Abstract
The diversity of the naïve T cell repertoire drives the replenishment potential and capacity of memory T cells to respond to immune challenges. Attrition of the immune system is associated with an increased prevalence of pathologies in aged individuals, but whether stem cell memory T lymphocytes (TSCM) contribute to such attrition is still unclear. Using single cells RNA sequencing and high-dimensional flow cytometry, we demonstrate that TSCM heterogeneity results from differential engagement of Wnt signaling. In humans, aging is associated with the coupled loss of Wnt/β-catenin signature in CD4 TSCM and systemic increase in the levels of Dickkopf-related protein 1, a natural inhibitor of the Wnt/β-catenin pathway. Functional assays support recent thymic emigrants as the precursors of CD4 TSCM. Our data thus hint that reversing TSCM defects by metabolic targeting of the Wnt/β-catenin pathway may be a viable approach to restore and preserve immune homeostasis in the context of immunological history.
Similar content being viewed by others
Introduction
The stem-cell-like self-renewal ability is a vital feature of immune memory T cells that preserves the lifetime health of an individual1,2. Naturally acquired immunity or the induction of long-term protection occurs during primary infection and vaccination3,4—even in the absence of re-exposure to the respective immunizing agent. The maintenance of immunological memory requires the preservation of T-cell stemness and the flexibility of various T-cell compartments to alternate between a persistent quiescent state and a proliferative state where specific clones divide asymmetrically to give rise to activated memory and effector T cells5,6, which are vital for pathogen clearance during antigen re-exposure7.
The recent discovery of stem-cell memory T lymphocytes (TSCM) fills an important void that had hitherto obscured our understanding of the ontogeny of memory T cells. TSCM represent a discrete but phenotypically detectable population in animal models (mice, nonhuman primates) and humans8,9,10. These cells have the ability not only to self-renew but also to differentiate into all subsets of memory and effector T cells11. Combined with their longevity, the preservation of TSCM plasticity may play a central role in maintaining immunologic competence with age12. Their essential role in the development of autoimmune disorders, chronic infections, and cancer have already identified them as putative targets for future vaccines or adaptive T-cell therapies13. Although the paucity of TSCM in peripheral blood limits the possibility of ex vivo studies, TSCM can evolve from naive T cells following in vitro stimulation with IL-7/IL-1514, activation of Wnt/β-catenin9,15 and Notch pathways16, or modulation of mTOR signaling17. Most studies on TSCM have focused on CD8 T cells, but the generation and programming of naive CD4 T cells into TSCM may occur through similar mechanisms. Whether the latter is achieved through specific transcriptional programming of putative TSCM-precursors, i.e., recent thymic emigrants (RTE), naive CD4 T cells with homeostatic proliferative history (CD31−), CD103+ naive T cells, memory T cells with a naive phenotype (TMNP)18, or virtual memory T cells19 remains unknown20,21,22. Moreover, it is unclear whether TSCM are preserved during the process of aging, where the processes of thymic involution23 and memory compartment inflation become prominent. Age-dependent thymic involution restricts the output of recent thymic emigrants (TRTE) that highly express PTK724 and CD3125,26,27. This process of immune aging summarizes a collection of immune defects that accumulates over the course of aging, resulting not only from compartmental changes in the representation of immune cells—such as the gradual decline of naive28 and accumulation of memory differentiated T cells—but also loss of function through immune signaling defects that evolve from intrinsic cellular remodeling.
In this paper, we test the hypothesis that human aging—either chronological or “inflammation induced by chronic HIV infection”—affects TSCM renewal capacity. A combination of flow cytometry phenotyping, single-cell RNA sequencing, confocal imaging, and functional assays supports our inquiry. Our results demonstrate (i) a reduction in TSCM frequencies with age and chronic inflammation; (ii) aging compromises the Wnt/β-catenin signature in CD4 TSCM; (iii) inflammation and aging promotes the production of DKK-1 (a natural inhibitor of the Wnt/β-catenin pathway); and (iv) CD4 RTE are the most likely source of peripheral CD4 TSCM cells. Collectively, our data thus reveal a potential for the rejuvenation of the CD4 T-cell compartment through therapeutic targeting of Wnt/β-catenin pathways. Specifically, we may restore loss of TSCM function and diversity that is impacted by immunological history through the calibrated use of Wnt/β-catenin agonists.
