Abstract
Rab5 is a master regulator for endosome biogenesis and transport while its in vivo physiological function remains elusive. Here, we find that Rab5a is upregulated in several in vivo and in vitro myogenesis models. By generating myogenic Rab5a-deficient mice, we uncover the essential roles of Rab5a in regulating skeletal muscle regeneration. We further reveal that Rab5a promotes myoblast differentiation and directly interacts with insulin receptor substrate 1 (IRS1), an essential scaffold protein for propagating IGF signaling. Rab5a interacts with IRS1 in a GTP-dependent manner and this interaction is enhanced upon IGF-1 activation and myogenic differentiation. We subsequently identify that the arginine 207 and 222 of IRS1 and tyrosine 82, 89, and 90 of Rab5a are the critical amino acid residues for mediating the association. Mechanistically, Rab5a modulates IRS1 activation by coordinating the association between IRS1 and the IGF receptor (IGFR) and regulating the intracellular membrane targeting of IRS1. Both myogenesis-induced and IGF-evoked AKT-mTOR signaling are dependent on Rab5a. Myogenic deletion of Rab5a also reduces the activation of AKT-mTOR signaling during skeletal muscle regeneration. Taken together, our study uncovers the physiological function of Rab5a in regulating muscle regeneration and delineates the novel role of Rab5a as a critical switch controlling AKT-mTOR signaling by activating IRS1.
Similar content being viewed by others
Introduction
Skeletal muscle plays critical roles in the regulation of the wider metabolism as well as driving locomotion [1, 2]. Impairment of muscle function, caused by injury or aging, severely influences life quality. In response to injury, muscle progenitor cells will be activated and give rise to a transient-amplifying population of myogenic cells called myoblasts [3, 4]. Subsequently, in the process of muscular reconstruction, committed myoblasts fuse either to each other to form new myofibers, or to their host’s existing myofibers [5, 6]. These cellular processes are coordinated by a variety of hormones and growth factors. In particular, insulin-like growth factor-1 (IGF-1) plays essential roles in the regulation of myoblast differentiation through the activation of AKT/PKB-mTOR signaling [7, 8].
The binding of IGF-1 to the IGF receptor (IGFR) stimulates the intrinsic tyrosine kinase activity of IGFR and subsequently activates insulin receptor substrate (IRS) [9]. Tyrosine phosphorylated IRS recruits and activates downstream Phosphatidylinositol 3-kinase (PI3K), AKT/PKB, and mechanistic target of rapamycin (mTOR) protein kinase complexes. Studies with skeletal muscle-specific ablation and transgenic mice show that the Akt-mTOR axis is essential for muscle regeneration [10,11,12,13]. Unlike other receptor tyrosine kinases, the signaling output of IGFR is determined mainly by IRS proteins, which act as docking sites for divergent SH2 domain-containing proteins [14,15,16]. Therefore, the fine-tuning of the IGFR-IRS interaction is required to ensure the accurate regulation of IGF-evoked AKT-mTOR activation. However, the molecular machinery controlling the IGFR-IRS association remains to be deciphered.
The small GTPase Rab5 is a master regulator of endosome fusion, tethering, and biogenesis in cells [17,18,19]. This has been validated by liver-specific knockdown of three Rab5 isoforms which results in a massive depletion of endosomes and lysosomes [20]. Furthermore, Rab5 modulates autophagosome closure [21], chromosome alignment and congression [22, 23], pinocytosis [24], mitochondrial quality control [25], and gluconeogenic gene expression [26]. Thus, the exploration of in vivo functions of Rab5, especially by tissue-specific ablation, represents a crucial step toward a better understanding of the physiological roles of Rab5. We recently demonstrated the co-localization between Rab5 and a promyogenic protein BNIP-2 [27], raising the possibility that Rab5 regulates myogenesis. Here, we showed that Rab5a is specifically upregulated in both in vivo and in vitro myogenesis and that the myogenic ablation of Rab5a impairs muscle regeneration. Unbiased screening revealed IRS1 as a novel binding partner of Rab5a. Moreover, we demonstrated that Rab5a governs the IRS1-IGFR association and the intracellular membrane localization of IRS1 to regulate AKT-mTOR signaling during muscle regeneration.
Results
Specific up-regulation of Rab5a during myoblasts differentiation and in vivo myogenesis
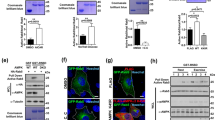
To analyze the function of Rab5 in myogenesis, we examined the temporal and spatial expression patterns of Rab5a, the best studied Rab5 isoform, in several in vitro and in vivo myogenesis systems. We employed the C2C12 mouse myoblast cell line, which fuse into multinucleated myotubes after their transfer to the differentiation medium (DM) from high-serum growth medium (GM). The expression levels of endogenous Rab5a increased during myoblast differentiation, similar to the profile of myogenic markers MyoG and myosin heavy chain (MHC) (Fig. 1a). The elevation of Rab5a expression during myoblast differentiation was further validated by immunostaining (Supplementary Fig. S1A). Consistently, Rab5a expression robustly increased during the differentiation of freshly isolated primary myoblasts (Fig. 1b). Prior to fusing into myotubes, myoblasts remodel from a fibroblast-like morphology to a spindle-like morphology [28]. Immunofluorescence staining displayed that endogenous Rab5 is specifically upregulated in spindle-like differentiating C2C12 myoblasts and primary myoblasts (Fig. 1c). Differentiating myoblasts with elevated expression of endogenous Rab5 also contain higher level of MyoG (Supplementary Fig. S1B). The up-regulation of Rab5a is specific since the expression levels of Rab7, Rab11, or Rab9 remained unchanged (Supplementary Fig. S1C). Interestingly, the expression levels of endosome markers including EEA1, Clathrin, and Caveolin remain unchanged in differentiating myoblasts (Supplementary Fig. S1D). Like other small GTPases, Rab5 cycles between an active GTP-bound state and an inactive GDP-bound state [29]. Rab5 activation assay showed that the concentration of GTP-bound (active) Rab5 rose when myoblasts were transferred into DM (Fig. 1d) or treated with IGF-1 (Fig. 1e), a key growth factor in promoting myogenesis [30].
a C2C12 myoblasts cultured to 80% confluence were transferred to DM for the indicated times. Total lysates were immunoblotted with anti-Rab5a and the indicated antibodies. DM: differentiation medium. b Lysates of freshly isolated primary myoblasts that were proliferating in GM or transferred to DM for two days were immunoblotted with the indicated antibodies. GM: growth medium. c C2C12 myoblasts or primary myoblasts that were transferred to DM for 36 h were fixed and stained for endogenous Rab5 as described in ‘Materials and Methods’. The actin-filaments were detected by direct staining with rhodamine-conjugated phalloidin. Bar: 50 μm. d Lysates of C2C12 myoblasts that were proliferating in GM or transferred to DM for 1.5 day were subjected to R5BD GST-pulldown, and then Western blotted with Rab5a antibody to measure Rab5a activation. e Lysates of C2C12 myoblasts that were untreated or treated with IGF1 for 5 min were subjected to R5BD GST-pulldown, and then Western blotted with Rab5a antibody to measure Rab5a activation. f Lysates of hindlimb muscles of two postnatal mice at the indicated ages were subjected to Western blot analysis with the indicated antibodies. P: postnatal. g Lysates of hindlimb muscles from two wild-type or two dystrophic mdx mice were subjected to Western blot analysis with the indicated antibodies. h Tibialis anterior (TA) muscles of eight-week-old C57 or mdx mice were subjected to cross-section and followed by staining with Rab5a. Hoechst and WGA were stained for nuclei and myofiber membrane boundaries respectively (n = 3 mice for each group). Bar: 50 μm.
We next investigated the in vivo expression of Rab5a during myogenesis. High levels of both Rab5a and embryonic/developmental MHC (eMHC), a marker for muscle development and regeneration, were observed in limb muscles of newborn mice (at the age of 3 and 5 days postnatal) in which active myogenesis occurred (Fig. 1f). When the neonatal myogenesis ceased after 2 weeks, the level of Rab5a and eMHC decreased and remained low in adult mice (Fig. 1f). Moreover, we compared wildtype mouse muscles and dystrophic muscles from mdx (X-linked muscular dystrophic) mice which were featured by a pathologically active muscle degeneration and regeneration [31]. Significant higher levels of Rab5a were detected in Tibialis anterior (TA) muscles from mdx mice (Fig. 1g, h). All the above in vitro and in vivo studies strongly suggest that Rab5a is associated with myogenesis.
Myogenic ablation of Rab5a impairs muscle regeneration
To test if Rab5a is critical for muscle regeneration, we first detected the expression profile of Rab5a during muscular post-injury regeneration. TA muscles of eight-week-old mice were subjected to a single cardiotoxin (CTX) injury and then allowed to recover for 3 to 14 days [32, 33]. During the acute phase of regeneration (3 days after injury), myoblasts fuse to form new myofibers which express eMHC. The expression levels of Rab5a were elevated on day 3 and day 7 when myoblasts fusion occurs and downregulated when active myogenesis has ceased on day 14 (Fig. 2a; Supplementary Fig. S2A).
