Abstract
There is justified concern about the impact of global warming on the persistence of tropical ectotherms. There is also growing evidence for strong selection on climate-relevant physiological traits. Understanding the evolutionary potential of populations is especially important for low dispersal organisms in isolated populations, because these populations have little choice but to adapt. Despite this, direct estimates of heritability and genetic correlations for physiological traits in ectotherms—which will determine their evolutionary responses to selection—are sparse, especially for reptiles. Here we examine the heritabilities and genetic correlations for a set of four morphological and six climate-relevant physiological traits in an isolated population of an Australian rainforest lizard, Lampropholis coggeri. These traits show considerable variation across populations in this species, suggesting local adaptation. From laboratory crosses, we estimated very low to moderate heritability of temperature-related physiological traits (h2 < 0.31), but significant and higher heritability of desiccation resistance (h2~0.42). These values contrasted with uniformly higher heritabilities (h2 > 0.51) for morphological traits. At the phenotypic level, there were positive associations among the morphological traits and between thermal limits. Growth rate was positively correlated with thermal limits, but there was no indication that morphology and physiology were linked in any other way. We found some support for a specialist–generalist trade-off in the thermal performance curve, but otherwise there was no evidence for evolutionary constraints, suggesting broadly labile multivariate trait structure. Our results indicate little potential to respond to selection on thermal traits in this population and provide new insights into the capacity of tropical ectotherms to adapt in situ to rapid climate change.
Similar content being viewed by others
Introduction
In a world undergoing unprecedented rates of climate change, there is increasing concern as to how rising temperatures will impact biodiversity (Thomas et al. 2004; Bellard et al. 2012). This is particularly so for tropical rainforest animal species, because species in these environments tend to be physiologically specialized and so may be more sensitive to changes in thermal (Deutsch et al. 2008; Tewksbury et al. 2008; Kingsolver 2009) and hydric conditions (Hoffman et al., 2003; Kellermann et al. 2006) than their temperate counterparts. The link between temperature and organismal performance is especially clear in tropical ectotherms (Angilletta 2009; Huey et al. 2009; 2010), and small increases are thus predicted to have especially large effects on thermal specialist ectothermic tropical rainforest species (Huey et al. 2009). In particular, heat waves have become more common globally, and especially in the tropics (Buckley and Huey 2016), directing attention to impacts on tropical populations subject to extreme temperature conditions and to their capacity to adapt to rapidly warming environments.
Climate change has already caused shifts in species’ ranges, both in latitude and altitude (Chen et al. 2011). For low dispersal species, however, adaptation is likely to be the most important mechanism by which populations can avoid extinction (Visser 2008; Hoffman & Sgró 2011). Sinervo et al. (2010), e.g., argue that up to 20% of lizard species may go extinct in the coming years due to increasing temperatures. This depends, however, on how rapidly species can adapt to changing environmental conditions. Our understanding of the potential capacity for populations to adapt to climate change is still in its infancy. For some terrestrial invertebrates (primarily Drosophila), there has been considerable work on adaptation to climate and the evolutionary dynamics of phenotypic traits relevant to dealing with changing temperatures (Hoffmann 2010). Because this work involves mostly model organisms, the genetic basis of trait shift in invertebrates is reasonably well understood. Only recently, however, have studies begun to show the potential for rapid trait change in terrestrial vertebrates in response to increasing environmental temperatures (e.g., Leal and Gunderson 2012; Muñoz et al. 2014; Campbell-Staton et al. 2016 and references therein). Further, whether such changes reflect genetic or plastic change is typically unknown (Merilä and Hendry 2014, but see Urban et al. 2014, Evans and Gustafsson. (2017)).
There is certainly evidence that climate can impose strong selection pressures on thermoregulatory traits. For example, Logan et al. (2014) showed that Anolis lizards transplanted to warmer environments experienced strong selection on optimum temperature (Topt) and performance breadth. This is despite the fact that individuals can buffer themselves from their thermal environment through behavioral thermoregulation (the “Bogert effect”: see Huey et al. 2003). On its own, however, selection is not enough to cause an adaptive evolutionary response: a response to selection requires heritable genetic variation underlying the phenotypic traits under selection (Stockwell et al. 2003; Falconer and Mackay 1996). The magnitude of this heritable variation is typically estimated via the narrow-sense heritability of a trait (h2), defined as the proportion of within-population phenotypic variance in a trait (VP) that is due to variance in additive genetic effects (VA), i.e., h2 = VA/VP. There is a large body of work estimating the heritability of many different traits in natural populations, from life history to morphology (reviewed in Mousseau and Roff 1987; Postma 2014), and increasing evidence of heritability of aspects of physiology such as metabolic rate across a range of taxa (e.g., fish, Munday et al. 2017; birds, Ronning et al. 2007; mammals, Nespolo et al. 2003; insects, Saastamoinen et al. 2013; see review in Burton et al. 2011). For traits associated with thermal and hydric physiology, however, to date only invertebrates have been well studied (reviewed in Hoffman et al., 2003, Angilletta 2009). Indeed, to our knowledge, while there is one estimate of heritability of thermal preference (Paranjpe et al. 2013), there are currently no estimates of the heritability of climate-relevant (thermal and hydric physiological) traits in reptiles.