Results
Depletion of TSCM CD4 cells during aging
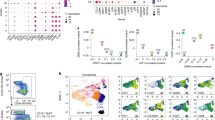
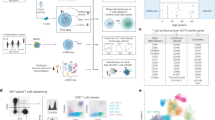
Despite an abundance of literature on the differentiation of CD4 T cells, the ontogeny of naive or early-stage memory CD4 T-cell subsets is poorly understood. Studies generally fail to appreciate their heterogeneity by grouping CD45RO−CCR7+CD27+CD62L+ CD4 T cells into a homogeneous TNAIVE cell compartment, despite their diverse expression of other functional T-cell markers (Supplementary Table 1). We hypothesize that compared with this global population of TNAIVE cells (CD45RO−CCR7+), TSCM, given their plasticity, are likely to be more heterogeneous and better sustained in older individuals to compensate for their reduced thymopoiesis. To illustrate this, we characterized T cells within the broad naive phenotype (Fig. 1; Supplementary Fig. 1A) into distinct populations using a combination of high-dimensional flow cytometry, molecular, and single-cell analysis with several analytical tools (including t-SNE, uMAP, Seurat, and diffusion map).
Workflow of high-dimensional analysis to characterize the heterogeneity of human CD4 TSCM cells. Arrows represent in vitro or in silico experiments, respectively. CCR7+CD45RO−CD27+ CD4 T cells from young (n = 4) and elderly donors (n = 4) were index sorted and distributed as single cells in 96-well plates (two plates per donor) (1). The MFI corresponding to surface protein expression for each individual cell was recorded for CD28, CD31, CD49d, CD95, CD122, CD127, CD150, CXCR3, CXCR4, and CCR7. A library of DNA sequences was established after the RNA extraction of each individual cell (2). The analysis of single-cell RNA-seq data was reported with t-SNE to identify the clusters of cells with similar profile of gene expression (3). Levels of CD95 protein expression were overlaid on the different clusters to identify clusters that were enriched in TSCM (4). The clusters corresponding to CD4 TSCM were extracted and re-analyzed by t-SNE (5). The heterogeneity of CD4 TSCM was validated with FACS Symphony high-dimensional flow cytometry staining (6).
First, CD4 TSCM frequencies demonstrated an even more pronounced age-associated trend than observed for TNAIVE cells (p < 0.0001, n = 43 and n = 166 for young and older donors, respectively, Fig. 2a), the latter may be linked to thymic atrophy as shown by the peripheral decrease of TRTE during aging (Supplementary Fig. 1B, C); we observed a similar trend for CD8 T cells (p < 0.0001, Supplementary Fig. 1D). Although both TSCM and TNAIVE frequencies were reduced, a correlation between the two population existed only in older individuals (Fig. 2b, n = 78, r = 0.7188, p < 0.0001), suggesting dysregulated homeostasis during aging.
a Depletion of TSCM CD4 cells during aging. Freshly isolated PBMCs were collected and stained for flow cytometry. The statistical analysis was performed on unpaired samples (U Mann–Whitney test) (**** for p < 0.0001). b Relationship between naive T-cell subsets during aging. The frequencies of TSCM and naive T cells were compared in young and old individuals (Spearman's rank-order test, p < 0.0001, r = 0.749). c Inflammation and aging. Pro-inflammatory molecules were measured in the plasma of young and older donors (n = 99 and n = 874, respectively). The statistical analysis was performed on unpaired samples (U Mann–Whitney test) (** and **** for p < 0.01 and p < 0.0001, respectively). d Rarefaction of CD31 expressing naive CD4 T cells and TSCM CD4 cells during aging and chronic HIV infection. Staining was performed on freshly collected blood of young (n = 28) and elderly donors (n = 70). Total CD4 T cells (right Y-axis) or CD4 T-cell subsets (left Y-axis) were enumerated during aging. Absolute counts were monitored in the peripheral blood of Malaysian cohort of healthy donors (n = 10) versus cART-treated HIV-infected patients (n = 6). The statistical analysis was performed on unpaired samples (U Mann–Whitney test; *, **, ***, and **** for p < 0.05, p < 0.01, p < 0.001, and p < 0.0001, respectively). e Restoration of CD4 T-cells distribution after successful HIV therapy. Longitudinal follow-up of CD4 T-cells subsets frequency was performed before and 48 weeks after the initiation of cART. The statistical analysis was performed on paired samples (Wilcoxon signed-rank test) (**, ***, and **** for p < 0.01, p < 0.001, and p < 0.0001, respectively). f Heterogeneity of CD4 TSCM by high-dimensional single-cell flow cytometry staining. CD4 TSCM cells of 20 donors were concatenated. CD4 TSCM clusters were visualized by phenograph and by a cold to hot heatmap, representing the intensity of each marker. Their distribution during aging was represented by the overlaid populations of CD4 TSCM from young and old patients. g Decreased of “RTE-like” CD4 TSCM cluster during aging. The frequency of CD31+PTK7+ TSCM CD4 cells was quantified by flow cytometry. The statistical analysis was performed on unpaired samples (Mann–Whitney, * for p < 0.05). Source data are provided as a Source Data file for all figures except (f).