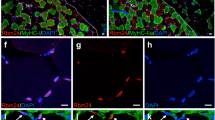
a Tibialis anterior (TA) muscles of eight-week-old mice were subjected to a single cardiotoxin (CTX) injury and then allowed to recover for 3, 7, or 14 days. Control mice were injected with PBS. TA muscle lysates were subjected to Western blot analysis with the indicated antibodies. b Lysates of liver, spleen, TA, or Gastrocnemius (GAS) muscles from Rab5amKO or control Rab5af/f mice were subjected to Western blot analysis with anti-Rab5a or Hsc70 indicated antibodies. (n = 3 mice for each group). c Cryosections of CTX-3 days regenerating TA muscles from Rab5amKO or control Rab5af/f mice were stained with Hoechst for nuclei and embryonic myosin heavy chain (eMHC) antibody for newly formed myofibers. Bar: 20 μm. Cross-sectional areas (CSAs) of regenerating myofibers from Rab5amKO or control Rab5af/f mice were analyzed at CTX-3 days using the Image-Pro Plus 6.0 program (n = 3 mice for each group). >300 fibers scored per sample. Data are presented as mean ± SD. **P < 0.01 (Student’s t-test). d PBS injected TA muscles or CTX-3 days regenerating TA muscles from Rab5amKO or Rab5af/f mice were subjected to Western blot analysis with anti-Rab5a or indicated myogenic protein antibodies (n = 3 mice for each group). e Cryosections of CTX-3 days or CTX-7 days regenerating TA muscles from Rab5amKO or control Rab5af/f mice were stained with Hoechst for nuclei, Desmin antibody for newly formed myofibers, and WGA for myofiber membrane boundaries. Bar: 20 μm. f Cryosections of CTX-14 days TA muscles from Rab5amKO or control Rab5af/f mice were stained with Hoechst for nuclei and WGA for myofiber membrane boundaries. Bar: 50 μm. CSAs of regenerating myofibers were analyzed (n = 3 mice for each group). >300 fibers scored per sample. Data are presented as mean ± SD. **P < 0.01 (Student’s t-test). g H&E staining of cross sections of Rab5amKO or control Rab5af/f TA muscles at CTX-14 days. Representative sections are shown (n = 3 mice for each group). Scale bars: 100 μm. h Representative Evans blue staining images of whole Rab5amKO or control Rab5af/f TA muscles at day 14 post-CTX injection (n = 3 mice for each group).
We next established a transgenic mouse model in which the Rab5a gene was conditionally knocked out in the skeletal muscle lineage. Mice harboring loxP-flanked Rab5a alleles (Rab5afloxed, henceforth Rab5af/f) were crossed to mice with a Cre recombinase gene knocked in to the MyoD locus (MyoD-Cre) [34]. For simplicity, we refer to MyoD-Cre; Rab5af/f as Rab5amKO (for Rab5a myogenic knockout) mice. Successful recombination of Rab5a was confirmed by PCR on genomic DNA (data not shown). Both Western blot and immunostaining analysis revealed a strong reduction of Rab5a in Rab5amKO skeletal muscles (Fig. 2b; Supplementary Fig. S2B). TA muscles of Rab5amKO and control Rab5af/f mice were given CTX injection and muscle regeneration was analyzed. Compared with the control Rab5af/f regenerating muscles, the newly formed myofibers in Rab5amKO regenerating muscles displayed smaller cross-section area (CSA) on day 3 post-CTX injection (CTX-3 days) (Fig. 2c). Both MyoG and eMHC were markedly upregulated in Rab5af/f mice muscles on day 3 after CTX injection (Fig. 2d, lane 1 versus 3). However, the injury-induced elevation of MyoG and eMHC was inhibited in Rab5amKO mice (Fig. 2d, lane 3 versus 4). Moreover, myogenic ablation of Rab5a also suppressed the elevation of Pax7, the marker for satellite cells, in regenerating muscles (Fig. 2d). These observations were supported by qPCR analysis for myogenic markers including MyoD, MyoG, Pax7, and Myf5 (Supplementary Fig. S2C).
We further assessed the expression of Desmin, an intermediate filament protein which is expressed in newly formed myofibers during muscle regeneration and myogenesis [35]. As shown in Fig. 2e, Desmin was strongly expressed in regenerating myofibers in Rab5af/f mice while Rab5amKO mice showed relatively lower expression of Desmin. On day 14 post injection, the CSA of myofibers in Rab5amKO mice were smaller than those in Rab5af/f mice (Fig. 2f), suggesting a defect on muscle regeneration. This was verified by H&E analysis (Fig. 2g). In addition, one day prior to harvesting muscle, the mice were intraperitoneally injected with Evans Blue dye, which accumulates in damaged muscle fibers and facilitates visualization of the nonregenerated myofibers [36]. Evans Blue staining in the CTX-treated Rab5amKO muscles was much more profound than in the control muscles on day 14 post injection (Fig. 2h), suggesting poor regeneration of Rab5a-null muscles. Taken together, these data strongly indicate that Rab5a contributes to the post injury regeneration of skeletal muscle.
Rab5a positively regulates myoblast differentiation
We next investigated the roles of Rab5a in myoblast differentiation which represents a pivotal step in muscle regeneration [5, 6]. Specific depletion of Rab5a by two independent sequences reduced the expression of MyoG and MHC in myoblasts (Fig. 3a) and resulted in shorter MHC-positive myotubes with fewer nuclei (Fig. 3b, c). RT-qPCR analysis also showed that the expression of myogenic markers decreased by Rab5a knockdown (Supplementary Fig. S3A). Gain-of-function assay was performed by stably overexpression of Rab5a in C2C12 myoblasts. Compared to the negative control, Rab5a-expressing myoblasts displayed enhanced levels of both MyoG and MHC (Fig. 3d). The MHC-positive myotubes derived from Rab5a-expressing C2C12 myoblasts were thicker and longer and contained a greater number of cell nuclei (Fig. 3e, f). Exogenous Rab5a were stained to monitor protein expression and verify its positive effects on myogenesis at single myotube level (Supplementary Fig. S3B).
a C2C12 myoblasts transfected with control or two Rab5a siRNAs were cultured in DM for 2 days followed by western blot analysis of Rab5, MyoG, MHC and tubulin proteins. b C2C12 myoblasts that transfected with Rab5a siRNA or an irrelevant (control) siRNA were cultured in DM, fixed, and stained with MHC antibody and Hoechst for immunofluorescence analysis. Bar: 50 μm. c Quantification of myoblast differentiation shown in A. MHC positive cells were scored as having a single nucleus, two to five nuclei, or greater than or equal to six nuclei. Data are presented as mean ± SD. ***P < 0.001 (Student’s t-test). d C2C12 myoblasts stably transfected with FLAG-tagged Rab5a or control FLAG plasmids were cultured in DM for 2 days. Cell lysates were subjected to Western blot analysis with the indicated antibodies. e C2C12 myoblasts stably transfected with FLAG-tagged Rab5a or control FLAG plasmids were cultured in DM for 3 days. Cell then were fixed and stained with MHC antibody for confocal fluorescence microscopy analysis. Bar: 50 μm. f Quantification of myoblast differentiation shown in F. MHC positive cells were scored as having a single nucleus, two to five nuclei, or greater than or equal to six nuclei. Data are presented as mean ± SD. *P < 0.05, ***P < 0.001 (Student’s t-test). g Lysates of C2C12 myoblasts transfected with control or two RabGDI1 siRNAs were subjected to R5BD GST-pulldown, and then Western blotted with Rab5 antibody. h C2C12 myoblasts that transfected with control or two RabGDI1 siRNAs were cultured in DM, fixed, and stained with MHC antibody and Hoechst for immunofluorescence analysis. Bar: 50 μm. i Quantification of myoblast differentiation shown in I. Data are presented as mean ± SD. ***P < 0.001 (Student’s t-test). j C2C12 myoblasts stably transfected with FLAG- Rab5a, FLAG-Rab5DN mutant, or control FLAG plasmid were cultured in DM for 3 days. Cell then were fixed and stained with MHC antibody and Hoechst for confocal fluorescence microscopy analysis. Bar: 50 μm. k C2C12 myoblasts stably transfected with FLAG-Rab5a, FLAG-Rab5aDN mutant, or control FLAG plasmid were cultured in DM for 2 days. Cell lysates were subjected to Western blot analysis with the indicated antibodies.
We further tested whether activation of Rab5 could enhance myoblast differentiation. Two siRNA sequences against RabGDI1 were transfected into C2C12 myoblasts since RabGDI inhibits Rab5 activation [37]. Both sequences effectively diminished RabGDI1 protein levels and increased GTP-bound Rab5 level (Fig. 3g). When cultured in DM, depletion of RabGDI1 led to longer and larger myotubes which contained more nuclei (Fig. 3h, i). We next altered Rab5a activity in myoblasts by transfecting a dominant negative (DN) construct of Rab5a (S34N). Overexpression of Rab5S34N resulted in shorter and thinner myotubes (Fig. 3j; Supplementary Fig. S3C) and reduced expression levels of MyoG and MHC (Fig. 3k). Collectively, these results indicate that Rab5a is critical for myoblast differentiation.
