin transgenic mice demonstrate extended B cell survival and follicular lymphoproliferation. Cell. 1989;57:79–88.
Han YC, Vidigal JA, Mu P, Yao E, Singh I, Gonzalez AJ, et al. An allelic series of miR-17 approximately 92-mutant mice uncovers functional specialization and cooperation among members of a microRNA polycistron. Nat Genet. 2015;47:766–75.
He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–33.
Mu P, Han YC, Betel D, Yao E, Squatrito M, Ogrodowski P, et al. Genetic dissection of the miR-17~92 cluster of microRNAs in Myc-induced B-cell lymphomas. Genes Dev. 2009;23:2806–11.
Olive V, Bennett MJ, Walker JC, Ma C, Jiang I, Cordon-Cardo C, et al. miR-19 is a key oncogenic component of mir-17-92. Genes Dev. 2009;23:2839–49.
Li Y, Choi PS, Casey SC, Dill DL, Felsher DW. MYC through miR-17-92 suppresses specific target genes to maintain survival, autonomous proliferation, and a neoplastic state. Cancer Cell. 2014;26:262–72.
Seibler J, Zevnik B, Kuter-Luks B, Andreas S, Kern H, Hennek T, et al. Rapid generation of inducible mouse mutants. Nucleic Acids Res. 2003;31:e12.
Villunger A, Michalak EM, Coultas L, Mullauer F, Bock G, Ausserlechner MJ, et al. p53- and drug-induced apoptotic responses mediated by BH3-only proteins puma and noxa. Science. 2003;302:1036–8.
Anastassiadis K, Glaser S, Kranz A, Berhardt K, Stewart AF. A practical summary of site-specific recombination, conditional mutagenesis, and tamoxifen induction of CreERT2. Methods Enzymol. 2010;477:109–23.
Brinkmann K, Grabow S, Hyland CD, Teh CE, Alexander WS, Herold MJ, et al. The combination of reduced MCL-1 and standard chemotherapeutics is tolerable in mice. Cell Death Differ. 2017;24:2032–43.
Pronk CJ, Rossi DJ, Mansson R, Attema JL, Norddahl GL, Chan CK, et al. Elucidation of the phenotypic, functional, and molecular topography of a myeloerythroid progenitor cell hierarchy. Cell Stem Cell. 2007;1:428–42.
Heng TS, Painter MW, Immunological Genome Project C. The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol. 2008;9:1091–4.
Morelli E, Biamonte L, Federico C, Amodio N, Di Martino MT, Gallo Cantafio ME, et al. Therapeutic vulnerability of multiple myeloma to MIR17PTi, a first-in-class inhibitor of pri-mir-17-92. Blood. 2018;132:1050–63.
Mi S, Li Z, Chen P, He C, Cao D, Elkahloun A, et al. Aberrant overexpression and function of the miR-17-92 cluster in MLL-rearranged acute leukemia. Proc Natl Acad Sci USA. 2010;107:3710–5.
de Graaf CA, Choi J, Baldwin TM, Bolden JE, Fairfax KA, Robinson AJ, et al. Haemopedia: an expression atlas of murine hematopoietic cells. Stem Cell Rep. 2016;7:571–82.
Choi J, Baldwin TM, Wong M, Bolden JE, Fairfax KA, Lucas EC, et al. Haemopedia RNA-seq: a database of gene expression during haematopoiesis in mice and humans. Nucleic Acids Res. 2019;47(D1):D780–D785.
Michalak EM, Jansen ES, Happo L, Cragg MS, Tai L, Smyth GK, et al. Puma and to a lesser extent Noxa are suppressors of Myc-induced lymphomagenesis. Cell Death Differ. 2009;16:684–96.
Michalak EM, Villunger A, Adams JM, Strasser A. In several cell types tumour suppressor p53 induces apoptosis largely via Puma but Noxa can contribute. Cell Death Differ. 2008;15:1019–29.
Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015;4:05005.
Christensen JL, Weissman IL. Flk-2 is a marker in hematopoietic stem cell differentiation: a simple method to isolate long-term stem cells. Proc Natl Acad Sci USA. 2001;98:14541–6.
Oguro H, Ding L, Morrison SJ. SLAM family markers resolve functionally distinct subpopulations of hematopoietic stem cells and multipotent progenitors. Cell Stem Cell. 2013;13:102–16.