A leading hypothesis is that enhanced inflammation and chronic infections such as HSV, CMV, dengue, or Helicobacter pylori during aging would affect immune homeostasis and contribute to pathology (Supplementary Table 2). Persistent stimulation of virus-specific TSCM CD4 cells might skew their differentiation toward an inflammatory-like state. Levels of pro-inflammatory molecules (Fig. 2c) are significantly elevated in older adults, which aligns with the concept of inflammaging; these elevations are also observed during HIV infection. We, respectively, demonstrate lower absolute CD31+ naive (including TRTE and TSCM) and TSCM CD4 T-cell counts in an independent aging (n = 98) and HIV-infected cohort (n = 16) (Fig. 2d; Supplementary Fig. 1E). This role of HIV in driving inflammation and CD4 depletion is supported by a reversal in the levels of systemic inflammation markers (Galectin-9, sCD163) and CD4 T-cell counts (and subsets)29 after HAART (Fig. 2e; Supplementary Fig. 1F). Although CD4 TSCM and TCM appeared most susceptible to HIV infection30, their recoveries were also most pronounced (p = 0.0004 and p < 0.0001, respectively; n = 14). Conversely, the frequencies of late-differentiated TEM was reduced (p < 0.0001, n = 14) by therapy. Overall, these results are consistent with the hypothesis that lifetime immunological history and levels of inflammation could alter the distribution of CD4 T-cell subsets, interfere with thymus activity31, and showed reversibility with the allieviation of inflammatory levels.
Next, we examined TSCM heterogeneity via high-dimensional flow cytometry. Here, several clusters of TSCM (from the pool of CCR7+CD45RA+CD27+CD95+) were identified in young and old donors (Fig. 2f, black/red plots, respectively). These clusters exhibited the differential expression of PTK7, CD31, CD127, CD150, and CXCR4. We observed that the representation of individual TSCM clusters was altered during aging. For instance, the population of TSCM co-expressing PTK7 and CD31 (RTE-like) was reduced in older donors (Fig. 2g, p < 0.05, n = 9 and n = 13 for young and elderly donors, respectively). The loss of TNAIVE, TRTE, and TSCM was exacerbated in older HIV patients, who also manifested the simultaneous enrichment of TMNP and TEM populations (Supplementary Fig. 1E). Finally, while HAART assists in the transient restoration of TSCM subsets, the absolute counts of TSCM did not normalize even after long-term therapy (Fig. 2d, e).
Heterogeneity of the TSCM and Wnt signaling pathway
In addition to the alteration of the above-mentioned markers during aging, we investigated the kinetics of additional markers using an unsupervised approach. Here, “naive” CD4 T cells (CD45RO−CCR7+CD27+CD62L+) were segregated into distinct populations based on the combinatorial expression of several differentiation-linked markers (Fig. 3a, b). Among ten clusters obtained from flow cytometry results, we were able to identify a cluster with a TSCM phenotype (CCR7+CD45RO−CD27+CD62L+CD122+CD95+, Cluster 5) that could be distinguished from immature TRTE (CCR7+CD45RO−CD27+CD62L+CD122−CD95−CD31high, Cluster 3), TNAIVE (CCR7+CD45RO−CD27+CD62L+CD122−CD95−CD31−, Cluster 10) and TMNP (CCR7+CD45RO−CD27+CD62L+CD122−CD95−CD31−CD49dhighCXCR3+, Cluster 8) cells. In order to demonstrate that these subsets were unique at the transcriptomic level, we performed RNA sequencing, including after single-cell index sorting. Regardless of age, CD4 TSCM appeared more genetically heterogeneous than other CD4 T-cell subsets (Fig. 3c, left panel and Supplementary Fig. 2A, B). TCD31negNAIVE, TRTE, TMNP, and TSCM CD4 T cells exhibited distinctive gene expression profiles (Fig. 3c, right panel), while non-RTE naive and TMNP clustered closely; the ontogenic proximity of the latter subsets can be corroborated by their coordinated modulation of CD28, CD122, CD150, and CXCR3 expression (Supplementary Figs. 1A, 2C, D).