Rab5a directly interacts with IRS1
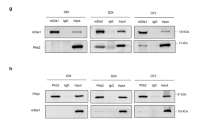
To further delineate the mechanism by which Rab5a promotes myogenesis, we employed a proteomics approach by subjecting purified recombinant glutathione S-transferase (GST)-Rab5a to the C2C12 cell for Pull-down assay to identify interaction partners of Rab5a in myoblasts. After silver-staining, the gel lanes were excised and subjected to mass spectrometry analysis, which revealed that the insulin receptor substrate 1 (IRS1), an essential scaffold protein of IGF signaling [14], is a potential binding proteins of Rab5a (Supplementary Table S1). To confirm the Rab5a-IRS1 association in vivo, we immunoprecipitated endogenous IRS1 in dissected mouse hindlimb muscle lysate and both Rab5a and IRS1 were present in the precipitate (Fig. 4a). To determine whether Rab5a-IRS1 interaction was mediated through their direct binding, we purified recombinant FLAG-IRS1 and GST-Rab5a proteins for pull down assay. GST-Rab5a, but not GST protein along, directly interacts with IRS1 (Fig. 4b). IRS1 contains a pleckstrin homology (PH) and a phosphotyrosine binding (PTB) domain in its amino terminus followed by an unstructured carboxyl-terminal tail sequence [14, 38]. A series of IRS1 mutants were prepared and tested for their ability to interact with Rab5a (Fig. 4c). We found that Rab5a directly interacts with the PTB domain of IRS1 (Fig. 4d). PTB domain recognizes and interacts with either phosphorylated or non-phosphorylated tyrosine resides [39]. We therefore prepared an IRS1-PTB domain mutant (IRS1-PTB2RA) by replacing arginine 207 and 222 residues, which create a basic pocket for binding negatively charged phosphate, to alanines. The two arginine-to-alanine mutations completely abolished the association of PTB domain to Rab5a (Fig. 4e). We further screening the tyrosine residues in Rab5a and found that replacing tyrosine 82, 89, and 90 to alanine (Rab5a3YA) impaired the interaction of Rab5a to the PTB domain of IRS1 (Fig. 4f).
a Lysates of mouse hindlimb muscle were immunoprecipitated with IRS1 antibody and then Western blotted with Rab5a antibody or IGFR antibody as a positive control. b FLAG vector or FLAG-IRS1 was transfected into 293T cell and immunoprecipitated with anti-FLAG-agarose beads followed by eluting with FLAG peptide. Control GST protein or GST-Rab5a fusion protein was incubated with purified FLAG-IRS, FLAG peptide, or BSA for direct Pull-down assay and then Western blotted with FLAG antibody (top panel). The amounts of GST and GST-Rab5a used in this experiment were indicated by the Coomassie blue staining (middle panel). c Schematic diagram of IRS1 and its mutants. d FLAG-tagged IRS1 or mutant constructs as shown in c was transfected into 293T cell and purified by using anti-FLAG-agarose beads (bottom panel). GST or GST-Rab5a fusion protein was incubated with purified FLAG-tagged proteins for direct Pull-down assay (top panel). The amounts of GST and GST-Rab5a were indicated by the Coomassie blue staining (middle panel). e GST or GST-Rab5a fusion protein was incubated with purified FLAG-tagged IRS1-PTB or PTB2RA mutant for direct Pull-down assay (top panel). The amounts of GST and GST-Rab5a were indicated by the Coomassie blue staining (middle panel). f GST, GST-Rab5a, or GST-Rab5a3YA fusion protein was incubated with purified FLAG-tagged IRS1-PTB protein for direct Pull-down assay (top panel). The amounts of GST proteins were indicated by the Coomassie blue staining (middle panel). g C2C12 myoblasts were cultured in GM or were transferred to DM to trigger differentiation for 2 days. Endogenous IRS1 was immunoprecipitated and coimmunoprecipitated Rab5a or IGFR was detected by Western blot analysis with specific antibody. h C2C12 myoblasts treated with or without 100 ng/ml IGF-1 for 10 min were subjected to immunoprecipitation with IRS1 antibody. Coimmunoprecipitated IGFR and Rab5a were detected by Western blot analysis with specific antibodies. i HEK 293 T cells were transfected with control FLAG plasmid or FLAG-IRS1. After 18 h post transfection, Cell lysates were incubated with immobilized GST-Rab5a loaded with GDP or GTP-γS for GST Pull-down assay. The beads were washed and analyzed by western blot for FLAG (IRS1). The amounts of GST proteins were indicated by the Coomassie blue staining. j C2C12 myoblasts stably transfected with control HA plasmid, HA-Rab5a, or HA-Rab5a3YA were cultured in DM for 2 days. Cell lysates were subjected to Western blot analysis with the indicated antibodies. k C2C12 myoblasts stably transfected with HA plasmid, HA-Rab5a, or HA-Rab5a3YA were cultured in DM for 3 days. Cell then were fixed and stained with MHC antibody for confocal fluorescence microscopy analysis and quantification. Bar: 100 μm. Data are presented as mean ± SD. *P < 0.05, ***P < 0.001 (Student’s t-test).
To determine the implication of Rab5a-IRS1 interaction in myogenesis, we performed the immunoprecipitation in proliferating (GM) or differentiating (DM) myoblasts. The association of endogenous Rab5a to IRS1 was much enhanced in differentiating myoblasts (Fig. 4g). In line with this, IGF-1 treatment also enhanced the Rab5a-IRS1 interaction (Fig. 4h). Additionally, we demonstrated that IRS1 preferentially bound to GTP-loaded GST-Rab5a, but not GDP-loaded GST-Rab5a (Fig. 4i). Moreover, the Rab5a3YA mutant, which could not associate with IRS1 (Fig. 4f), failed to enhance myoblast differentiation (Fig. 4j, k; Supplementary Fig. S4). Taken together, we verified that Rab5a and IRS1 form a novel physiologic complex in both myoblasts and muscle.
Rab5a coordinates the interaction between IGFR and IRS1 during myoblast differentiation
IRS1 was regulated by multi-phosphorylation sites [14]. The Tyr phosphorylation sites of IRS1 recruit effector molecules while Ser/Thr phosphorylation of IRS1 inhibits its downstream signaling [14]. We thus examined if Rab5a modulates the phosphorylation status of IRS1. Interestingly, depletion of Rab5a impaired Tyr phosphorylation and enhanced the Ser phosphorylation of IRS1, but had little effect on the tyrosine phosphorylation of IGFR (Fig. 5a). This indicates that Rab5a positively regulates myoblast differentiation at a site downstream of IGFR, which could be IRS1. To test this postulation, we checked the effect of IRS1 on myoblast differentiation under Rab5a knockdown condition. IRS1 enhanced MHC expression in control siRNA-transfected, but not in Rab5a siRNA-transfected, differentiating myoblasts (Fig. 5b). Immunofluorescence analysis confirmed that the stimulatory effects of IRS1 on myoblast differentiation is Rab5a-dependent (Fig. 5c; Supplementary Fig. S5A). Moreover, IRS12RA mutant, whose PTB domain loss the interaction with Rab5a (Fig. 4e), failed to enhance myoblast differentiation and led to shorter myotubes with less nuclei (Fig. 5d, e; Supplementary Fig. S5B). These data indicate that IRS1 promotes myogenesis in a Rab5a-dependent manner.
a C2C12 myoblasts transfected with control or Rab5a siRNAs were triggered to differentiation for 2 days. Cell Lysates were western blotted with anti-IRS1, anti-pIRS1 (Y895, Y608, and S318), anti-IGFR, anti-pIGFR and other indicated antibodies. b C2C12 myoblasts stably expressing FLAG-IRS1 were transfected with control or Rab5a siRNA followed by culturing in DM for 48 h. Lysates were western blotted with indicated antibodies. c C2C12 myoblasts stably expressing control FLAG vector or FLAG-IRS1 were transfected with control or Rab5a siRNA followed by culturing in DM for 3 days. Cell then were fixed and stained with MHC antibody and Hoechst for confocal fluorescence microscopy analysis. Bar: 100 μm. Myoblast differentiation was quantified by calculating the ratio of nuclei within MHC-positive myotubes. Data are presented as mean ± SD. **P < 0.01, ***P < 0.001 (Student’s t-test). d C2C12 myoblasts stably transfected with control FLAG plasmid, FLAG-IRS1, or FLAG-IRS12RA were cultured in DM for 2 days. Cell lysates were subjected to Western blot analysis with the indicated antibodies. e C2C12 myoblasts stably transfected with control FLAG plasmid, FLAG-IRS1, or FLAG-IRS12RA were cultured in DM for 3 days. Cell then were fixed and stained with MHC antibody for confocal fluorescence microscopy analysis and quantification. Bar: 100 μm. Data are presented as mean ± SD. *P < 0.05, **P < 0.01 (Student’s t-test). f C2C12 myoblasts transfected with control or Rab5a siRNA were cultured in GM or triggered to differentiation for 2 days. Endogenous IRS1 was immunoprecipitated and coimmunoprecipitated IGFR, p110 subunit of PI3K, and APPL1 was detected by Western blot analysis with specific antibody. g C2C12 myoblasts stably expressing control FLAG or FLAG-Rab5aDN mutant plasmid were cultured in GM or triggered to differentiation for 2 days. Endogenous IRS1 was immunoprecipitated and coimmunoprecipitated IGFR was detected by Western blot analysis with specific antibody. h C2C12 myoblasts stably expressing control FLAG or FLAG-Rab5a plasmid were cultured in GM or triggered to differentiation for 2 days. Endogenous IRS1 was immunoprecipitated and coimmunoprecipitated IGFR was detected by Western blot analysis.
