Abstract
TP53 is a critical tumor suppressor that is mutated in approximately 50% of human cancers. Unveiling the downstream target genes of TP53 that fulfill its tumor suppressor function is an area of intense investigation. Zmat3 (also known as Wig-1 or PAG608) is one such downstream target of p53, whose loss in hemopoietic stem cells lacking the apoptosis and cell cycle regulators, Puma and p21, respectively, promotes the development of leukemia. The function of Zmat3 in tumorigenesis however remains unclear. Here, to investigate which oncogenic drivers co-operate with Zmat3 loss to promote neoplastic transformation, we utilized Zmat3 knockout mice in models of c-MYC-driven lymphomagenesis and KrasG12D-driven lung adenocarcinoma development. Interestingly, unlike loss of p53, Zmat3 germline loss had little impact on the rate of tumor development or severity of malignant disease upon either the c-MYC or KrasG12D oncogenic activation. Furthermore, loss of Zmat3 failed to rescue KrasG12D primary lung tumor cells from oncogene-induced senescence. Taken together, we conclude that in the context of c-MYC-driven lymphomagenesis or mutant KrasG12D-driven lung adenocarcinoma development, additional co-occurring mutations are required to resolve Zmat3 tumor suppressive activity.
Similar content being viewed by others
Introduction
P53 is a pivotal suppressor of cancer development and progression. The importance of p53 tumor suppression function and response to anti-cancer agents has driven the development of novel strategies to target the p53 pathway in cancer therapy1. Although the role of p53 in preventing tumor development has provoked intense investigation, the underlying mechanisms crucial in tumor suppression downstream of p53 have remained elusive2,3,4. Cell cycle arrest, cell senescence and apoptosis have been implicated in p53-mediated tumor suppression2,5,6. However, no spontaneous tumors arise in mice lacking Puma, Noxa, and p21, the critical mediators of p53-induced apoptosis and G1/S cell cycle arrest7. Likewise, in several mouse tumor models in different cellular compartments, it has been demonstrated that p53 mutants defective in the induction of apoptosis, cell cycle arrest and cell senescence retain tumor-suppressor activity8,9,10. Collectively, these studies demonstrate that p53 must suppress tumor development through currently underappreciated processes that act apart from, or in addition to, the induction of apoptosis, cell cycle arrest and cell senescence7,11,12,13.
Utilizing an in vivo shRNA screening approach, we recently demonstrated that Zmat3 acts as a tumor suppressor gene downstream of p53, with its loss cooperating with inactivation of cell cycle arrest, cell senescence and apoptosis inducers (PUMA and p21) in the development of hematopoietic malignancy14. Zmat3 has been described as an RNA-binding zinc-finger protein that is involved in post-transcriptional regulation of gene expression and as such is found expressed in a broad range of tissues15. Moreover, akin to being a p53 target gene, Zmat3 has been reported to exert bona fide roles in the control of cell proliferation and cell survival16, where it acts through the regulation of p53 and p21 mRNA17,18,19,20. Interestingly, while both decreased and increased Zmat3 mRNA expression have been reported in human tumors15 the exact role of Zmat3 in tumorigenesis remains elusive. We therefore hypothesized that the function of Zmat3 in tumorigenesis may be context dependent, influenced by genetic background, cell type or oncogenic drivers specific to each tumor. To further investigate the functions of Zmat3 in tumor suppression, we evaluated the impact of its loss in c-MYC-driven lymphomagenesis and mutant KrasG12D-driven non-small cell lung cancer (NSCLC)21, in which the p53 pathway has been shown to play a critical tumor suppressive role22,23,24.
Materials and methods
Mice
Animal experiments were conducted according to the regulatory standards approved by the Animal Ethics Committees of the Walter and Eliza Hall Institute of Medical Research (WEHI) and the Barcelona Biomedical Research Park (PRBB). Eµ-Myc24, p5325,26, KrasLSL-G12D/+ mice21 and Zmat314 mice have been previously described. All animals were maintained on a C57BL/6 background, and equal proportions of males and females were used in all experiments. Seven- to eight- week-old KrasLSL-G12D/+ compound mice were intranasally (i.n.) infected with 20 μL of 1 × 1010 PFU/mL Ad5-CMV-Cre virus (University of Iowa Gene Transfer Core Facility) according to standard procedures27. Lungs were harvested at defined time points (6-, 10- and 16-weeks post i.n. infection) or when mice showed signs of morbidity for Kaplan-Meier survival analysis.