a Heterogeneity of naive CD4 T cells revealed by high-dimension flow cytometry. Naive CD4 T cells of eight donors (four young and four old donors), defined as CCR7+CD45RO−CD27+, were concatenated and analyzed by t-SNE. Clusters were visualized by phenograph. b Phenotype of naive CD4 T-cell clusters. The intensity of fluorescence for each marker was visualized by a normalized heatmap (gradient of increased expression from blue to red). c Preservation of specific gene signature of CD4 TSCM cells during aging. Naive, TSCM, TCM, and TEM CD4 T cells (n = 5 for all subsets by age except TEM, n = 3) were sorted, and analyzed for their gene expression by nanostring. Naive T-cell subsets were sorted as naive, TRTE, TMNP, and TSCM CD4 cells and analyzed by RNA-seq (n = 5). PCA analysis of mRNA expression was performed to evaluate the specificity and preservation of T-cell subsets signature during aging. d Heterogeneity of the CD4 TSCM and Wnt/β-catenin signaling pathway. Identification of CD4 TSCM clusters in young and old donors by scRNAseq. Single cells corresponding to CD4 TSCM in all naive CD4 T cells subsets from young (n = 946) or old donors (n = 993) were analyzed by t-SNE, and clusters were automatically identified. The expression of genes coding for the Wnt/β-catenin pathway was quantified and normalized for each cluster in young and old donors. e Canonical and noncanonical Wnt signaling signatures in CD4 TSCM clusters from young donors. The enrichment of gene expression detected in CD4 TSCM clusters for each pathway was calculated in comparison with TRTE signature. Enrichment plot of the gene set reported by GSEA as most enriched among all canonical and noncanonical signaling pathway gene sets (GO:0016055). The profile shows running enrichment score (green curve) and positions of gene set members (black vertical bars) on the rank ordered list of differential gene expression. f Inflammatory signature in CD4 TSCM clusters from young and old donors. Enrichment plot of the gene set reported by GSEA as most enriched among all inflammatory gene sets (GO:0006954). The profile was displayed as in (e).
Based on this established TSCM phenotype (based on protein expression measured by flow cytometry during index sorting), we investigated whether genes associated with TSCM generation—specifically the canonical Wnt/β-catenin pathway (Fig. 3d)—were differentially expressed by different TSCM subsets and whether these patterns of expression were altered during aging. First, we observed that TSCM from the young separated into three clusters based on the expression of molecules associated with canonical Wnt/β-catenin signaling (Fig. 3d); this pattern of gene expression is also drastically altered during aging (Fig. 3d). In the gene set enrichment analysis (GSEA, Fig. 3e; Supplementary Fig. 3A) of TSCM transcripts from young donors (n = 4), we identified the enrichment of genes that overlapped with noncanonical Wnt–calcium and planar cell polarity (PCP) pathways (Supplementary Fig. 3B) in Clusters 0 and 1, while the expression of RNA transcripts within Cluster 2 overlapped most with canonical signaling (high TCF7, LRP5, GSK3B, CTNNB1, and FZD4 expression). The specific expression of NFATC1 and NLK or of RAC1 coupled with CDC42 and FZD6 suggested an increased engagement of the calcium and PCP pathway in Clusters 0 and 1, respectively. In addition to those listed in Fig. 2e, others genes are also involved in noncanonical signaling (Supplementary Fig. 3A). Notably, we found that TSCM from elderly donors (n = 4) exhibited relatively weaker canonical Wnt signatures (Supplementary Fig. 3B), as key molecules such as CTNNB1, FZD4, NFATC1, RAC1, CDC42, and FZD6 were not highly expressed in TSCM clusters from old donors. The complete analysis of gene signatures within TSCM clusters reveals pathways, which are not only unrelated to Wnt signaling (Supplementary Data File 1) but are also altered during aging.