Adaptive responses to selection will also depend on the genetic architecture of multivariate phenotypes. Even if traits under selection are heritable, they may not be able to evolve because evolutionary constraints (shaped by genetic covariances between multiple traits) can operate to slow or halt adaptive responses (Merilä et al. 2001; Davis and Shaw 2001; Walsh and Blows 2009). According to some studies, these constraints may hamper adaptation in terrestrial ectotherms (such as lizards) such that they may not be able to evolve to track climate change (Sinervo et al. 2010). Several such constraints have been posited. For example, the specialist–generalist trade-off implies that individuals can not have both high maximum performance and a wide thermal breadth of performance; and the hotter-is-better hypothesis suggests that higher performance should accrue at higher optimal temperatures (Angilletta 2009). Similarly, considerations arising from Jensen’s inequality suggest that high-optimum temperatures might be associated with thermal specialization (Martin and Huey 2008; Vickers et al. 2011). What support exists for these and other trade-offs, however, typically stems from macroevolutionary patterns and has rarely been tested within populations. To assess whether these trade-offs exist as fundamental evolutionary constraints requires information on genetic covariances; information that is, to our knowledge, currently absent for terrestrial vertebrates.
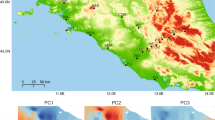
Here, we measure heritabilities and genetic correlations of a set of climate-relevant traits—those that mediate the relationship between climate and fitness—using lab-bred families of a population of an Australian lizard, the rainforest sunskink Lampropholis coggeri. This species shows considerable climate-related variation in thermal physiology traits both within and among populations (Phillips et al. 2016; Llewelyn et al. 2016a, 2018). Because of the increasing interest in the capacity of populations to adapt to extreme heat conditions, we focus on an isolated, lowland population that experiences especially hot and dry conditions relative to the species’ range (Llewelyn et al. 2016a; Fig. 1). The traits measured include those relevant to both thermal and hydric physiology, and also morphological characters. These latter traits were included in order to compare h2 values and to test for genetic covariance between physiology and morphology. Our results show that key physiological traits exhibit a range of heritabilities. They also reveal phenotypic and genetic covariance structures and, for physiological traits, point to a general lack of consonance between covariances at phenotypic and genetic levels. Real constraints to adaptation—low heritability and strong genetic correlations—do appear to exist, but the picture is complex and, for covariances in particular, remains poorly resolved.
Material and methods
Study species
The rainforest sunskink is a small (up to 45-mm snout to vent length), oviparous, scincid lizard endemic to the Wet Tropics region of northeastern Australia (Wilson and Swan 2008). For this study, we collected and tested sunskinks from one population, Hervey range (HR), towards the southern limit of the region’s rainforest (Fig. 1a). This population occupies a very small rainforest fragment at low elevation (182 m above sea level) at the southern end of the species’ distribution (Fig. 1a) and is genetically distinct and ecologically isolated from nearby low-mid elevation populations (Bell et al. 2010). Relative to most of the species’ range, this population experiences drier conditions and a high seasonal variance in temperature with more extreme heat conditions (Fig. 1b; Llewelyn et al. 2016b), and also shows an elevated heat-hardening response (rapid acclimation to heat shock) compared to a nearby montane population (Phillips et al. 2016). There is also considerable phenotypic plasticity in thermal physiological traits in the population associated with seasonal climate in the field (Llewelyn et al. 2016a) and acclimation in the laboratory (Llewelyn et al. 2018).
Collection and breeding
Animals were collected in the field between June 2013 and February 2014. Following collection, sunskinks were housed individually in a temperature-controlled room (set at 21 °C) for the first 6 months following collection before being paired. We did this because females can store sperm for an undetermined amount of time, and we wanted only F1s with known parentage. A temperature gradient within cages allowed the lizards to thermoregulate behaviorally. To produce offspring, 6 months after collection, we randomly paired field-collected sunskinks, which simply involved placing pairs into shared housing containers. Eggs were collected as they were produced by checking containers daily. After 3 months, pairs were re-assorted in order to pair females with different males. The breeding design was initially constructed as a half-sib design in which each male was mated to multiple females and each female to only one male. However, this turned out to be complicated by the females’ ability to store sperm for long periods of time (see below). Eggs were produced in clutches ranging in size from 1 to 3 eggs. The eggs were placed individually in air-tight containers (84 ml) that were two-thirds filled with moist vermiculite (50:50 ratio of vermiculite to water by weight), and the egg containers were put in incubators set at one of two temperatures (23 °C or 26 °C). These different incubation temperatures have no effect on the adult phenotype (see Llewelyn et al. 2018 for details). Eggs from the same clutch were split between these incubation treatments. The hatchling F1s were housed individually in tubs in the same way as the field-collected adults. We measured physiological traits in their laboratory-reared offspring (F1 generation) at adult size (>0.75 g total mass; see Table 1).
Traits and sample size
The phenotypic traits measured are listed in Table 1. Full details on physiological methods are presented elsewhere (Phillips et al. 2016; Llewelyn et al. 2016b). In brief, for thermal traits, we estimated the minimum (CTmin) and maximum (CTmax) thermal limits: temperatures at which lizards could no longer right themselves after being flipped on to their backs, with a ramp speed of 1 °C/min. We also conducted sprint trials at nine different temperatures (one temperature per day in ascending order over 9 days). For each individual, we took eight measures of speed at each temperature and used the maximum of this set as the sprint speed at that temperature. We recorded maximum sprint speeds at temperatures, ranging from 12 to 38 °C. We fitted a generic phenomenological curve to each sunskink’s sprint data using the curve-fitting method described in Phillips et al. (2014), treating sprint speed as “performance”, and extracted the sunskink’s thermal optimum Topt (the temperature at which it reached maximum speed), maximal performance Pmax (the maximum speed reached), and performance breadth Pbreadth80 (the breadth of temperatures over which a lizard’s sprint speed is above 80% of its Pmax) from the curve (Fig. 2). Desiccation rate was measured as an individual’s rate of weight loss when exposed to standardized desiccating conditions (Llewelyn et al. 2016b).