qRT-PCR analysis
Pre-B cells were treated with 5 Gy γ-irradiation or with 10 µM Nutlin3a (Nut3a), in the presence of 25 µM QVD-OPH (Sigma-Aldrich, St. Louis, MO, USA) in culture. WT and Zmat3−/− mice, 2–5 months old, were either exposed to 8 Gy γ-irradiation or left untreated and lung tissue was harvested after 6 h. RNA was extracted using Trizol reagent (Invitrogen, Carlsbad, CA, USA) and converted to cDNA using the SuperScript First Strand Synthesis Kit (Thermo Fisher Scientific). Quantitative RT-PCR was performed using TaqMan probes (Thermo Fisher Scientific) for Zmat3 (Mm01292424_m1), Puma (Mm00519268_m1), p21 (Mm00432448_m1) and analyzed by the ΔΔCT method relative to HMBS (Mm01143545_m1).
Flow cytometry
B lymphoid cells were isolated from bone marrow collected from both femurs and tibias of 4 week-old Eµ-Myc mice and stained with the following antibodies: B220-PE (clone RA3-6B2), IgM-FITC (clone 5.1) and IgD-FITC (clone 11-26C). Viable pre-B cells (B220+IgM-IgD-PI-) were isolated by flow cytometry using an ARIA flow cytometer (Becton Dickinson, San Jose, CA, USA). 1 × 106 cells/mL were cultured in DMEM (Thermo Fisher Scientific, Waltham, MA, USA), 10% FBS (Sigma-Aldrich), 100 U/mL penicillin (Thermo Fisher Scientific), 100 µg/mL streptomycin (Sigma-Aldrich), 50 μM 2-Mercaptoethanol and 100 μM asparagine (Thermo Fisher Scientific) during the time-course. Antibodies were produced in-house.
Immunophenotyping of lymphomas was performed on single-cell suspensions stained with surface-marker-specific antibodies: CD43-BV650 (BD Biosciences; #740464; dilution 1:800); B220-APC-Cy7 (BD Biosciences; #552094; dilution 1:200); IgD-BV510 (BD Biosciences; #563110; dilution 1:200); IgM-FITC (BD Biosciences; #553437; dilution 1:100) to define pro/pre B cells (B220+CD43+) and Ig+ B cells (IgM and IgD). Samples were run on the LSR II (BD Biosciences). Dead cells (PI+) were excluded from analysis.
For immune cell analysis of lung tissue, the left lobe of each lung was weighed and harvested in PBS for digestion to generate a single cell suspension and blocked as described previously28. Blocked cells were incubated with fluorochrome-conjugated antibodies to define key immune cell populations from the lymphoid and myeloid compartments: B cells (CD45+CD3−CD19+), T cells (CD45+CD3+), NK cells (CD45+CD3−NKp46+DX5+), alveolar macrophages (CD45+CD11c+CD103−), CD103 dendritic cells (CD45+CD11c+CD103+), monocytes (CD45+CD11b+Ly6G−SSClo), neutrophils (CD45+CD11b+Ly6G+) and eosinophils (CD45+CD11b+Ly6G−SSChi) as described previously29.
Histology and immunohistochemistry
Lungs were perfused and fixed with 4% paraformaldehyde (Sigma-Aldrich) for 24 h at 4 °C and embedded in paraffin. Sections 2 μm thick were stained with hematoxylin and eosin (H&E), as described previously28. Immunohistochemistry was performed as previously described28, using antibodies against Ki67 (Cell Signaling, Beverly, MA, USA; #12202), Nkx2.1/TTF-1 (Dako/Agilent Technologies, Santa Clara, CA, USA; #M3575) and Hmga2 (BioCheck, Foster City, CA, USA; #59170AP). H&E stained lung sections were imaged at low power and abnormal regions were masked on Image J software (Softonic International, Barcelona, Spain). Abnormal regions were classified as hyperplastic or adenoma/adenocarcinoma (ADC) based on morphology, and lesion number per lung was quantified. Hmga2 and Ki67 stained images were scored automatically using a custom Image J30 pipeline which utilized the Stardist plugin for nuclear segmentation31. Nuclei were then scored based on their intensity in the brown channel after color deconvolution. Tumor burden was determined using Image J through a combination of rank and morphology filters, followed by an auto-threshold to select the whole tissue, and the regions of dense nuclei comprising the tumor.