Since Tyr phosphorylation of IRS1 is activated by IGFR [9], we investigated if Rab5a facilitates the IGFR-IRS1 interaction. The IRS1 interaction with IGFR was enhanced in differentiating myoblasts (lane 2 versus 3) while this interaction was greatly reduced when Rab5a was depleted (lane 3 versus 5). Depletion of Rab5a also reduced the association of IRS1 to p110 subunit of PI3K and APPL1 (Fig. 5f), two IRS1 downstream effectors [40, 41]. Moreover, the Rab5aS34N DN mutant impaired the differentiation-stimulated binding between IGFR and IRS1 (Fig. 5g; lane 4 versus 5), while overexpression of Rab5a markedly enhanced the association of IGFR to IRS1 in differentiating myoblasts (Fig. 5h; lane 3 versus 5). Thus, we concluded that Rab5a regulates the activation of IRS1 by coordinating its association with IGFR.
Rab5a targeted IRS1 to the intracellular membrane
The Tyr-phosphorylated IRS1 is localized in membrane fractions, high-speed pellet (HSP) and low-speed pellet (LSP), while Ser-phosphorylated IRS1 is in cytosol [42,43,44]. We therefore examined if Rab5 regulates the intracellular membrane association of IRS1. The majority of IRS1 targets to the membrane-containing fractions (HSP and LSP) in control myoblasts, while knockdown of Rab5a much increased the cytosolic IRS1 (Fig. 6a). This is in accordance with the data showing that ectopic expression of Rab5a enhanced IRS1 levels in HSP and reduced IRS1 levels in cytosolic fraction (Fig. 6b). In contrast, altering the expression levels of Rab5a has no effect on the membrane localization of IGFR (Fig. 6a, b). Rab5a DN mutant also reduced HSP-localized IRS1 (Fig. 6c). These data thus suggest that the intracellular membrane localization of IRS1 is dependent on Rab5a.
a C2C12 myoblasts transfected with control or Rab5a siRNA were homogenized and extracts were fractionated into cytosol, HSP, and LSP. Fractions were analyzed by western blotting with 20 µg of protein loaded in each lane. b C2C12 myoblasts stably expressing control FLAG vector or FLAG-Rab5a were processed as in a. c C2C12 myoblasts stably expressing control FLAG vector or FLAG-Rab5a DN mutant were processed as in a. d C2C12 myoblasts stably expressing control FLAG vector or FLAG-Rab5aSSSN mutant were processed as in a. e C2C12 myoblasts stably expressing control FLAG vector, FLAG-Rab5a, or FLAG-Rab5aSSSN were cultured in DM for 3 days. Bar: 100 μm. Myoblast differentiation was quantified by calculating the ratio of nuclei within MHC-positive myotubes. Data are presented as mean ± SD. *P < 0.05, (Student’s t-test). f C2C12 myoblasts stably expressing control FLAG vector, FLAG-Rab5a, or FLAG-Rab5aSSSN were cultured in DM for 3 days. Lysates were western blotted with indicated antibodies.
Rab5a can target to membrane in a prenylation-dependent manner [45]. We prepared a Rab5aSSSN nonprenylatable mutant, which failed to target to membrane [45], to determine if Rab5a directly targets IRS1 to the intracellular membrane. As shown in Fig. 6d, Rab5aSSSN mutant increased the cytosolic IRS1 and reduced the IRS1 levels in HSP fraction. Consistently, Rab5aSSSN mutant failed to enhance myoblast differentiation (Fig. 6e, f; Supplementary Fig. S6A). We also examined the cellular localization of endogenous IRS-1 with Rab5a wildtype or mutants. As shown in Fig. S6B, endogenous IRS1 displayed punctate structures which partially colocalized with the endosomal wildtype Rab5a puncta. In contrast, few colocalization was observed between IRS1 puncta and Rab5a DN or Rab5aSSSN mutants (Supplementary Fig. S6B). Moreover, co-transfection of Rab5a DN or Rab5aSSSN led to more diffusive distribution of IRS1 (Supplementary Fig. S6B). All these above results indicate that Rab5a coordinates the intracellular membrane localization of IRS1.
Rab5a-IRS1 interaction is essential for AKT-mTOR signaling during myoblast differentiation and muscle regeneration
IGF-1-activated IRS1 coordinates muscle growth via activation of PI3K-AKT/PKB-mTOR pathway [46]. We therefore investigated if Rab5a regulates AKT-mTOR signaling during myoblast differentiation. Overexpression of HA-AKT restored MyoG and MHC expression and rescued defective differentiation in Rab5a-depleted myoblasts (Fig. 7a, b; Supplementary Fig. S7A). The differentiating C2C12 myoblasts with increased levels of endogenous Rab5 contained elevated mTOR (Ser2448) and S6 phosphorylation (Fig. 7c; Supplementary Fig. S7B). In line with this, depletion of Rab5a reduced phosphorylation of AKT-mTOR signaling proteins in differentiating myoblasts (Fig. 7d). Furthermore, the AKT-mTOR signaling was suppressed by the Rab5a DN mutant (Supplementary Fig. S7C) and enhanced by stably overexpression of Rab5a (Fig. 7e). Consistently, IGF-1-stimulated or IRS1-enhanced activation of AKT-mTOR signaling was diminished in Rab5a-depleted myoblasts (Fig. 7f, g; Supplementary Fig. S7D). In contrast to wildtype IRS1, either IRS12RA or Rab5a3YA mutant failed to induce AKT-mTOR activation (Fig. 7h, i), indicating that IRS1-Rab5a interaction is essential for AKT-mTOR axis.
a C2C12 myoblasts transiently transfected with HA vector or HA-AKT were transfected with control or Rab5a siRNA sequence. These cells were induced to differentiate for 2 days followed by Western blot analysis with the indicated antibodies. b C2C12 myoblasts transiently transfected with HA vector or HA-AKT were transfected with control or Rab5a siRNA sequence. The cells were then induced to differentiate for 3 days. Bar: 50 μm. Myoblast differentiation was quantified by calculating the ratio of nuclei within MHC-positive myotubes. Data are presented as mean ± SD. **P < 0.01 (Student’s t-test). c C2C12 myoblasts that were transferred to DM for 1.5 day were fixed and stained for endogenous Rab5 and pmTOR (Ser2448). Bar: 50 μm. d Lysates of C2C12 myoblasts transfected with control or Rab5a siRNAs and cultured for 48 h in DM were western blotted with anti-Rab5, pAKT (S473 and T308), pmTOR (S2481 and S2448), and other indicated AKT-mTOR signaling protein antibodies. e Lysates of C2C12 myoblasts stably transfected with control or FLAG-Rab5a and cultured for 48 h in DM were western blotted with indicated AKT-mTOR signaling protein antibodies. f C2C12 myoblasts transfected with control or Rab5a siRNAs were treated with or without 100 ng/ml IGF-1 for 15 min. Lysates were western blotted with indicated AKT-mTOR signaling protein antibodies. g C2C12 myoblasts stably expressing HA vector or HA-IRS1 were transfected with control or Rab5a siRNAs and induced for differentiation for 3 days. Lysates were western blotted with indicated AKT-mTOR signaling protein antibodies. h Lysates of C2C12 myoblasts stably transfected with control, FLAG-IRS1, or FLAG-IRS12RA plasmid and cultured for 48 h in DM were western blotted with indicated AKT-mTOR signaling protein antibodies. i Lysates of C2C12 myoblasts stably transfected with HA control, HA-Rab5a, or HA-Rab5a3YA plasmid and cultured for 48 h in DM were western blotted with indicated AKT-mTOR signaling protein antibodies. j TA muscles from Rab5amKO or Rab5af/f mice were injected with PBS or CTX for 3 days and subjected to western blot analysis with anti-Rab5a or indicated AKT-mTOR signaling protein antibodies (n = 3 mice for each group). k Schematic model for Rab5a-mediated IRS1 activation to trigger myoblast differentiation during muscle regeneration. We found that IGF-1 activates Rab5a in myoblasts in response to muscle injury. Rab5a directly binds and targets IRS1 to intracellular membrane where IGFR interacts with and phosphorylates IRS1. The downstream AKT-mTOR signaling is therefrom activated to promote myoblast differentiation for muscle regeneration.