Morphological traits
To estimate growth rate we weighed skinks once per month from hatching until they reached a threshold size for physiological testing (0.75 g, at ~4 months of age, range 2–7 months). We calculated the change in mass per time over each of these intervals, and removed the effect of initial mass by taking the residuals of a quadratic regression of change in mass per time to give an estimate of individual growth rate. Thus, individual growth is measured relative to the mean growth function for the population. Additionally, we also measured morphological traits for hatchlings on the day of hatching (snout vent length, tail length, and total body mass).
Overall, we measured phenotypes of 152 F1 sunskinks generated by crossing field-collected F0 adults, though exact sample sizes varied slightly between the different traits (Table 1). The F1 data set comprised offspring from 33 F0 mothers paired to 30 F0 males; the average number of males per female was 1.11, and the average size of each full-sib family was 4.37. The complete familial design and number of offspring is presented as a table in the supplementary material.
Paternity assignment
To check paternity, we used the commercial genotyping services of Diversity Arrays (DArT) who have developed a widely used technique called DArTseq™. This method is an implementation of sequencing complexity-reduced representations (Altshuler et al. 2000) on next generation sequencing platforms (Baird et al. 2008, Elshire et al. 2011). Sequences generated from each lane were processed using proprietary DArT analytical pipelines. This clean-up process resulted in a comprehensive data set of ~18,000 SNPs with an average call rate of 93.5% and a reproducibility rate of 98.8%. We used the 5000 markers with the best coverage (mean depth 46.6; call rate and reproducibility of 100%, and that were represented in all sampled individuals) to build a Hamming distance matrix between individuals (supplementary material). Recent studies show that as few as 30 optimized SNPs are sufficient to differentiate among 100,000 individuals based on Hamming Distance values (Hu et al. 2015). All F1 individuals were lined up against their mother and siblings and the Hamming Distance values evaluated to cross check for sample mix-ups. Hamming Distance values were then compared against each of the potential sires given the known mother, and the sire/F1 with the lowest value was considered a match. We also checked the values for the mothers to look for potential misassignments. We could unambiguously assign paternities for 131 out 152 F1 individuals; for the remaining 21, the father was clearly not among the sampled F0 males, indicating the female had stored sperm for more than 6 months from a pre-capture mating. Overall this resulted in a combination of full siblings, maternal half-siblings and paternal half-siblings among the offspring. The 21 individuals were used in the subsequent analysis but with father’s identity treated as missing.
Statistical analyses
CTmin, CTmax, Pmax, Pbreadth80, and DR were all log-transformed prior to analyses to conform to assumptions of residual normality. Growth rate was multiplied by 1000 to improve scaling.
Phenotypic variation (VP) in our data set was partitioned using an “animal model” (Henderson 1950; Lynch and Walsh 1998; Kruuk 2004) to partition the total phenotypic variance in a trait into its additive genetic (VA), maternal (Vm, when applicable) and environmental or residual variance (Ve). All the analyses were implemented using the statistical package ASReml version 3.0 (https://www.vsni.co.uk/software/asreml/). We compared models with different random effects structure by implementing likelihood-ratio tests (LRTs) between the nested models, using a threshold for significance of p < 0.05. We estimated heritability values as h2 = VA/VP, with significance of heritability assessed by the likelihood-ratio test for VA. Maternal effects—defined as the effects of a mother on her offspring’s phenotype over and above those due to the direct effects of genotype—are recognized as important factors in phenotypic variation in a number of organisms, and their role has been studied extensively in lizards. Maternal effects are known to influence body size (mass), performance traits, and growth rates (Shine and Harlow 1993; Paranjpe et al. 2013), and failure to account for maternal effects would likely inflate heritability estimates (Kruuk and Hadfield 2007). We therefore also tested for maternal effects by fitting the mother’s identity as an additional random effect. Where this was not found to be significant, based on a likelihood-ratio test comparison of a model with and without the maternal effect component, it was dropped from the final analysis. All models included sex as a fixed effect, and models for the physiological traits also included (log) mass at time of measuring as a covariate (to account for variation due to allometry).
Second, we considered associations between traits using a multivariate model. All traits were standardized to unit variance before analyses, to improve model convergence, and the output for the multivariate analyses therefore represents standardized values. We first quantified the phenotypic covariances between traits by fitting a multivariate model for all ten traits to generate a full phenotypic variance–covariance matrix, P. A full 10-trait animal model estimating the full genetic variance–covariance matrix G between all traits would not converge, so we generated individual covariance estimates on a pairwise, bivariate basis (see e.g., Coltman et al. 2003). From these pairwise models, we estimated both a genetic correlation and a residual correlation between each pair of traits, the latter accounting for all remaining aspects of covariation not captured by the genetic correlation. For completeness, we ran bivariate models on all pairs of traits, though we did not expect to find genetic correlations for pairs of traits with low estimated heritability. The phenotypic data as well as the pedigree file are available in the supplementary material.
Results
Trait variation and heritabilities
All traits showed substantial phenotypic variance (Table 2). For morphological traits—size-related traits of hatchlings—estimates of heritability were high (h2 = 0.51 ± 0.20SE to 0.61 ± 0.21SE; Table 2). Growth rate showed lower estimated heritability (h2 = 0.19 ± 0.17SE). Heritability estimates were low to moderate (0.03–0.26) across the thermal physiology traits (Table 2). Our parameter uncertainty was such that none of the thermal physiological traits achieved statistical significance (against the null hypothesis that heritability is zero). By contrast, two other physiological traits—the maximum performance measure, Pmax, and desiccation resistance, DR—had higher heritability estimates (Pmax: h2 = 0.31 ± 0.19SE, p = 0.014 and DR: 0.42 ± 0.19SE, p = 0.001).