Cell growth analysis
Cell lines were seeded from a single cell suspension derived from the left lobe of the lung. At each passage, cells were counted and 2 × 105 cells were re-seeded into one well of a 6-well plate in primary cell medium (DMEM/F-12+GlutaMAX (Thermo Fisher Scientific), 10% FBS (Sigma-Aldrich), 100 U/mL penicillin (Thermo Fisher Scientific), 100 μg/mL streptomycin (Thermo Fisher Scientific), 0.04 mg/mL hydrocortisone (Sigma-Aldrich), 1× Insulin-Transferrin-Selenium-Ethanolamine (ITS-X; Thermo Fisher Scientific) and 5 ng/mL epidermal growth factor (Sigma-Aldrich)).
Statistical analysis
Prism (Version 8; GraphPad) software was used for all statistical analyses. Two-group comparisons were made using two-tailed Student’s t-tests assuming equal variances and multivariate comparisons were performed using one-way ANOVA (Kruskal–Wallis test) with Tukey’s multiple comparisons. Animal survival data were plotted using Kaplan–Meier curves and compared using log-rank (Mantel–Cox) test.
Results and discussion
Loss of Zmat3 does not accelerate Myc driven lymphoma development
To interrogate the role of Zmat3 in tumorigenesis in vivo, we investigated the collaboration of loss of Zmat3 with deregulated expression of the oncogene c-Myc, using the Eµ-Myc model of pre-B/B lymphoma development24. Critically, Eμ-Myc mice have been extensively used to study the defects in the p53-pathway (e.g. impact of loss of Puma or loss of one allele of p53) on tumorigenesis22,24,32. First, we examined the expression of Zmat3 in pre-leukemic Eµ-Myc pre-B lymphoid cells isolated from the bone marrow of 4-week old Eµ-Myc mice, with or without activation of p53. Indeed, Zmat3 mRNA expression was induced in pre-leukemic pre-B lymphoid cells in response to both the MDM2 inhibitor Nutlin3a33 and γ-irradiation (Fig. 1a), confirming that Zmat3 is a bona fide p53 target gene. It has been shown that loss of one allele of p53 significantly accelerates lymphoma development in Eμ-Myc mice3,11,24. Therefore, we next investigated whether loss of Zmat3 would accelerate lymphoma development in Eµ-Myc mice. To address this question, Zmat3 knockout (Zmat3−/−) mice14 were crossed with Eµ-Myc transgenic mice24 and monitored for lymphoma development. Strikingly, neither loss of one allele or even both alleles of Zmat3 altered the onset of lymphoma in Eµ-Myc mice (Fig. 1b). Post-mortem analysis demonstrated no significant difference in leukemic burden, the overall proportion of pre-B vs mature B-cell tumors, or in the weight of hematopoietic organs between sick Eµ-Myc;Zmat3−/− vs Eµ-Myc mice (Fig. 1c, d). Taken together, these results reveal that, unlike loss of p53, germline loss of Zmat3 fails to accelerate c-MYC-driven lymphoma development.
a Zmat3 mRNA expression in pre-leukemic Eµ-Myc B lymphoid cells 0, 6, 12 and 24 h following 5 Gy γ-irradiation (IR) or treatment with 10 µM Nutlin3a (Nut3a). Mean ± SEM. b Lymphoma-free survival of Eµ-Myc (n = 23), Eµ-Myc;Zmat3+/− (n = 21) and Eµ-Myc;Zmat3−/− (n = 17) mice. Median lymphoma-free survival: Eµ-Myc: 125 days; Eµ-Myc;Zmat3+/−: 94 days; Eµ-Myc;Zmat3−/−: 93 days. Eµ-Myc vs Eµ-Myc;Zmat3+/− p = 0.0718; Eµ-Myc vs Eµ-Myc;Zmat3−/− p = 0.3231. c The proportions of Ig+ B cell lymphomas and pro-B/pre-B cell lymphomas in sick Eµ-Myc;Zmat3−/− (n = 4) and sick Eµ-Myc (n = 3) mice. No significant differences were observed; p = 0.5 calculated by two-tailed t-test. d Spleen, thymus and lymph node weight (grams) in sick Eµ-Myc (n = 7), Eµ-Myc;Zmat3+/− (n = 15) and Eµ-Myc;Zmat3−/− (n = 9) mice at ethical endpoint. Mean ± SEM.