We next tested if Rab5a modulates AKT-mTOR axis in vivo. Three days after CTX injection, the activation of AKT, S6K, and S6 was significantly elevated in regenerating Rab5af/f mice muscles (Fig. 7j; lane 1 versus 3; Supplementary Fig. S8). However, the injury-induced upregulation of AKT-mTOR signaling was inhibited in Rab5amKO mice (Fig. 7j; lane 3 versus 4; Supplementary Fig. S8). Thus, we concluded that Rab5a interacts with IRS1 to regulate AKT-mTOR signaling during myogenesis in vivo and in vitro.
Discussion
Whilst extracellular signals are triggered on the plasma membrane, the following aspects of signal extension and propagation have been seen to be dependent on endosomes and trafficking proteins [47, 48]. Rab5, in particular, is known to participate in the spatial-temporal modulation of various signaling pathways [47]. This includes Rab5-mediated APPL translocation regulating chromatin structure and gene expression [40] and Rab5-activated Rac being delivered to the plasma membrane during cell migration [49], to give just two examples. We previously revealed that Rab5 also colocalizes with BNIP-2, which promotes p38MAPK signaling during myoblast differentiation [27]. However, the exact role of Rab5 in mTOR activation has remained elusive, despite previous in vitro analysis in both PC3 and 293 A cells demonstrating that the overexpression of either the constitutive active mutant or the DN mutant of Rab5 led to inhibited mTORC1 activity [50,51,52]. Here, we have provided substantial evidence demonstrating that Rab5a enhances the AKT-mTOR axis to promote myoblast differentiation (Fig. 7a–g). This was strongly supported by our in vivo study showing that Rab5a deficiency in skeletal muscle inhibited the AKT-mTOR axis and subsequently led to impaired muscle regeneration (Figs. 2 and 7j). To our knowledge, this is the first description of a rab5a ablation mouse. Correspondingly, knockdown of Rab5 in the liver has been shown to lead to metabolic abnormalities [26], highlighting the key roles of Rab5 in metabolic regulation. Given that insulin signaling shares the same downstream components with the IGF pathway [53, 54], our study raises the possibility that Rab5a might play a role in coordinating insulin sensitivity and related actions in skeletal muscles. Further investigation on this issue is now underway.
As a critical node in Insulin/IGF signaling, the activation of AKT needs to be tightly regulated [7, 9, 10, 55]. Evidence for a role of Rab5 in the control of AKT-mTOR activation emerged from the observation of PI3K as an effector of Rab5 [56,57,58]. Likewise, knockdown of Rab5 in NIH3T3/hIR cells inhibited insulin-stimulated Akt activation, but not tyrosine phosphorylation of IRS1, via the impairment of the binding of IRS1 to the p85 subunit of PI3K [59]. These studies suggest that PI3K is a target for Rab5-dependent regulation of AKT. In addition, another Rab5 effector, APPL1 [40, 41], has also been recognized as regulating AKT activation through its interaction with IRS1 [60]. Here, we demonstrated that, in addition to regulating the IRS1 downstream signaling proteins, Rab5a also directly targets and activates IRS1 to elicit stimulatory effects on IGF-evoked AKT activation. In the current paper, several lines of evidence have been presented. Firstly, the knockdown of Rab5a not only reduced the tyrosine phosphorylation of IRS1 (Fig. 5a), but also inhibited IRS1-activated myogenesis (Fig. 5b, c) and AKT-mTOR signaling (Fig. 7g). Secondly, Rab5a determined the appearance of IRS1 in the HSP fraction (Fig. 6), which represents the active fraction of IRS1 [42]. Moreover, Rab5a modulates the association of IRS1 and IGFR (Fig. 5f–h), which is the key event relating to IGF function. Importantly, both Rab5a3YA and IRS12RA mutants, having lost their association with IRS and Rab5a respectively (Fig. 4e, f), fail to enhance AKT-mTOR signaling and myogenesis (Figs. 4, 5 and 7; Supplementary Fig. S8). Therefore, our findings, together with those of previous work [40, 41, 56], indicate that Rab5a coordinates different effectors to elicit cooperative roles in the regulation of AKT-mTOR signaling. Our study thus highlights the pivotal roles of Rab5 as a “critical node” in governing IGF-evoked AKT-mTOR signaling during myogenesis (Fig. 7k).
The inhibition of IRS1 contributes to reduced insulin/IGF1 responsiveness and type 2 diabetes [38, 61]. The localization of IRS1 to intracellular membrane compartments is a pivotal event for insulin/IGF action [43]. However, the underlying molecular mechanism is not well understood since no transmembrane domain has been identified in IRS1. It has been reported that the membrane skeleton [62] and the adaptor protein (AP) complexes [44] are involved in this process, but the detailed mechanism remains elusive. In this study, we revealed a previously unappreciated role of Rab5a in promoting the intracellular membrane targeting of IRS1 by altering the expression of Rab5a (Fig. 6). Moreover, the nonprenylatable Rab5aSSSN mutant, led to an impairment of the membrane compartment of IRS1 and a failure to promote myoblast differentiation (Fig. 6d–f). These results place Rab5a as a determinant for the spatial regulation of IRS1.
Materials and methods
Mice and animal care
All the mouse strains were obtained from Jackson Laboratory (Bar Harbor, ME) under these stock numbers: #014140 (MyoDCre, Myod1tm2.1(icre)Glh) and #022387 (Rab5aflox/flox, Rab5atm1b(KOMP)Wtsi). PCR genotyping was done by using protocols described by the supplier. Male mice with same age and without stress or discomfort signs (including stereotyped behaviors, hair loss) were employed to minimize physiological variability and to reduce S.D. from mean. In every experiment, at least three mice were analyzed for each time point after CTX-injection and were sufficient to obtain statistical differences. Animal experiments were blinded using number codes till the final data analyses were performed. All mice used in this study had a C57BL/6J genetic background and were housed in a pathogen-free facility in the University Laboratory Animal Center. All animal experiment protocols were approved by the Review Committee of Zhejiang University School of Medicine and were in compliance with ethical regulations and institutional guidelines.
Culture and maintenance of cells
C2C12 cell line and HEK293T cell line were from American Type Culture Collection, USA. Cell lines were authenticated by examination of morphology and growth characteristics and tested for mycoplasma contamination by the standard PCR method. C2C12 cells were grown in DMEM (high glucose) supplemented with 15% (v/v) fetal bovine serum and Human 293T cells were grown in RPMI-1640 medium supplemented with 10% (v/v) fetal bovine serum, 2 mM l-glutamine, 100 U/ml penicillin and 100 mg/ml streptomycin (all from Hyclone Laboratories, Logan, UT).
Primary myoblasts were isolated from skeletal muscles of 1-week-old mice as described before [63]. Briefly, total hindlimb muscles were minced and digested with 5 mg/ml type IV collagenase (Life Technologies, Carlsbad, CA, USA) and 1.4 mg/ml dispase II (Life Technologies) for 1 h, and cell suspensions were filtered through 70 and 40 µm cell strainer, respectively, then pre-plated for 3 h. Non-adherent cells were centrifuged and cultured on Collagen-coated plates in F10 medium (Life Technologies) supplemented with 20% FBS and basic fibroblast growth factor (bFGF) (Life Technologies, 2.5 ng/ml). After removing fibroblasts by pre-plating, primary myoblast cells were cultured in F10/DMEM medium (1:1) supplemented with 20% FBS and bFGF.
Myoblast differentiation
To induce differentiation of C2C12 cells, cultures were transferred from DMEM containing 15% FBS (GM) to DMEM containing 2% horse serum (DM). The myogenic differentiation was quantified as the ratio of the nuclei within myotubes and MHC-positive mononucleated cells.
Stable overexpression studies in myoblasts
For stable overexpression studies in myoblasts, pXJ40 vectors encoding FLAG-tagged plasmids were cotransfected with pBabePuro into C2C12 myoblasts with lipofectamine 2000 (Life Technologies), and cultures were selected in puromycin-containing medium. Drug-resistant cells were pooled and analyzed. Multiple such pools were studied in each case.
Antibody
Antibodies from Santa Cruz Biotechnology were the following: anti-Myogenin (sc-576), anti-MyoD (sc-32758), anti-AKT (sc-8312), anti-RabGDI (sc-374649), anti-Rab5 (sc-46692). Antibodies from Cell Signaling Technology (Danvers, MA) were the following: anti-pAKT (S473) (#4060), anti-pAKT (T308) (#2965), anti-mTOR (#2983), anti-pmTOR (S2448) (#5536), anti-pmTOR (S2481) (#2974), anti-P70S6K (#2708), anti-pP70S6K (T389) (#9234), anti-S6 (#2217), anti-pS6 (Ser235/236) (#4858), anti-PI3K (P85) (#4257), anti-PI3K (P110β) (#3011), anti-IGFRβ (#9750), anti-IGFRβ (Y1135/1136) (#3024), anti-IRS1 (#3407), anti-pIRS1 (Y895) (#3070), anti-pIRS1 (S612) (#3023), anti-pIRS1 (S318) (#5610), anti-APPL1 (#3858), anti-FOXO3a (#12829), anti-pFOXO3a (S253) (#13129), anti-Rab5 (#3547), anti-Rab7 (#9367), anti-Rab9 (#5118), anti-Rab11 (#2413), anti-Desmin (#5332), anti-EEA1 (#3288), anti-Caveolin (#3267), anti-Clathrin (#4796). Antibodies from Developmental Studies Hybridoma Bank were the following: anti-MHC (MF-20), anti-myogenin (F5D), anti-Pax7 (Pax7-s), anti-embryonic MHC (F1.652.b). Other antibodies were the following: anti-FLAG epitope (Sigma-Aldrich, F7425), anti-HA epitope (HuaAn Biotechnology, 0906-1), anti-Tubulin (HuaAn Biotechnology, M1305-2), anti-Hsc70 (Stressgen, SPA815), anti-GAPDH (Home Made), anti-Dystrophin (Abcam, ab15277).