Phenotypic covariances between traits
The phenotypic variance–covariance matrix, P, for all ten phenotypic traits is presented in Table 3. Correlations between traits were generally low, but there were several exceptions. First, all (hatchling) size-related measures had significant positive phenotypic correlations (r = 0.44–0.58, all p < 0.001). This is unsurprising: hatchlings with higher body mass also tended to have larger snout vent lengths and longer tails. Residual hatchling growth rate was not, however, correlated with any of the other morphological traits.
Second, there were significant phenotypic correlations between hatchling growth rate and thermal limits (CTmin and CTmax) (Table 3). In both cases animals with higher thermal limits tended to grow faster. Growth rate was also correlated with other aspects of physiology: animals that grew faster had broader thermal performance Pbreadth80, but lower Topt (Table 3). Growth rate was also negatively associated with desiccation rate: animals that grew slower had higher rates of desiccation, DR.
Third, among physiological traits, there were very few strong phenotypic correlations. The two exceptions were an unambiguous negative correlation between Pmax and Pbreadth80 (r = −0.294, p < 0.001; Table 3); and a positive correlation between Topt and Pbreadth80 (r = 0.38, p < 0.001; Table 3).
Genetic and residual covariances between traits
We estimated pairwise genetic and residual covariances for all pairs of traits (Table 4). For morphological traits, the directions of genetic correlations were concordant with the observed phenotypic correlations (i.e., all positive): whatever genetic variation causes some individuals to have high body mass also causes them to have longer bodies and tails. The positive phenotypic correlation between growth rate and the critical thermal maximum CTmax was also reflected at the genetic level. There were also significant positive genetic correlations between Pmax and two of the hatchling size measures: snout vent length and tail length (Table 4a). These correlations were unexpected given that they were not apparent at the phenotypic level, presumably due to the fact that the corresponding residual correlations were negative (Table 4b). However the unusually high correlation (r ~ 1) between Pmax and snout vent length leads us to interpret the result with caution as a potential statistical anomaly.
As expected given the generally low heritabilities for physiological traits, the phenotypic correlations between these traits were typically not reflected at the genetic level; i.e., there were no significant genetic correlations between physiological traits. Uncertainty in our estimates of residual covariance was more homogenous across physiological traits, however. Two of the phenotypic correlations—those between performance breadth Pbreadth80 and optimum temperature Topt, and between optimum temperature Topt and growth rate—appeared to be driven by residual effects (environmental or non-additive genetic) rather than additive genetic effects.
Discussion
Our analyses of laboratory-reared families of the rainforest sunskink L. coggeri showed substantial phenotypic variation in all traits, as expected from previous field and laboratory observations (Phillips et al. 2016; Llewelyn et al. 2016b, 2018). All of the measures of hatchling size-related traits were highly heritable, as were desiccation rate and maximum running performance. Growth rate and thermal physiological traits exhibited lower heritabilities. Standard errors related to our estimates were often large, which may be attributable to our modest sample sizes, but our estimates remain valuable because they are currently the only estimates of heritability (and genetic covariances) for a set of thermal physiological traits in a vertebrate and so fill a void in understanding potential for adaptation of tropical ectotherms to climate change.
Although we include p-values as a useful heuristic of the weight of evidence against a heritability of zero, we emphasize parameter estimates over p-values throughout, noting that a heritability of zero is a priori unlikely to be true. We use an animal model to estimate additive genetic variance. Our breeding design contained a mixture of full siblings, maternal half-siblings, and paternal half-siblings, and so the animal model provides the most efficient means of using data from all relationships. However, as with many quantitative genetic analysis, there is the risk that estimates of additive variance are inflated by maternal effects (which may themselves be environmental or genetic in origin; Kruuk and Hadfield 2007). A large proportion of females in the data set had only one or two offspring, which makes separating off maternal effects variance difficult. Adding a maternal effects term to our model did not result in any significant improvement of log-likelihood. Furthermore, models with just maternal effects returned very similar estimates of variance as models fitting variance between sires, which strongly suggests a direct genetic rather than maternal basis to differences between families. However, the possibility remains that our estimates of heritability are inflated by maternal effects, and thus that the differences between families in morphological traits, Pmax, and dessication resistance (see below) are in part driven by maternal effects. Irrespective of these issues, our main conclusion, that we see only small differences between families for the thermal physiological traits, regardless of which model was fitted, stands.
There are, of course, two explanations for low heritability estimates: (1) low-additive genetic variance, or (2) large measurement error or plasticity, causing high-residual variation which dwarfs the additive genetic variance. In our case low-genetic variance for these traits is a distinct possibility. Strong directional selection will reduce genetic variation, so all else being equal, traits under strong selection may be expected to have lower heritability (Fisher 1930). Given that the population in question is both geographically and environmentally marginal to the rest of the species’ range, and experiences high variance in maximum temperature (Llewelyn et al. 2016a), strong selection on thermal physiological traits is plausible. It is also relevant that Logan et al. (2014) found positive selection on Topt and Pbreadth80 in Anolis lizards, but not on Pmax—the one thermal performance-related trait for which we did find significant heritability in L. coggeri. Erosion of genetic diversity through drift is also possible in this small and genetically isolated population. However, such drift should affect all traits, whereas we find low heritability for just some physiological traits (see also Hoffmann et al. 2003).