The impact of Zmat3 loss on adult lung development
To further investigate the role of Zmat3 in oncogene driven tumorigenesis, we chose a lung adenocarcinoma (LUAD) model, in which p53 has been shown to play a critical tumor suppressive role34. In this model, activated KrasG12D drives the development of lung adenomas, which progress to adenocarcinomas in the absence of p5334. First, we examined the expression of Zmat3 in lungs isolated from wt and Zmat3−/− mice, with or without activation of p53 by γ-irradiation. Indeed, Zmat3 mRNA expression was markedly induced in wt lung tissue in response to DNA damage, but absent in the lung tissues from Zmat3−/− mice (Supplementary Fig. 1a). To determine whether the loss of Zmat3 impacted normal development and homeostasis of the lung, we performed an in-depth examination of the cellular composition and immune microenvironment of the lung tissue from Zmat3 deficient mice. As Zmat3−/− mice are viable and healthy14, we aged Zmat3−/− and Zmat3+/+ (wt) littermate controls to adulthood (16 to 18 weeks old) and evaluated the histopathology and cellularity (epithelial and immune) of the lungs of these mice. The lungs of Zmat3−/− mice were histologically indistinguishable from the lungs of p53−/− or wt control animals (Fig. 2a). Moreover, no differences in lung cellularity or weight were observed between Zmat3−/− and controls (Supplementary Fig. 1b, c). Furthermore, the immune cell composition of the lungs was unchanged, with the levels of resident myeloid and lymphoid cell populations comparable between Zmat3−/−, p53−/− and wt mice (Fig. 2b). To further investigate the role of Zmat3 in lung epithelium we generated primary cell cultures from Zmat3−/−, p53−/− and wt adult lungs and examined the impact of the loss of Zmat3 on cell proliferation and cellular morphology in vitro. Cumulative cell counts (over 8 passages) revealed that Zmat3−/− lung epithelial cells, similar to those from wt mice but unlike p53−/− epithelial cells, failed to bypass replicative senescence (Fig. 2c–e). Consequently, similar to wt control cells, Zmat3−/− cells underwent growth arrest exhibiting flattened appearance and multi-nucleation, characteristic of cells undergoing senescence (Fig. 2e). Collectively, these results show that the loss of Zmat3 has no impact on the development, homeostasis or immune cell composition of the murine lung and does not allow lung epithelial cells to evade cellular senescence in culture.
a Representative hematoxylin and eosin (H&E) staining of lungs from Zmat3−/−, littermate Zmat3+/+ (wt) and p53−/− control mice aged 16-18 weeks. Scale, 200 μm. b Flow cytometric analysis of the lymphoid and myeloid cells present in the lungs from Zmat3−/− (n = 4), p53−/− (n = 2) and wt (n = 4) mice. Immune cell populations are normalized to wt control mice. c Growth analysis of primary cell lines derived from the lung epithelium of wt (n = 8), Zmat3−/− (n = 8) and p53−/− (n = 4) mice. Mean ± SEM. d Accumulative cell number following 8 passages of primary wt (n = 8), p53−/− (n = 4) and Zmat3−/− (n = 8) cell lines. Mean ± SD, ****p < 0.0001. e Representative light microscope images of Zmat3+/+ (wt), p53−/− and Zmat3−/− cell lines 8 passages following their derivation. Scale, 10 μm.