R5BD pulldown (Rab5 activation) assay
The Rab5-binding domain (R5BD, residues 739–862) of Rabaptin5 was cloned into the pGEX4T1 vector (Amersham Pharmacia Biosciences, Piscataway, NJ). The constructs were subsequently transformed in to Escherichia coli. BL21 cells for expression and purified by glutathione-sepharose 4B beads (Amersham Pharmacia Biosciences). Subsequently GST control or GST-R5BD beads were incubated with whole cell lysate at 4 °C for 2–3 h, finally proceed to western blot using anti-Rab5 antibody.
Direct binding assay
293 T lysates transfected with FLAG-tagged expression plasmids were subjected to immunoprecipitation with anti-FLAG M2 affinity gel. The desired proteins were eluted from the beads with 200 ng/μl FLAG peptide. Subsequently GST control or GST fusion protein-beads were incubated with eluted FLAG-tagged proteins at 4 °C for 2–3 h in binding buffer (200 mM sodium chloride, 50 mM Tris, pH 7.3, 0.25 mM EDTA, 1% (w/v) sodium deoxycholate, 1% (v/v) Triton X-100, 0.2% sodium fluoride, 0.1% sodium orthovanadate, and a mixture of protease inhibitors from Roche Applied Science, Indianapolis, IN). The bound proteins were separated by SDS-PAGE for Western blot using anti-FLAG antibody.
In vitro GTP/GDP loading and binding assay
As previously described [64], a total of 10 μg of GST-Rab5a fusion protein were preloaded with 5 mM GTPγS or GDP in binding buffer (20 mM Tris-HCl, pH 7.5, 25 mM NaCl, 10 mM EDTA, and 0.1 mM DTT) at 30 °C for 20 min. The reaction was stopped with MgCl2 to a final concentration of 10 mM. The beads were incubated with cell lysates at 4 °C in lysis buffer (150 mM NaCl, 50 mM HEPES pH 7.4, 1.5 mM MgCl2, 10% glycerol, 5 mM EGTA, 1% Triton X-100, 5 mM sodium orthovanadate, and a mixture of protease inhibitors). Samples were analyzed by Western blotting.
Identification of Rab5a interacting partners by mass spectrometry analysis
Cells were lysed in lysis buffer (50 mM HEPES, pH 7.4, 150 mM sodium chloride, 1.5 mM magnesium chloride, 5 mM EDTA, 10% (vol/vol) glycerol, 1% (vol/vol) Triton X-100, a mixture of protease inhibitors (Roche Diagnostics)), 5 mM sodium orthovanadate, and 25 mM glycerol phosphate (Sigma-Aldrich, St. Louis, MO). The glutathione S-transferase (GST)-Rab5a proteins, coupled to glutathione beads, were incubated with precleared cell lysates for 4 h at 4 °C. The bound proteins were resolved by SDS-PAGE and were visualized by silver-staining. The gel lanes were excised and cut from top to bottom into small pieces and subjected to mass spectrometry analysis.
Immunoprecipitation studies and western blot analyses
Control cells or cells transfected with expression plasmids were lysed in lysis buffer (150 mM sodium chloride, 50 mM Tris, pH 7.3, 0.25 mM EDTA, 1% (w/v) sodium deoxycholate, 1% (v/v) Triton X-100, 0.2% sodium fluoride, 0.1% sodium orthovanadate, and a mixture of protease inhibitors from Roche Applied Science). Lysates were immunoprecipitated (IP) with anti-FLAG M2 beads (Sigma-Aldrich, St. Louis, MO) or protein A/G agarose. Samples were run in SDS/PAGE gels and analyzed by Western blotting with anti-FLAG or indicated antibodies.
Immunofluorescence and direct fluorescence studies
Cells were seeded on coverslips in a six-well plate and transfected with various expression constructs for 24–36 h and then stained for immunofluorescence detection using confocal fluorescence microscopy or directly visualized for cells expressing GFP-tagged proteins as previously described [65]. FLAG-tagged proteins were detected with monoclonal anti-FLAG followed by Texas Reds or FITCs dye-conjugated goat anti-mouse IgG (Life Technologies). Filamentous actin was detected by rhodamine-phalloidin (Life Technologies). The images were collected with a 63 × 1.4 NA or 20 × objective lens using appropriate laser excitation on a LSM880 Meta laser-scanning confocal microscope (Carl Zeiss, Germany) or an Olympus FV3000 OSR laser-scanning confocal microscope. The detector gain was first optimized by sampling various regions of the coverslip and then fixed for each specified channel. Once set, the detector gain value was kept constant throughout the image acquisition process. Images were analyzed with Zeiss LSM Image Examiner Software or FV3000-FV31S-SW 2.1 Viewer.
Subcellular fractionation
Cells were washed with ice-cold PBS and homogenized in buffer (0.5 mM EDTA, 10 mM Tris (pH 7.4), 25 mM sucrose, supplemented with protease and phosphatase inhibitors) by passing through a 27.5 G needle 15 times. The homogenate was centrifuged for 10 min at 1000g at 4 °C to obtain LSP and the supernatant was recentrifuged for 1 hr at 200,000 g at 4 °C to acquire cytosol and HSP fractions. The HSP and LSP were resuspended with IP buffer (20 mM Tris (pH 7.5), 1 mM EDTA, 150 mM NaCl, 1% Triton X-100, 1 mM EGTA) and RIPA, respectively.
Evans blue staining
Evans blue dye (EBD) was dissolved in PBS (150 mM NaCl, 10 mM phosphate buffer, pH 7.4) and sterilized by passage through membrane filters with a 0.2μm pore size. The concentration of the injected dye was 0.5 mg EBD/0.05 ml PBS. Animals were injected with 50 μl of this solution per 10 g body weight. 24 h prior to harvesting muscle, the mice were given intraperitoneally injected with Evans blue dye, which accumulates in damaged muscle fibers and facilitates visualization of the nonregenerated myofibers to assess muscle damage.
RNA interference
C2C12 myoblasts at 30–40% confluency were transfected with 100 nM of siRNA (Ribobio company, Guangzhou, China) using Lipofectamine RNAiMAX (Life Technologies) according to the manufacturer’s protocol.
Gene | Sequence (5′ → 3′) |
|---|---|
siRNA Rab5a#1 | GCCAAAUACUGGAAAUAAA |
siRNA Rab5a#2 | GCCAAAUACUGGAAAUAAA |
siRNA RabGDI1#1 | GAACAUACAUGCUGAACAA |
siRNA RabGDI1#2 | CCAAAUUCCUCAUGGCCAA |
Control siRNA | UUCUCCGAACGUGUCACGU |
Quantitative RT-qPCR
Total RNA was isolated with RNeasy Kit (Qiagen, Germany). Reverse transcription was performed with SuperScript III reverse transcriptase kit (Life Technologies). qPCR reactions using KAPA SYBR FAST qPCR MasterMix kit (kapabiosystems, Wilmington, MA) were performed in triplicate. PCR was performed in triplicate for each gene and three independent samples were prepared under the same conditions. The values were calculated with the second derivative method and normalized to actin expression. The relative levels of mRNAs were calculated according to the 2−(▵▵Ct) equation. All quantitative results are presented as mean ± SD. Statistical significance was set at P<0.05. Asterisks indicated as follows: *P < 0.05, **P < 0.01, and ***P < 0.001.