An alternative explanation for low heritability in the thermal physiological traits is high-residual variance, leading to low heritability. Such variance could derive from measurement error, and/or phenotypic plasticity. With regard to measurement error, there is no doubt that these thermal physiological traits are challenging to measure with high accuracy. For example, CTmin and CTmax are known to have substantial measurement error (Phillips et al. 2016, Terblanche et al. 2007). Unfortunately, it is impractical to quantify this measurement error because rapid acclimation means that the act of measuring some of these traits (notably CTmin and CTmax) actually temporarily changes the trait, and repeat measures likely cause longer-term acclimation to occur (Phillips et al. 2016). Despite these general issues, there is a wealth of literature that not only focuses on these traits, but also regularly finds meaningful associations between traits and between traits and the environment (e.g., Angilletta 2009; Hoffmann and Parsons 1997), including for our focal species (Llewelyn et al. 2016b). Reversible plasticity in physiological traits in L. coggeri has been demonstrated experimentally, and physiological traits have also been observed to shift seasonally in natural and laboratory populations of this species (Llewelyn et al. 2016a, b, 2018) and many others (see Kingsolver and Huey 1998; Hoffman, 2010). The measurements in the present study were from laboratory acclimated individuals raised in the same manner and kept in the same conditions, so we would expect this source of variance to be small.
One trait from the thermal performance curve—maximum sprint speed, Pmax—did exhibit a significant estimate of heritability (h2 = 0.31 ± 0.19SE; LRT χ2(1) = 6.07, p = 0.014; we note that the SE is relatively high despite the significance of the likelihood-ratio test). Sprint speed (at a fixed temperature) is often used as a proxy for whole-organism performance (Arnold 1983). In some lizard species, sprint speed is also known to be correlated with survival (Miles 2004; Calsbeek and Irschick 2007; Warner and Andrews 2002; Le Galliard and Ferriére 2008), so we might expect this trait to be under strong selection and so to have low heritability. L. coggeri is, however, semi-fossorial in its habits, escaping predators by vanishing into the leaf litter on which it lives rather than fleeing, and foraging for small insect prey on and below the leaf litter. As a consequence, sprint speed over bare ground may be only weakly related to individual survival in L. coggeri. Substantial heritability of sprint speed has also been reported for several other squamates (Tsuju et al. 1989; Brodie 1989; Garland 1988), indicating that this trait may generally have the potential to evolve in response to selection.
The one physiological trait not related to temperature, desiccation rate, displayed a substantial (and significant) estimate of heritability (h2 = 0.42 ± 0.19SE). This result is interesting considering that the population studied here occupies a very small rainforest isolate at the very edge of the species’ distribution; animals living in small rainforest fragments are expected to be subject to temperature extremes and high desiccation rates due to strong edge effects (Turton and Freiburger 1997; Didham and Lawton 1999; Huey et al. 2003). Desiccation is also likely to be especially problematic for a very small animal like L. coggeri. Our observed high heritability in this case is opposite to the pattern observed in Drosophila from nearby rainforest, in which realized heritability for desiccation resistance was effectively zero (Hoffmann et al. 2003), but similar to a recent study in two Drosophila species that used less stringent desiccation conditions (van Heerwaarden and Sgro 2014). In L. coggeri, substantial heritability for desiccation rate suggests that this population could respond to selection imposed, e.g., by increasing severity of dry seasons.
Our analysis also revealed various correlations between traits, at both phenotypic and genotypic levels. At the phenotypic level, there was a clear positive association between morphological traits, simply because the morphological measures are all measures of size. Interestingly, however, these correlations are almost perfectly accounted for at the genetic level (Table 4), suggesting that there is heritable variation for overall size at hatching. Whether such variation is driven by genetic differences in incubation time, embryonic growth rate or other factors is unknown. Whatever the mechanism, it is likely that hatchling size will have impacts on fitness because size will affect numerous aspects of individual performance, from running speed to desiccation rate and prey availability (e.g., Elphick and Shine 1998; Kingsolver and Huey 2008).
There was also a clear positive phenotypic correlation between individual growth rate, and several elements of physiology. Animals that grew faster had higher critical thermal limits, broader thermal performance breadth, lower desiccation rates, and lower optimum performance temperatures. These relationships were less clear at the genetic level. Generally the genetic correlations were in the same direction as the phenotypic correlation, but the low precision of the estimates makes conclusions difficult. There remained, however, a clear positive genetic correlation between the upper critical thermal limit, CTmax and growth rate. Interestingly, CTmax in L. coggeri has been shown to respond plastically to the temperatures an animal experiences, and there is clear variation between individuals and across populations in the preferred body temperature (Llewelyn et al. 2016b). Thus, the positive genetic correlation observed here between CTmax and growth rate may reflect a positive correlation between preferred body temperature—a variable that we did not measure directly—and growth rate: individuals that prefer warmer body temperatures regularly achieve higher temperatures and, as a consequence, grow faster and have higher CTmax.
Another interesting outcome of our analysis is a very clear positive genetic correlation between hatchling body size (SVL and tail length) and maximum performance of adults. This correlation is not apparent at the phenotypic level, but emerges as a genetic effect that is obscured in the phenotype, by a counteracting negative residual correlation. We might first think that the genetic correlation is driven by body size (larger hatchlings grow into larger adults, and larger adults run faster), but our analysis of maximum performance explicitly accounted for adult body size as a covariate, so we are left with the intriguing correlation in which larger hatchlings appear to grow into adults with a genetic tendency towards higher performance independent of body size.
With regard to evolutionary constraints on the evolution of climate-relevant traits, our data are intriguing though also limited by low precision of some estimates. There are three posited constraints on the evolution of thermal performance to which our data can speak: (1) the specialist–generalist trade-off (that maximum performance comes at a cost to breadth); (2) that higher optimal temperatures requires thermal specialization (Martin and Huey 2008) and (3) that hotter is better—with higher thermal optimum will tend also to have higher performance (Huey and Kingsolver 1989; Angilletta 2009).