Germline loss of Zmat3 does not recapitulate p53-mediated acceleration of KrasG12D-induced tumorigenesis
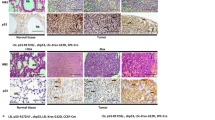
Next, to functionally evaluate the significance of Zmat3 in KRAS-mutant LUAD, we crossed Zmat3−/− mice with KrasLSL-G12D/+ mice21. Cohorts of KrasLSL-G12D/+ (hereafter; K), KrasLSL-G12D/+;Zmat3−/− (hereafter; KZ) mice and KrasLSL-G12D/+;p53−/− (hereafter; KPnull) mice were infected by intranasal inhalation with the ubiquitous Ad5-CMV-Cre adenovirus to recombine and activate mutant KrasG12D expression in the respiratory epithelium27 (Fig. 3a). Consistent with previous reports utilizing a p53 conditional allele21, germline loss of p53 significantly accelerated lung cancer development in KrasG12D mice (median survival K: 151 days vs KPnull: 72 days; p < 0.0001) (Fig. 3b). In contrast, loss of Zmat3 did not significantly alter the survival of KrasG12D mice (median survival: KZ: 138 days versus K: 151 days) (Fig. 3b). Consistent with the animal survival data, we observed no differences in tumor histology between the lungs from the K vs KZ mice, nor the proliferation index, as determined by Ki67 staining (Fig. 3c, Supplementary Fig. 2a). The tumors from both K and KZ mice were defined as highly differentiated, reflected by high Nkx2.1 expression and low to absent expression of Hmga2 (Fig. 3c). In contrast, tumors from KPnull mice displayed increased numbers of Ki67 and Hmga2 positive nuclei, indicative of more aggressive and more poorly differentiated adenocarcinoma lesions (Fig. 3c, Supplementary Fig. 2b), consistent with the crucial tumor suppressive function of p53 in mutant KRAS-driven LUAD.
a Schematic of experimental design. KrasLSL-G12D/+ (K) mice21 were crossed with p53−/− mice37 (KPnull) or Zmat3−/− mice14 (KZ). b Kaplan–Meier tumor-free survival analysis of K (n = 6), KPnull (n = 6) and KZ (n = 12) mice following i.n. infection of Ad5-CMV-Cre virus. c Representative H&E staining and immunohistochemistry of Ki67, Nkx2.1/TTF-1 and Hmga2 in end-stage lung tumors from KrasG12D, KrasG12D;p53−/− and KrasG12D;Zmat3−/− mice. Scale, 100 μm.
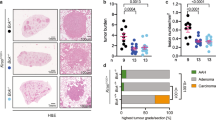
To evaluate whether Zmat3 loss impacts tumor initiation following oncogenic KrasG12D activation, we performed a time course study, whereby K, KZ and KPnull mice were analyzed 6, 10 and 16 weeks following Ad5-CMV-Cre infection. While hyperplasic and adenomatous lesions were only apparent in lungs of KPnull mice 6 weeks following Ad5-CMV-Cre infection (Fig. 4a), sporadic lung lesions were observed in both K and KZ mice by 10 weeks, with further progression seen only in the lungs of KPnull mice (Fig. 4a). Quantification of the histopathology of lesions in KZ, KPnull and K mice 10 weeks following Ad5-CMV-Cre administration failed to reveal significant differences in tumor burden between K and KZ mice suggesting that Zmat3 loss does not accelerate KrasG12D-induced tumorigenesis (Fig. 4b, Supplementary Fig. 2c). These findings were further verified in mice analyzed 16 weeks following Ad5-CMV-Cre infection, with no difference in tumor burden detected between K and KZ mice (Supplementary Fig. 2d).
a Representative low magnification H&E stained lungs of KrasG12D/+ (K), KrasG12D/+;p53−/− (KPnull) and KrasG12D/+;Zmat3−/− (KZ) mice 6- and 10-weeks following i.n. administration of Ad5-CMV-Cre. Scale, 1 mm. b Lesion classification in the lungs of K (n = 4), KPnull (n = 4) and KZ (n = 5) mice 10 weeks following i.n. Ad5-CMV-Cre infection. c Wet weight (mg) of the left lung lobe of K (n = 3), KPnull (n = 3) and KZ (n = 8) mice 6 weeks following i.n. Ad5-CMV-Cre infection. Violin plot indicates the median weight and distribution.
Next, we assessed the tumor immune microenvironment using multiparametric flow cytometry. Consistent with KrasG12D-driven lung adenocarcinoma models, alveolar macrophages were abundant in the microenvironment of all genetic backgrounds tested, suggesting no alterations in this key feature of mutant KrasG12D-driven adenocarcinoma development due to the loss of Zmat335. Furthermore, no differences were identified in the lymphoid or myeloid compartments between the K, KZ and KPnull cohorts (Supplementary Fig. 3a). Consistent with the presence of tumors in the lungs 10-weeks post Ad5-CMV-Cre administration, CD8+ T cells in the lungs of tumor-bearing mice were activated and displayed elevated PD-1 expression (Supplementary Fig. 3b). These findings show that loss of Zmat3 does not alter the key factors shown to impact the immune milieu under homeostatic conditions or in response to oncogene activation.