Loss of Wnt signaling signature in TSCM CD4 and inflammation
A comparison of the mRNA libraries of CD4 TRTE and TSCM from young and old donors indicates that these subsets may be inprinted with a pro-inflammatory signature with age (Supplementary Fig. 3C–E; elevated levels of TNF, CD40, and NFκB); this was further validated by nanostring (p = 0.0067 and p = 0.0165, n = 10 for NFκB2 and NFκBIZ, Supplementary Fig. 2A). We further observed that ontology clusters that reflect modulations in activation and functional capacity were differentially enriched in TRTE and TSCM with age (Supplementary Fig. 4A, B). These results led us to postulate that a heightened inflammatory signature, demonstrated by changes in NFκB and TNF signaling for all clusters from older donors (Fig. 3f), could contribute to dysregulated CD4 TSCM homeostasis through altered Wnt signaling.
Together with observations from HIV-infected donors, we found evidence which suggest that heightened levels of inflammation and concomitant changes in the levels of homeostatic cytokines32,33,34 could disrupt the genetic signatures of TSCM and their further differentiation. Thus, we examined whether the inflammatory environment—induced by aging or chronic inflammation—interferes with CD4 T-cell development or disrupts TSCM homeostasis. As shown above, CD4 TSCM, and TRTE frequencies were positively correlated and could therefore be responsive to overlapping homeostatic mediators. In studying the effect of homeostatic, inflammatory, and effector mediators in healthy and HIV donors (Supplementary Fig. 4C, D), we observed that the expression of inflammatory molecules was negatively associated with the prevalence of CD4 TSCM (n = 113, p = 0.046, r = −0.21 for IL-8; n = 23, p = 0.0292, r = −0.4364 for IL-21; n = 99, p = 0.0063, r = −0.2727 for sCD163; n = 113, p = 0.0065, r = −0.2546 for sCD14, n = 98, p = 0.0118, r = −0.2787 for neopterin). Among the different pro-inflammatory cytokines measured, only IFN-γ was positively associated with CD4 TSCM frequencies (n = 76, p = 0.0004, r = 0.3967). As previously described for IL-7 and naive T cells during HIV infection32, IL-21 concentration was also negatively associated with percentages of CD4 TRTE (n = 23, r = −0.5332 and r = −0.5953, respectively). This difference in correlation values between TSCM and TRTE suggests a difference in sensitivity to the systemic inflammatory environment, which is also highlighted by the increased expression of activation markers by these subsets (HLA-DR and Ki-67, Supplementary Fig. 4E). We next investigated how these molecular changes could be dynamically translated into functional or developmental features.
Functional erosion of TSCM CD4 cells during aging
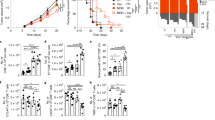
In light of our findings, we tested whether TSCM-specific functional adaptations occur during aging. While the main transcriptomic signatures of TSCM, TCM, TEM, and TNAIVE cells remain consistent during aging35, genes encoding transcription factors (LEF1, TCF7, TBX21, FOXP3, IRF4, BATF, RORC, and NOTCH1), cytokines (IL-5, IL-21, IL-17A, and TGF-β), cytokine receptors (IL7R, SOCS1, TNF, and IL2R), chemokines (CCL3, CCL4, CCL15, and CCL20) and effector molecules (GNLY, GZMB, and PRF1) were modulated (Supplementary Fig. 2A, B) with age. Thus, we measured the ability of young and aged CD4 T-cell subsets to proliferate in response to TCR or homeostatic stimulation. As expected, the highest proliferative potential in healthy young donors was observed in TSCM cells (Fig. 4a), but TSCM cells from older adults showed compromised proliferative capacity (p = 0.0205, n = 15)—even when compared with TCM cells (Fig. 4b). Moreover, the induction of TSCM proliferation by IL-7, and the secretion of IL-2, IFN-γ, and TNF by proliferating TSCM, were impaired in older donors (Supplementary Fig. 5A).