Gene | Primer pair used | Accession no. |
|---|---|---|
Myf5 | Forward: 5′-ACAGCAGCTTGACAGCATC-3′ Reverse: 5′-AGCTGGACACGGAGCTTTTA-3′ | NM_008656.3 |
MyoD | Forward: 5′-TTCTTCACCACACCTCTGACA-3′ Reverse: 5′-GCCGTGAGAGTCGTCTTAACTT-3′ | NM_010866.1 |
MyoG | Forward: 5′-CTGGGGACCCCTGAGCATTG-3′ Reverse: 5-ATCGCGCTCCTCCTGGTTGA-3′ | NM_031189.2 |
MHC | Forward: 5′-CCAAGGGCCTGAATGAGGAG-3′ Reverse: 5′-GCAAAGGCTCCAGGTCTGAG-3′ | NM_080728.2 |
Rab5a | Forward: 5′-GCTAATCGAGGAGCAACAAGAC-3′ Reverse: 5′-CCAGGCTTGATTTGCCAACAG-3′ | NM_025887.4 |
Actin | Forward: 5′-ATGCTCCCCGGGCTGTAT-3′ Reverse: 5′-CATAGGAGTCCTTCTGACCCATTC-3′ | NM_007393.1 |
Pax7 | Forward: 5′-TTGGGGAACACTCCGCTGTGC-3′ Reverse: 5′-CAGGGCTTGGGAAGGGTTGGC-3′ | NM_011039 |
Muscle injury and regeneration analysis
To induce muscle regeneration, Rab5amKO and control Rab5af/f mice (randomly selected, eight-week-old, male) were anesthetized using pentobarbital sodium solution and cardiotoxin (CTX) was injected (50 μl of 10 μM solution) into tibialis anterior (TA) muscles. Muscles were then harvested at different time points post injection. To assess the completion of regeneration and repair, isolated TA muscles were frozen in liquid nitrogen, then frozen in Tissue-Tek CRYO-OCT compound (Fisher Scientific) and cut in cryostat transverse sections of 8 μm as previously described [66]. H&E staining of muscle cryosections was performed according to procedures provided by the Hematoxylin and Eosin Staining Kit (Beyotime Institute of Biology, Suzhou, China). For immunofluorescence studies, muscle cryosections were fixed with 4% paraformaldehyde, after washing with PBS, antigen retrieval was performed by heating slides in citrate buffer (1.8 mM citric acid and 8.2 mM sodium citrate) at 70 °C for one hour and incubated in blocking buffer (5% goat serum for 1 h) followed by incubation with the primary antibodies overnight. After washing with PBS, the samples were incubated with Flour Alexa 488- and 546-conjugated secondary antibody (1:200) for one hour at room temperature.
Statistical analysis
Statistical analyses were performed with GraphPad Prism 8.0.1 (GraphPad Software). Values are presented as mean ± SD of three independent experiments as shown in each legend. For the comparisons between two groups, unpaired student’s two-tailed t-tests were used. The statistically significant differences were considered at P < 0.05. *P < 0.05; **P < 0.01; ***P < 0.001.
References
Egerman MA, Glass DJ. Signaling pathways controlling skeletal muscle mass. Crit Rev Biochem Mol. 2014;49:59–68.
Izumiya Y, Hopkins T, Morris C, Sato K, Zeng L, Viereck J, et al. Fast/Glycolytic muscle fiber growth reduces fat mass and improves metabolic parameters in obese mice. Cell Metab. 2008;7:159–72.
Buckingham M. Myogenic progenitor cells and skeletal myogenesis in vertebrates. Curr Opin Genet Dev. 2006;16:525–32.
Bentzinger CF, Wang YX, Rudnicki MA. Building muscle: molecular regulation of myogenesis. Cold Spring Harbor Perspect Biol. 2012;4:a008342.
Abmayr SM, Pavlath GK. Myoblast fusion: lessons from flies and mice. Development. 2012;139:641–56.
Tedesco FS, Dellavalle A, Diaz-Manera J, Messina G, Cossu G. Repairing skeletal muscle: regenerative potential of skeletal muscle stem cells. J Clin Investig. 2010;120:11–9.
Tureckova J, Wilson EM, Cappalonga JL, Rotwein P. Insulin-like growth factor-mediated muscle differentiation: collaboration between phosphatidylinositol 3-kinase-Akt-signaling pathways and myogenin. J Biol Chem. 2001;276:39264–70.
Hribal ML, Nakae J, Kitamura T, Shutter JR, Accili D. Regulation of insulin-like growth factor-dependent myoblast differentiation by Foxo forkhead transcription factors. J cell Biol. 2003;162:535–41.
Taniguchi CM, Emanuelli B, Kahn CR. Critical nodes in signalling pathways: insights into insulin action. Nat Rev Mol Cell Biol. 2006;7:85–96.
Bodine SC, Stitt TN, Gonzalez M, Kline WO, Stover GL, Bauerlein R, et al. Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nat cell Biol. 2001;3:1014–9.
Ohanna M, Sobering AK, Lapointe T, Lorenzo L, Praud C, Petroulakis E, et al. Atrophy of S6K1(-/-) skeletal muscle cells reveals distinct mTOR effectors for cell cycle and size control. Nat cell Biol. 2005;7:286–94.
Risson V, Mazelin L, Roceri M, Sanchez H, Moncollin V, Corneloup C, et al. Muscle inactivation of mTOR causes metabolic and dystrophin defects leading to severe myopathy. J Cell Biol. 2009;187:859–74.
Ge Y, Chen J. Mammalian target of rapamycin (mTOR) signaling network in skeletal myogenesis. J Biol Chem. 2012;287:43928–35.
Boura-Halfon S, Zick Y. Phosphorylation of IRS proteins, insulin action, and insulin resistance. Am J Physiol Endocrinol Metab. 2009;296:E581–91.
Copps KD, White MF. Regulation of insulin sensitivity by serine/threonine phosphorylation of insulin receptor substrate proteins IRS1 and IRS2. Diabetologia. 2012;55:2565–82.
Yoneyama Y, Lanzerstorfer P, Niwa H, Umehara T, Shibano T, Yokoyama S, et al. IRS-1 acts as an endocytic regulator of IGF-I receptor to facilitate sustained IGF signaling. eLife. 2018;7:e32893.
Wandinger-Ness A, Zerial M. Rab proteins and the compartmentalization of the endosomal system. Cold Spring Harb Perspect Biol. 2014;6:a022616.
Zhen Y, Stenmark H. Cellular functions of Rab GTPases at a glance. J Cell Sci. 2015;128:3171–6.
Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Bio. 2009;10:513–25.
Zeigerer A, Gilleron J, Bogorad RL, Marsico G, Nonaka H, Seifert S, et al. Rab5 is necessary for the biogenesis of the endolysosomal system in vivo. Nature. 2012;485:465–70.
Zhou F, Zou SS, Chen Y, Lipatova Z, Sun D, Zhu XL, et al. A Rab5 GTPase module is important for autophagosome closure. Plos Genet. 2017;13:e1007020.
Capalbo L, D’Avino PP, Archambault V, Glover DM. Rab5 GTPase controls chromosome alignment through Lamin disassembly and relocation of the NuMA-like protein Mud to the poles during mitosis. Proc Natl Acad Sci USA. 2011;108:17343–8.
Serio G, Margaria V, Jensen S, Oldani A, Bartek J, Bussolino F, et al. Small GTPase Rab5 participates in chromosome congression and regulates localization of the centromere- associated protein CENP-F to kinetochores. Proc Natl Acad Sci USA. 2011;108:17337–42.
Schnatwinkel C, Christoforidis S, Lindsay MR, Uttenweiler-Joseph S, Wilm M, Parton RG, et al. The Rab5 effector rabankyrin-5 regulates and coordinates different endocytic mechanisms. PLoS Biol. 2004;2:1363–80.
Hsu F, Spannl S, Ferguson C, Hyman AA, Parton RG, Zerial M. Rab5 and Alsin regulate stress-activated cytoprotective signaling on mitochondria. eLife. 2018;7:e32282.
Zeigerer A, Bogorad RL, Sharma K, Gilleron J, Seifert S, Sales S, et al. Regulation of liver metabolism by the endosomal GTPase Rab5. Cell Rep. 2015;11:884–92.
Yi P, Chew LL, Zhang Z, Ren H, Wang F, Cong X, et al. KIF5B transports BNIP-2 to regulate p38 mitogen-activated protein kinase activation and myoblast differentiation. Mol Biol Cell. 2015;26:29–42.
Nowak SJ, Nahirney PC, Hadjantonakis AK, Baylies MK. Nap1-mediated actin remodeling is essential for mammalian myoblast fusion. J Cell Sci. 2009;122(Pt 18):3282–93.
Liu J, Lamb D, Chou MM, Liu YJ, Li G. Nerve growth factor-mediated neurite outgrowth via regulation of Rab5. Mol Biol Cell. 2007;18:1375–84.
Engert JC, Berglund EB, Rosenthal N. Proliferation precedes differentiation in IGF-I-stimulated myogenesis. J Cell Biol. 1996;135:431–40.
Nowak KJ, Davies KE. Duchenne muscular dystrophy and dystrophin: pathogenesis and opportunities for treatment. EMBO Rep. 2004;5:872–6.
Charge SB, Rudnicki MA. Cellular and molecular regulation of muscle regeneration. Physiological Rev. 2004;84:209–38.
Fan W, Gao XK, Rao XS, Shi YP, Liu XC, Wang FY, et al. Hsp70 interacts with mitogen-activated protein kinase (MAPK)-activated protein kinase 2 to regulate p38MAPK stability and myoblast differentiation during skeletal muscle regeneration. Mol Cell Biol. 2018;38:e00211–18.
Kanisicak O, Mendez JJ, Yamamoto S, Yamamoto M, Goldhamer DJ. Progenitors of skeletal muscle satellite cells express the muscle determination gene, MyoD. Developmental Biol. 2009;332:131–41.