We have previously shown that lung weight is a robust readout of tumor burden in KrasG12D-driven GEMMs35. Interestingly, while lung weights were consistently in a relatively narrow range within the K (uniformly low lung weight) and KPnull cohorts (uniformly high lung weights), large variations in lung weights were observed between individual KZ mice (Fig. 4c). This may indicate some tumor suppressor function of Zmat3, perhaps dependent on oncogenic lesions cooperating with mutant KrasG12D and the loss of Zmat3 that are present in some but not all KZ mice.
Zmat3 loss fails to rescue KrasG12D lung adenocarcinoma cells from oncogene-induced senescence
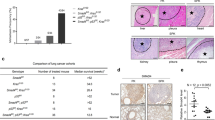
Loss of p53 function prevents cell cycle arrest and enables evasion from cellular senescence in mutant KRAS-driven tumorigenesis. To assess whether loss of Zmat3 could promote evasion from oncogene-induced senescence (OIS) in KrasG12D mutant lung epithelial cells, primary cell lines generated from KZ and KPnull lung tumors were assessed for proliferation over 8 passages. Consistent with previous findings9, cumulative cell counts demonstrated that while KPnull cells replicated indefinitely, and KZ cells underwent growth arrest that was evident after 4 passages (Fig. 5a–c). These results suggest that the loss of Zmat3, unlike the loss of p53, does not permit evasion from OIS in KrasG12D driven lung tumorigenesis.
a Growth analysis of primary cell lines derived from the lung epithelium of KPnull (n = 8) and KZ (n = 12) mice 10 weeks following i.n. Ad5-CMV-Cre infection. Mean ± SEM. b Accumulative cell number following 8 passages of primary KPnull (n = 8) and KZ (n = 12) cell lines. Mean ± SD, ***p < 0.0001. c Representative light microscope images of KrasG12D/p53−/− (KPnull) and KrasG12D/Zmat3−/− (KZ) cell lines 8 passages following their derivation. Scale, 10 μm. d Schematic representation of p53 target genes and their role in tumor suppression.
Taken together, we have demonstrated that in murine models of Myc-driven lymphoma and KrasG12D-driven LUAD, the germline loss of Zmat3 did not have a considerable effect on the rate of onset, overall incidence or severity of tumor development, unlike p53 loss. In humans, it has been shown that Zmat3 is expressed ubiquitously in all tissues and its deregulation has been implicated in various cancers, including leukemia as well as lung and breast cancer15,16,17. More importantly, our previous findings have shown that loss of Zmat3 causes leukemia only in overlap with concomitant loss of the p53 targets, Puma and p2114, that are critical for p53-mediated apoptosis and cell cycle arrest, respectively. Hence, it is possible that the tumor suppressive function of Zmat3 is more pronounced in other tissues and oncogene induced models. The understanding of the molecular mechanisms underlying p53s’ function in tumor suppression is constantly expanding. It is now clear that for p53-mediated tumor suppression other functions in the cell are critical in addition to induction of cell cycle arrest, apoptosis or senescence4,5,6,7,8,9. Zmat3 is a p53 target gene that is involved in activation of cell cycle arrest or apoptosis indirectly, and as it regulates gene expression in a post-translational manner could modulate the p53 response towards other still unknown biological effector processes15,36. The inability of loss of Zmat3 to cooperate with c-MYC and KRAS expression in tumorigenesis may be explained by the finding that Zmat3 regulates a large set of targets at the mRNA level36 and that its’ function is dependent on additional co-occurring mutations other than deregulated c-MYC expression, mutations in KRAS and cellular stress (Fig. 5d). Further studies will be required to investigate the function and biological significance of Zmat3 in epithelial and lymphoid tumorigenesis.
References
Attardi, L. D. & Jacks, T. The role of p53 in tumour suppression: lessons from mouse models. Cell. Mol. Life Sci. 55, 48–63 (1999).