Nakano K, Vousden KH. PUMA, a novel proapoptotic gene, is induced by p53. Mol Cell. 2001;7:683–94.
Yu J, Zhang L, Hwang PM, Kinzler KW, Vogelstein B. PUMA induces the rapid apoptosis of colorectal cancer cells. Mol Cell. 2001;7:673–82.
Erlacher M, Michalak EM, Kelly PN, Labi V, Niederegger H, Coultas L, et al. BH3-only proteins Puma and Bim are rate-limiting for gamma-radiation- and glucocorticoid-induced apoptosis of lymphoid cells in vivo. Blood. 2005;106:4131–8.
Guo Z, Mu X, Ouyang H, Cheng Z, Wang Z, Xu B. Operative treatment of huge adult frontal nasoethmoid meningoencephalocele. J Craniofac Surg. 2013;24:1669–70.
Xu Z, Sharp PP, Yao Y, Segal D, Ang CH, Khaw SL, et al. BET inhibition represses miR17-92 to drive BIM-initiated apoptosis of normal and transformed hematopoietic cells. Leukemia. 2016;30:1531–41.
Clybouw C, Merino D, Nebl T, Masson F, Robati M, O’Reilly L, et al. Alternative splicing of Bim and Erk-mediated Bim(EL) phosphorylation are dispensable for hematopoietic homeostasis in vivo. Cell Death Differ. 2012;19:1060–8.
Herold MJ, Rohrbeck L, Lang MJ, Grumont R, Gerondakis S, Tai L, et al. Foxo-mediated Bim transcription is dispensable for the apoptosis of hematopoietic cells that is mediated by this BH3-only protein. EMBO Rep. 2013;14:992–8.
Acknowledgements
We thank C Stivala, S Russo, J Mansheim, T Baldinger, M Patsis and G Siciliano for expert animal care; B Helbert and K Mackwell for genotyping; J Corbin and J McManus for automated blood analysis; S Monard and his team for help with flow cytometry; S Mifsud for assistance with colony assays and Dr P Bouillet for providing Bim−/− mice. This work was supported by grants and fellowships from the Deutsche Krebshilfe (Dr Mildred Scheel post-doctoral fellowship to KB) the Australian National Health and Medical Research Council (NHMRC) (Project Grants 1145728 to MJH, 1143105 to MJH and AS, 1122783 to APN, Program Grants 1016701 to AS and 1113577 to WSA and Fellowships 1020363 to AS, 1058344 to WSA, 1156095 to MJH, GNT1035229 to CAdG and 1124788 to NDH), the Leukemia and Lymphoma Society of America (LLS SCOR 7001-13 to AS and MJH), the Cancer Council of Victoria (project grant 1052309 to AS and Venture Grant to MJH and AS), a Victorian Cancer Agency Fellowship (ECSG18020 to JR) as well as by operational infrastructure grants through the Australian Government Independent Research Institute Infrastructure Support Scheme (361646 and 9000220) and the Victorian State Government Operational Infrastructure Support Program.
Author information
Authors and Affiliations
Contributions
KB, MJH and AS designed and conceived the study, planned experiments and prepared the paper. KB conducted and analysed the experiments. CAdG helped with the analysis of the RNA expression data (Fig. 1b) CH and WSA provided advice for the analysis of the LT-HSC compartment, helped with these experiments and data analysis and provided the antibodies for staining cells (Figs. 3–5). APN and LDR performed the colony formation assays (Fig. 6a). JR and NDH gave advice on experiments with human CD34+ cord blood cells and provided the cells and media (Fig. 6). EM provided MIR17PTi and helped with planning the colony formation assays (Fig. 6a).
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Brinkmann, K., Ng, A.P., de Graaf, C.A. et al. miR17~92 restrains pro-apoptotic BIM to ensure survival of haematopoietic stem and progenitor cells. Cell Death Differ 27, 1475–1488 (2020). https://doi.org/10.1038/s41418-019-0430-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-019-0430-6
This article is cited by
-
Regulation of programmed cell death by Brd4
Cell Death & Disease (2022)
-
The transcription factor TAL1 and miR-17-92 create a regulatory loop in hematopoiesis
Scientific Reports (2020)