At the phenotypic level we see clear evidence of a specialist–generalist trade-off in the temperature performance curve: maximum performance is negatively correlated with thermal breadth. This relationship has been observed in other taxa (see Angilletta 2009 and references therein) and we would expect this specialist–generalist trade-off to manifest within populations as well as across them (Phillips et al. 2014). In our case, the association does persist at the genetic level, although estimation uncertainty means we cannot conclude that it is a true genetic constraint.
With regard to high-optimum temperatures requiring thermal specialization, in our data we observe the opposite: individuals with higher optimum temperatures tended to have broader thermal performance curves. And this association appears to be driven, not through a genetic correlation but, rather, a residual correlation. Although we cannot make any real conclusion here as to the mechanism, it seems likely that, if the correlation is not genetic, there may be elements of thermoregulatory behavior underpinning this result instead.
We find no evidence for the idea that “hotter is better”, in that there is no relationship between Topt and Pmax at either phenotypic or genetic levels. This hypothesis stems from the basic physical principle that chemical reactions tend to go faster as temperatures increase (Huey and Kingsolver 1989; Angilletta 2009), and there is evidence to support this idea at the macroevolutionary level. There are, however, clear design trade-offs at the enzymatic level such that for hotter to really be better requires a suite of coadapted enzymes to effectively utilize the additional heat energy (Hochachka and Somero 2002; Somero 2004). Because of this, we might only expect the hotter is better pattern to play out at the macroevolutionary level (Phillips et al. 2014).
Overall, our analysis, while constrained by low precision, suggests low to moderate heritability of thermal physiology traits. The only physiological trait with substantial heritability was desiccation resistance. Low-genetic variance aside, evidence for fundamental evolutionary constraints on climate-relevant traits in our species is weak, with the possible exception of a specialist–generalist trade-off in thermal traits. While our results are restricted to just one population, they suggest that with the exception of desiccation resistance, climate-relevant traits in this species have low heritability and any in situ responses to rapid environmental change will need to come through behavioral plasticity rather than genetic adaptation. Should these observations hold for other populations, the limited capacity to evolve higher heat tolerance could pose a threat to persistence in isolated populations of this rainforest species. If this low within-population heritability is driven by local adaptation, however, genetic trait variation will be found across populations (rather than within them) such that targeted gene flow (Kelly and Phillips 2016) could be a powerful tool for mitigating impact. Clearly, however, estimates of genetic variances for key physiological traits across taxa and populations in peripheral and core areas will be required to assess the generality of the situation described here.
References
Altshuler D, Pollara VJ, Cowles CR, Van Etten WJ, Baldwin J, Linton L, Lander ES (2000) An SNP map of the human genome generated by reduced representation shotgun sequencing. Nature 407:513–516
Angilletta MJ (2009) Thermal adaptation: a theoretical and empirical synthesis. Oxford University press, Oxford
Arnold SJ (1983) Morphology, performance, and fitness. Am Zool 23:347–361
Baird NA, Etter PD, Atwood TS, Currey MC et al. (2008) Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS ONE 3:e3376
Bell RC, Parra JL, Tonione M, Hoskin CJ, Williams SE, Moritz C (2010) Patterns of persistence and isolation indicate resilience to climate change in montane rainforest lizards. Mol Ecol 19:2531–2544
Bellard C, Bertelsmeier C, Leadley P, Thuiller W, Courchamp F (2012) Impacts of climate change on the future of biodiversity. Ecol Lett 15:365–377
Brodie ED (1989) Genetic correlations between morphology and antipredator behaviour in natural populations of the garter snake Thamnophis ordinoides. Nature 342:542–543
Buckley LB, Huey RB (2016) Temperature extremes: geographic patterns, recent changes, and implication for organismal vulnerabilities. Glob Chang Biol 22:3829–3842
Burton T, Killen SS, Armstrong JD, Metcalfe NB (2011) What causes intraspecific variation in resting metabolic rate and what are its ecological consequences? Proc R Soc B 278:3465–3473
Calsbeek R, Irschick DJ (2007) The quick and the dead: correlational selection on morphology, performance, and habitat use in island lizards. Evolution 61:2493–2503
Campbell-Staton SC, Edwards SV, Losos JB (2016) Climate-mediated adaptation after mainland colonization of an ancestrally subtropical island lizard, Anolis carolinensis. J Evol Biol 29:2168–2180
Chen I-C, Hill JK, Ohlemüller R, Roy DB, Thomas CD (2011) Rapid range shifts of species associated with high levels of climate warming. Science 19:1024–1026
Coltman DW, O’Donoghue P, Jorgenson JT, Hogg JT et al. (2003) Undesirable evolutionary consequences of trophy hunting. Nature 426:655–658
Davis MB, Shaw RG (2001) Range shifts and adaptive responses to quaternary climate change. Science 292:673–679
Didham RK, Lawton JH (1999) Edge structure determines the magnitude of changes in microclimate and vegetation structure in tropical forest fragments. Biotropica 31:17–30
Deutsch CA, Tewksbury JJ, Huey RB, Sheldon KS et al. (2008) Impacts of climate warming on terrestrial ectotherms across latitude. PNAS 105:6668–6672
Elphick MJ,Shine R (1998) Longterm effects of incubation temperatures on the morphology and locomotor performance of hatchling lizards (Bassiana duperreyi, Scincidae). Biol J Linn Soc 63:429–447
Elshire RJ, Glaubitz JC, Sun Q, Poland JA et al. (2011) A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 6:e19379
Evans, SR & Gustafsson. L 2017. Climate change upends selection on ornamentation in a wild bird. Nat Ecol Evol 1:0039. https://doi.org/10.1038/s41559-016-0039.