Bieging, K. T., Mello, S. S. & Attardi, L. D. Unravelling mechanisms of p53-mediated tumour suppression. Nat. Rev. Cancer 14, 359–370 (2014).
Aubrey, B. J., Kelly, G. L., Janic, A., Herold, M. J. & Strasser, A. How does p53 induce apoptosis and how does this relate to p53-mediated tumour suppression? Cell Death Differ. 25, 104–113 (2018).
Kastenhuber, E. R. & Lowe, S. W. Putting p53 in context. Cell 170, 1062–1078 (2017).
Mello, S. S. & Attardi, L. D. Deciphering p53 signaling in tumor suppression. Curr. Opin. Cell Biol. 51, 65–72 (2018).
Brady, C. A. & Attardi, L. D. p53 at a glance. J. Cell Sci. 123, 2527–2532 (2010).
Valente, L. J. et al. p53 Efficiently suppresses tumor development in the complete absence of its cell-cycle inhibitory and proapoptotic effectors p21, puma, and noxa. Cell Rep. 3, 1339–1345 (2013).
Brady, C. A. et al. Distinct p53 transcriptional programs dictate acute DNA-damage responses and tumor suppression. Cell 145, 571–583 (2011).
Li, T. et al. Tumor suppression in the absence of p53-mediated cell-cycle arrest, apoptosis, and senescence. Cell 149, 1269–1283 (2012).
Mello, S. S. et al. A p53 super-tumor suppressor reveals a tumor suppressive p53-Ptpn14-Yap axis in pancreatic cancer. Cancer Cell 32, 460–473.e6 (2017).
Valente, L. J., Grabow, S., Vandenberg, C. J., Strasser, A. & Janic, A. Combined loss of PUMA and p21 accelerates c-MYC-driven lymphoma development considerably less than loss of one allele of p53. Oncogene 35, 3866–3871 (2016).
Li, T. et al. Tumor suppression in the absence of p53-mediated cell cycle arrest, apoptosis, senescence. Cell 149, 1269–1283 (2012).
Brady, C. A. et al. Distinct p53 transcriptional programs dictate acute DNA-damage responses and tumor suppression. Cell 145, 571–583 (2011).
Janic, A. et al. DNA repair processes are critical mediators of p53-dependent tumor suppression. Nat. Med. 24, 947–953 (2018).
Vilborg, A., Bersani, C., Wilhelm, M. T. & Wiman, K. G. The p53 target Wig-1: a regulator of mRNA stability and stem cell fate? Cell Death Differ. 18, 1434–1440 (2011).
Bersani, C., Xu, L.-D., Vilborg, A., Lui, W.-O. & Wiman, K. G. Wig-1 regulates cell cycle arrest and cell death through the p53 targets FAS and 14-3-3σ. Oncogene 33, 4407–4417 (2014).
Hellborg, F. et al. Human wig-1, a p53 target gene that encodes a growth inhibitory zinc finger protein. Oncogene 20, 5466–5474 (2001).
Vilborg, A. et al. The p53 target Wig-1 regulates p53 mRNA stability through an AU-rich element. Proc. Natl Acad. Sci. USA 106, 15756–15761 (2009).
Vilborg, A., Wilhelm, M. T. & Wiman, K. G. Regulation of tumor suppressor p53 at the RNA level. J. Mol. Med. 88, 645–652 (2010).
Kim, B. C. et al. Wig1 prevents cellular senescence by regulating p21 mRNA decay through control of RISC recruitment. EMBO J. 31, 4289–4303 (2012).
Jackson, E. L. et al. The differential effects of mutant p53 alleles on advanced murine lung cancer. Cancer Res. 65, 10280–10288 (2005).
Garrison, S. P. et al. Selection against PUMA gene expression in Myc-driven B-cell lymphomagenesis. Mol. Cell. Biol. 28, 5391–5402 (2008).
Michalak, E. M. et al. Puma and to a lesser extent Noxa are suppressors of Myc-induced lymphomagenesis. Cell Death Differ. 16, 684–696 (2009).
Adams, J. M. et al. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature 318, 533–538 (1985).
Donehower, L. A. et al. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature 356, 215–221 (1992).
Jacks, T. et al. Tumor spectrum analysis in p53-mutant mice. Curr. Biol. 4, 1–7 (1994).