Goldfarb LG, Dalakas MC. Tragedy in a heartbeat: malfunctioning desmin causes skeletal and cardiac muscle disease. J Clin Investig. 2009;119:1806–13.
Hamer PW, McGeachie JM, Davies MJ, Grounds MD. Evans Blue Dye as an in vivo marker of myofibre damage: optimising parameters for detecting initial myofibre membrane permeability. J Anat. 2002;200(Pt 1):69–79.
Audhya A, Desai A, Oegema K. A role for Rab5 in structuring the endoplasmic reticulum. J Cell Biol. 2007;178:43–56.
White MF. IRS proteins and the common path to diabetes. Am J Physiol-Endoc M. 2002;283:E413–22.
Uhlik MT, Temple B, Bencharit S, Kimple AJ, Siderovski DP, Johnson GL. Structural and evolutionary division of phosphotyrosine binding (PTB) domains. J Mol Biol. 2005;345:1–20.
Miaczynska M, Christoforidis S, Giner A, Shevchenko A, Uttenweiler-Joseph S, Habermann B, et al. APPL proteins link Rab5 to nuclear signal transduction via an endosomal compartment. Cell. 2004;116:445–56.
Schenck A, Goto-Silva L, Collinet C, Rhinn M, Giner A, Habermann B, et al. The endosomal protein Appl1 mediates Akt substrate specificity and cell survival in vertebrate development. Cell. 2008;133:486–97.
Shah OJ, Hunter T. Turnover of the active fraction of IRS1 involves Raptor-mTOR- and S6K1-dependent serine phosphorylation in cell culture models of tuberous sclerosis. Mol Cell Biol. 2006;26:6425–34.
Hakuno F, Fukushima T, Yoneyama Y, Kamei H, Ozoe A, Yoshihara H, et al. The novel functions of high-molecular-mass complexes containing insulin receptor substrates in mediation and modulation of insulin-like activities: emerging concept of diverse functions by IRS-associated proteins. Front Endocrinol. 2015;6:73.
Yoneyama Y, Matsuo M, Take K, Kabuta T, Chida K, Hakuno F, et al. The AP-1 complex regulates intracellular localization of insulin receptor substrate 1, which is required for insulin-like growth factor I-dependent cell proliferation. Mol Cell Biol. 2013;33:1991–2003.
Gomes AQ, Ali BR, Ramalho JS, Godfrey RF, Barral DC, Hume AN, et al. Membrane targeting of Rab GTPases is influenced by the prenylation motif. Mol Biol Cell. 2003;14:1882–99.
Serra C, Palacios D, Mozzetta C, Forcales SV, Morantte I, Ripani M, et al. Functional interdependence at the chromatin level between the MKK6/p38 and IGF1/PI3K/AKT pathways during muscle differentiation. Mol Cell. 2007;28:200–13.
Villasenor R, Kalaidzidis Y, Zerial M. Signal processing by the endosomal system. Curr Opin Cell Biol. 2016;39:53–60.
Platta HW, Stenmark H. Endocytosis and signaling. Curr Opin Cell Biol. 2011;23:393–403.
Palamidessi A, Frittoli E, Garre M, Faretta M, Mione M, Testa I, et al. Endocytic trafficking of Rac is required for the spatial restriction of signaling in cell migration. Cell. 2008;134:135–47.
Li L, Kim E, Yuan H, Inoki K, Goraksha-Hicks P, Schiesher RL, et al. Regulation of mTORC1 by the Rab and Arf GTPases. J Biol Chem. 2010;285:19705–9.
Bridges D, Fisher K, Zolov SN, Xiong T, Inoki K, Weisman LS, et al. Rab5 proteins regulate activation and localization of target of rapamycin complex 1. J Biol Chem. 2012;287:20913–21.
Duran RV, Hall MN. Regulation of TOR by small GTPases. EMBO Rep. 2012;13:121–8.
Boucher J, Tseng YH, Kahn CR. Insulin and insulin-like growth factor-1 receptors act as ligand-specific amplitude modulators of a common pathway regulating gene transcription. J Biol Chem. 2010;285:17235–45.
Cai W, Sakaguchi M, Kleinridders A, Gonzalez-Del Pino G, Dreyfuss JM, O’Neill BT, et al. Domain-dependent effects of insulin and IGF-1 receptors on signalling and gene expression. Nat Commun. 2017;8:14892.
Cong XX, Rao XS, Lin JX, Liu XC, Zhang GA, Gao XK, et al. Activation of AKT-mTOR signaling directs tenogenesis of mesenchymal stem cells. Stem Cells. 2018;36:527–39.
Christoforidis S, Miaczynska M, Ashman K, Wilm M, Zhao L, Yip SC, et al. Phosphatidylinositol-3-OH kinases are Rab5 effectors. Nat Cell Biol. 1999;1:249–52.
Shin HW, Hayashi M, Christoforidis S, Lacas-Gervais S, Hoepfner S, Wenk MR, et al. An enzymatic cascade of Rab5 effectors regulates phosphoinositide turnover in the endocytic pathway. J Cell Biol. 2005;170:607–18.
Lodhi IJ, Bridges D, Chiang SH, Zhang Y, Cheng A, Geletka LM, et al. Insulin stimulates phosphatidylinositol 3-phosphate production via the activation of Rab5. Mol Biol Cell. 2008;19:2718–28.
Su X, Lodhi IJ, Saltiel AR, Stahl PD. Insulin-stimulated Interaction between insulin receptor substrate 1 and p85alpha and activation of protein kinase B/Akt require Rab5. J Biol Chem. 2006;281:27982–90.
Ryu JY, Galan AK, Xin XB, Dong F, Abdul-Ghani MA, Zhou LJ, et al. APPL1 potentiates insulin sensitivity by facilitating the binding of IRS1/2 to the insulin receptor. Cell Rep. 2014;7:1227–38.
Lavin DP, White MF, Brazil DP. IRS proteins and diabetic complications. Diabetologia. 2016;59:2280–91.
Clark SF, Martin S, Carozzi AJ, Hill MM, James DE. Intracellular localization of phosphatidylinositide 3-kinase and insulin receptor substrate-1 in adipocytes: Potential involvement of a membrane skeleton. J Cell Biol. 1998;140:1211–25.
Zhou L, Wang L, Lu L, Jiang P, Sun H, Wang H. A novel target of microRNA-29, Ring1 and YY1-binding protein (Rybp), negatively regulates skeletal myogenesis. J Biol Chem. 2012;287:25255–65.
Zhou YT, Chew LL, Lin SC, Low BC. The BNIP-2 and Cdc42GAP homology (BCH) domain of p50RhoGAP/Cdc42GAP sequesters RhoA from inactivation by the adjacent GTPase-activating protein domain. Mol Biol Cell. 2010;21:3232–46.
Ling Zheng L, Wang FY, Cong XX, Shen Y, Rao XS, Huang DS, et al. Interaction of heat shock protein Cpn10 with the cyclin E/Cdk2 substrate nuclear protein Ataxia-Telangiectasia (NPAT) is involved in regulating histone transcription. J Biol Chem. 2015;290:29290–300.
He MY, Xu SB, Qu ZH, Guo YM, Liu XC, Cong XX, et al. Hsp90 beta interacts with MDM2 to suppress p53-dependent senescence during skeletal muscle regeneration. Aging Cell. 2019;18:e13003.
Acknowledgements
We thank Ms. Yanwei Li, Junli Xuan and Guifeng Xiao from the core facility platform of Zhejiang university school of medicine for their technical support. This work was supported by the National Key R&D Program of China (2018YFA0800403), the National Natural Science Foundation of China (31571402, 31671417, 31671479, 31371476), the International Science & Technology Cooperation Program of China (the Ministry of Science and Technology of PRC, 2015DFG32130), grants from Science and Technology Commission of Zhejiang Province (LZ20H060001), China Postdoctoral Science Foundation (2019M662034).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by R. Johnstone
Rights and permissions
About this article
Cite this article
Cong, X.X., Gao, X.K., Rao, X.S. et al. Rab5a activates IRS1 to coordinate IGF-AKT-mTOR signaling and myoblast differentiation during muscle regeneration. Cell Death Differ 27, 2344–2362 (2020). https://doi.org/10.1038/s41418-020-0508-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-020-0508-1
This article is cited by
-
Adipose-derived stem cells promote the repair of chemotherapy-induced premature ovarian failure by inhibiting granulosa cells apoptosis and senescence
Stem Cell Research & Therapy (2023)
-
VAPB-mediated ER-targeting stabilizes IRS-1 signalosomes to regulate insulin/IGF signaling
Cell Discovery (2023)
-
Phase separation of insulin receptor substrate 1 drives the formation of insulin/IGF-1 signalosomes
Cell Discovery (2022)
-
Identification of prognostic lipid droplet-associated genes in pancreatic cancer patients via bioinformatics analysis
Lipids in Health and Disease (2021)
-
The ESCRT-0 subcomplex component Hrs/Hgs is a master regulator of myogenesis via modulation of signaling and degradation pathways
BMC Biology (2021)