Falconer DS, Mackay TFC (1996) Introduction to quantitative genetics. Longman, Essex
Fisher RA (1930) The genetical theory of natural selection. Clarendon Press, Oxford
Garland TJ (1988) Genetic basis of activity metabolism. I. Inheritance of speed, stamina, and antipredator displays in the gartersnake Thamnophis sirtalis. Evolution 42:335–350
Henderson CR (1950) Estimation of genetic parameters. Ann Math Stat 21:309–310
Hochachka PW, Somero GN (2002) Biochemical adaptation: mechanism and process in physiological evolution. Oxford University Press, Oxford
Hoffmann AA (2010) Physiological climatic limits in Drosophila: patterns and implications. J Exp Biol 213:870–880
Hoffmann AA, Hallas RJ,Dean JA, Schiffer M (2003) Low potential for climatic stress adaptation in a rainforest Drosophila species. Science 301:100–102
Hoffmann AA, Sørensen JG,Loeschcke V (2003) Adaptation of Drosophila to temperature extremes: bringing together quantitative and molecular approaches. J Therm Biol 28:175–216
Hoffmann AA, Parsons PA (1997) Extreme environmental change and evolution. Cambridge University Press, Cambridge, New York
Hoffmann AA, Sgrò CM (2011) Climate change and evolutionary adaptation. Nature 470:479–485
Hu H, Liu X, Jin W, Ropers HH, Wienker TF (2015) Evaluating information content of SNPs for sample-tagging in re-sequencing projects. Sci Rep 5:10247
Huey RB, Deutsch CA, Tewksbury JJ, Vitt LJ et al. (2009) Why tropical forest lizards are vulnerable to climate warming. Proc R Soc B 276:1939–1948
Huey RB, Hertz PE, Sinervo B (2003) Behavioral drive versus behavioral inertia in evolution: a null model approach. Am Nat 161:357–366
Huey RB, Kingsolver JG (1989) Evolution of thermal sensitivity of ectotherm performance. Trends Ecol Evol 4:131–135
Huey RB, Losos JB, Moritz C (2010) Are lizards toast? Science 328:832–833
Kellermann VM,Heerwaarden BV,Hoffmann AA, Sgró CM (2006) Very low additive genetic variance and evolutionary potential in multiple populations of two rain-forest Drosophila species. Evolution 60:1104–1108
Kelly EL, Phillips BL (2016) Targeted gene flow for conservation. Conserv Biol 30:259–267
Kingsolver JG (2009) The well-temperatured biologist. Am Nat 174:755–768
Kingsolver JG, Huey RB (1998) Evolutionary analyses of morphological and physiological plasticity in thermally variable environments. Am Zool 38:545–560
Kingsolver JG, Huey RB (2008) Size, temperature, and fitness: three rules. Evol Ecol Res 10:251–268
Kruuk LEB (2004) Estimating genetic parameters in wild populations using the ‘animal model’ Philos Trans R Soc Lond B Biol Sci 359:873–890
Kruuk LEB, Hadfield JD (2007) How to separate genetic and environmental causes of similarity between relatives. J Evol Biol 20:1890–1903
Leal M, Gunderson AR (2012) Rapid change in the thermal tolerance of a tropical lizard. Am Nat 180:815–822
Le Galliard J-F, Ferrière R (2008) Evolution of maximal endurance capacity: natural and sexual selection across age classes in a lizard. Evol Ecol Res 10:157–176
Llewelyn, J, MacDonald, SL, Hatcher, A, Moritz, C & Phillips, BL 2016a. Thermoregulatory behaviour explains countergradient variation in the upper thermal limit of a rainforest skink. Oikos 12:748–757. https://doi.org/10.1111/oik.03933.
Llewelyn J, MacDonald SL, Hatcher A, Moritz C, Phillips BL (2016b) Intraspecific variation in climate-relevant traits in a tropical rainforest lizard. Divers Distrib 22:1000–1012
Llewelyn J, MacDonald SL, Moritz C, Martins F, Hatcher A, Phillips BL (2018) Adjusting to climate: acclimation, adaptation, and developmental plasticity in physiological traits of a tropical rainforest lizard. Int Zool 13:1–13
Logan ML, Cox RM, Calsbeek R (2014) Natural selection on thermal performance in a novel thermal environment. PNAS 111:14165–14169
Lynch M, Walsh B (1998) Genetics and analysis of quantitative traits. Sinauer, Sunderland, MA
Martin TL, Huey RB (2008) Why “Suboptimal” is optimal: Jensen’s inequality and ectotherm thermal preferences. Am Nat 171:E102–E118
Merilä J, Hendry AP (2014) Climate change, adaptation, and phenotypic plasticity: the problem and the evidence. Evol Appl 7:1–14
Merilä J, Sheldon B, Kruuk L (2001) Explaining stasis: microevolutionary studies in natural populations. Genetica 112:199
Miles D,B (2004) The race goes to the swift: fitness consequences of variation in sprint performance in juvenile lizards. Evol Ecol Res 6:63–75
Mousseau TA, Roff DA (1987) Natural selection and the heritability of fitness components. Heredity 59:181–197
Munday PL, Donelson JM, Domingos JA (2017) Potential for adaptation to climate change in a coral reef fish. Glob Chang Biol 23:307–317
Muñoz MM, Stimola MA, Algar AC, Conover A et al. (2014) Evolutionary stasis and lability in thermal physiology in a group of tropical lizards. Proc R Soc B 281:20132433
Nespolo RF, Bacigalupe LD, Bozinovic F (2003) Heritability of energetics in a wild mammal, the leaf-eared mouse (Phyllotis darwini). Evolution 57:1679–1688
Paranjpe DA, Bastiaans E, Patten A, Cooper RD, Sinervo B (2013) Evidence of maternal effects on temperature preference in side-blotched lizards: implications for evolutionary response to climate change. Ecol Evol 3:1977–1991