DuPage, M., Dooley, A. L. & Jacks, T. Conditional mouse lung cancer models using adenoviral or lentiviral delivery of Cre recombinase. Nat. Protoc. 4, 1064–1072 (2009).
Best, S. A., Kersbergen, A., Asselin-Labat, M. L. & Sutherland, K. D. Combining cell type-restricted adenoviral targeting with immunostaining and flow cytometry to identify cells-of-origin of lung cancer. In Methods in Molecular Biology 15–29 (Humana Press Inc., 2018).
Best, S. A. et al. Synergy between the KEAP1/NRF2 and PI3K pathways drives non-small-cell lung cancer with an altered immune microenvironment. Cell Metab. 27, 935–943.e4 (2018).
Schindelin, J. et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Schmidt, U., Weigert, M., Broaddus, C. & Myers, G. Cell detection with star-convex polygons. In Medical Image Computing and Computer Assisted Intervention – MICCAI 2018. 265–273 (2018).
Michalak, E. M. et al. Puma and to a lesser extent Noxa are suppressors of Myc-induced lymphomagenesis. Cell Death Differ. 16, 684–696 (2009).
Vassilev, L. T. et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 303, 844–848 (2004).
Jackson, E. L. et al. The differential effects of mutant p53 alleles on advanced murine lung cancer. Cancer Res. 65, 10280–10288 (2005).
Best, S. A. et al. Distinct initiating events underpin the immune and metabolic heterogeneity of KRAS-mutant lung adenocarcinoma. Nat. Commun. 10, 4190 (2019).
Bersani, C. et al. Genome-wide identification of Wig-1 mRNA targets by RIP-Seq analysis. Oncotarget 7, 1895–1911 (2016).
Jacks, T. et al. Tumor spectrum analysis in p53-mutant mice. Curr. Biol. 4, 1–7 (1994).
Acknowledgements
We are grateful to C. Gatt, K. McKenzie, S. Oliver and G. Siciliano for expert animal care; B. Helbert, K. Mackwell, and R. Chan for genotyping; E. Tsui in the WEHI Histology Facility and S. Monard in the WEHI Flow Cytometry Facility for expert support; E. Michalak for providing animal data and discussions. This work was supported by the Australian Phenomics Network (APN); the Australian National Health and Medical Research Council (NHMRC) Project Grant to M.J.H. (1145728), to M.J.H. and A.S. (1143105), Program Grant to A.S. (1016701); the Leukemia and Lymphoma Society of America to A.S. and M.J.H. (LLS SCOR 7001-13); the Cancer Council of Victoria Project grant to A.S. (1052309) and Venture Grant to M.J.H. and A.S.; support to A.S. from the estate of Anthony (Toni) Redstone OAM and a Spanish Ministry of Economy and Development Grant to A.J. (RTI2018-099017-A-I00). K.D.S. is supported by a Victorian Cancer Agency Mid-Career Research Fellowship (18003) and the Peter and Julie Alston Centenary Fellowship, A.J. is supported by Ramon y Cajal Fellowship (RYC2018-025244-I), A.S. and M.J.H. are supported by NHMRC Fellowships (1020363 and 1156095), M.S.B is supported by Cancer Council Victoria Postdoctoral Fellowship. This work was made possible through the Victorian Government Operational Infrastructure Support and Australian Government, a “Maria de Maeztu” Programme for Units of Excellence in R&D MDM-2014-0370 (Government of Spain) and La Caixa banking foundation (51110009).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors share senior authorship: Kate D. Sutherland, Ana Janic
Edited by U. Moll
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Best, S.A., Vandenberg, C.J., Abad, E. et al. Consequences of Zmat3 loss in c-MYC- and mutant KRAS-driven tumorigenesis. Cell Death Dis 11, 877 (2020). https://doi.org/10.1038/s41419-020-03066-9
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41419-020-03066-9
This article is cited by
-
Combined absence of TRP53 target genes ZMAT3, PUMA and p21 cause a high incidence of cancer in mice
Cell Death & Differentiation (2024)
-
Nutlin-3 suppresses tumorigenesis and progression of oral squamous cell carcinoma and enhances chemosensitivity to cisplatin
Cytotechnology (2023)
-
Of the many cellular responses activated by TP53, which ones are critical for tumour suppression?
Cell Death & Differentiation (2022)