Phillips BL, Llewelyn J, Hatcher A, Macdonald S, Moritz C (2014) Do evolutionary constraints on thermal performance manifest at different organizational scales? J Evol Biol 27:2687–2694
Phillips BL, Muñoz MM, Hatcher A, Macdonald SL et al. (2016) Heat hardening in a tropical lizard: geographic variation explained by the predictability and variance in environmental temperatures. Funct Ecol 30:1161–1168
Postma E (2014) Four decades of estimating heritabilities in wild vertebrate populations: improved methods, more data, better estimates? In: Charmantier A, Garant D, Kruuk LEB (eds) Quantitative genetics in the Wild. Oxford University Press, Oxford, pp 16–33
Rønning B, Jensen H, Moe B, Bech C (2007) Basal metabolic rate: heritability and genetic correlations with morphological traits in the zebra finch. J Evol Biol 20:1815–1822
Saastamoinen M, Brommer JE, Brakefield PM, Zwaan BJ (2013) Quantitative genetic analysis of responses to larval food limitation in a polyphenic butterfly indicates environment- and trait-specific effects. Ecol Evol 3:3576–3589
Shine R, Harlow P (1993) Maternal thermoregulation influences offspring viability in a viviparous lizard. Oecologia 96:122–127
Sinervo B, Méndez-de-la-Cruz F, Miles DB, Heulin B, Bastiaans E et al. (2010) Erosion of lizard diversity by climate change and altered thermal niches. Science 328:894–899
Somero GN (2004) Adaptation of enzymes to temperature: searching for basic “strategies”. Comp Biochem Physiol B 139:321–333
Stockwell CA, Hendry AP, Kinnison MT (2003) Contemporary evolution meets conservation biology. Trend Ecol Evol 18:94–101
Terblanche JS, Deere JA, Clusella-Trullas S, Janion C, Chown SL (2007) Critical thermal limits depend on methodological context. Proc R Soc B 274:2935–2943
Tewksbury J,J, Huey RB, Deutsch CA (2008) Putting the heat on tropical animals. Science 320:1296–1297
Thomas CD, Cameron A, Green RE, Bakkenes M, Beaumont LJ, Collingham YC et al. (2004) Extinction risk from climate change. Nature 427:145–148
Tsuju JS, Huey RB, Van Berkum FH, Garland Jr TJ, Shaw RG (1989) Locomotor performance of hatchling fence lizards (Sceloporus occidentalis): quantitative genetics and morphometric correlates. Evol Ecol 3:240–252
Turton, SM, Freiburger, HJ 1997. Edge and aspect effects on the microclimate of a small tropical forest remnant on the Atherton Tableland, Northeastern Australia. In: WF Laurance & RO Bierregard (eds) Tropical forest remnants, ecology, management, and conservation of fragmented communities. The University of Chicago Press, Chicago, pp 45–54
Urban MC, Richardson JL, Freidenfelds NA (2014) Plasticity and genetic adaptation mediate amphibian and reptile responses to climate change. Evol Appl 7:88–103
van Heerwaarden B, Sgro CM (2014) Is adaptation to climate change really constrained in niche specialists? Proc R Soc B 281:20140396
Vickers M, Manicom C, Schwarzkopf L (2011) Extending the cost-benefit model of thermoregulation: high-temperature environments. Am Nat 177:452–461
Visser ME (2008) Keeping up with a warming world: assessing the rate of adaptation to climate change. Proc R Soc B 275:649–659
Walsh B, Blows MW (2009) Abundant genetic variation plus strong selection=multivariate genetic constraints: a geometric view of adaptation. Annu Rev Ecol Evol Syst 40:41–59
Warner DA, Andrews RM (2002) Laboratory and field experiments identify sources of variation in phenotypes and survival of hatchling lizards. Biol J Linn Soc 76:105–124
Wilson S, Swan G (2008) A complete guide to reptiles of Australia, 2nd edn. New Holland Publishers, Sydney
Acknowledgements
We thank Amberlee Hatcher for her unstinting assistance with husbandry and data collection. Naomi Harney, Kyana Pike, and Margaret Murray for assisting with the animal husbandry. Joe Holtum for providing access to useful equipment. Stewart Macdonald assisted with the collection of animals and figures. Thanks to Leo Tedeschi and Mark Hubbe for helping with the figures. We thank Andrzej Killian and the DArT crew for their help and support. We also thank Scott Keogh for suggestions on the manuscript. Funding for this research was provided by the Australian Research Council (DP 1094646, 130100318 for BP and CM; FT110100453 for LK) and the CSIRO/JCU Tropical Landscapes Joint Venture. Our study conformed to the legal requirements regarding animal research in Australia (JCU animal ethics permits: A1976 and A1726; collection permits: WISP10659412, WISP14625914, WITK15224114, and WITK10659112).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Martins, F., Kruuk, L., Llewelyn, J. et al. Heritability of climate-relevant traits in a rainforest skink. Heredity 122, 41–52 (2019). https://doi.org/10.1038/s41437-018-0085-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41437-018-0085-y
This article is cited by
-
Heritability and developmental plasticity of growth in an oviparous lizard
Heredity (2024)
-
Tadpoles Develop Elevated Heat Tolerance in Urban Heat Islands Regardless of Sex
Evolutionary Biology (2024)
-
Quantitative genetics of breeding coloration in sand lizards; genic capture unlikely to maintain additive genetic variance
Heredity (2023)