a Proliferation profile of CD4 T-cell subsets during aging. Representative histogram of CFSE dilution from sorted T-cell subsets collected in young or older donors and stimulated with anti-CD3/CD28 microbeads or IL-7 (10 ng/ml) during 5 days. b Alteration of proliferative potential of TSCM CD4 cells in response to TCR stimulation as measured in (a). T-cells subsets were freshly isolated from blood of young and older donors (n = 8 and 9, respectively). The statistical analysis was performed on unpaired samples (U Mann–Whitney test) (* for p < 0.05). Source data are provided as a Source Data file. c Decreased secretion of homeostatic and effector cytokines by TSCM CD4 cells during aging. Sorted CD4 T-cell subsets were polyclonally stimulated with PMA/Ionomycin. The cytokines concentration was represented by an heatmap to visualize the acquisition of effector functions during differentiation and the specific signature associated with aging (n = 6 for young and old donors). Source data are provided as a Source Data file. d Increased engraftment of human TSCM CD4 cells from aged donors in humanized NOD SCID gamma chain (NSG) mice. TSCM CD4 cells from young (n = 2) or old donors (n = 2) were differentiated from T-naive precursors, expanded in vitro before their xenotransplantation into NSG mice (n = 13 and n = 9, respectively). At 21 (Exp#1) or 28 (Exp#2) days after the human TSCM CD4 cells transfer, animals were killed and euthanized by CO2, and tissues were collected (spleen, lungs). N = 2 independent experiments were performed and labeled by the color code on the graph (filled circles: Exp#1; open circles: Exp#2; filled gray circle represents the mouse with GVHD signs and killed at day 16). The statistical analysis was performed on unpaired samples (U Mann–Whitney test) (** for p < 0.01). e Reduced CDR3 diversity in naive CD4 T cells. Naive T-cell subsets were sorted as naive, TRTE, TMNP, and TSCM CD4 cells. The extraction of mRNA was performed just after T-cell sorting, and analyzed by RNA-seq. CDR3 composition was compared between cell subsets and during aging. A connective arc represented high degree of homology (80%) between CDR3 sequences during differentiation and aging.
Finally, we studied the expression of three key markers of T-cell differentiation/activation—Ki-67, T-bet, and Eomes (Supplementary Fig. 5B). Here, we found that TCR stimulation of TSCM induced similar T-bet expression (corresponding to their effector functions), but lower Eomes and Ki-67 acquisition than in TCM. Moreover, TSCM from FACS-sorted and polyclonally stimulated CD4 subsets from elderly (Fig. 4c) revealed diminished IL-2 and IL-21 secretion (p = 0.0101 and p = 0.0303, respectively, n = 10); the release of effector cytokines (IFN-γ and TNF) (p = 0.0415 and p = 0.0186, respectively) and pro-inflammatory molecules (IL-17A and CCL20) were also significantly reduced (p = 0.0496 and p = 0.0062, respectively).
Preservation of TSCM proliferation is unrelated to clonality
Since the in vitro functionality of CD4 TSCM was modified with age, we rationalized that the capacity for asymmetric division and proliferation by CD4 TSCM could also be affected in vivo. NOD SCID Gamma (NSG) mice were separately transplanted with human CD4 TSCM that were generated, in vitro, from TNAIVE CD4 precursors from young and old donors (Fig. 4d; Supplementary Fig. 5C). Surprisingly, the engraftment of human CD4 TSCM cells was more efficient for both lymphoid (spleen, p < 0.01) and nonlymphoid tissue (lung, p < 0.01) when cells were obtained from old rather than young donors (Fig. 4d)—this suggested that intrinsic age-associated factors related to tissue migration or homeostatic function in CD4 TSCM could affect the frequency and location of peripheral TSCM. We tracked the time-sensitive development of transplanted cells (31 days after TSCM transplantation) and observed that the fate of transplanted TSCM cells located within three main cellular clusters, corresponding to TCM, TSCM, and late transitional memory cells (Supplementary Information). Altogether, our data show that transplanted CD4 TSCM from both young and old donors could persist in their hosts and differentiate into both TTM and TCM—albeit with the possible requirement that they are inducted into a younger noninflammatory environment.
Finally, since TCR diversity is typically lost in T-cell subsets with age36,37, we wanted to test whether CD4 TSCM are subjected to the the same fate. Here, we fitted RNA-Seq data into a Circos plot to visualize changes in β-chain V–(D)–J rearrangement of TSCM with age (Fig. 4e) through the use of Circos plots. Here, we observed that the greatest TCR diversity was observed in CD4 TMNP—independently of age—and that the loss of heterogeneity in TCR clonotypes was most significant in both CD4 TRTE and TSCM from elderly donors.
Regulation of TSCM CD4 cells homeostasis: the Wnt/DKK-1 axis
The dysregulation of the Wnt/β-catenin pathway during aging was further validated at the proteomic level. TCF-1, a main transcription factor within this signaling pathway has been associated with the stemness of CD8 T cells38,39,40 and SLAMF-6 (CD352) expression41,42. We attempted to validate the relationship between TCF-1 and SLAMF-6 and observed positive correlations between TCF-1 and SLAMF-6 expression only in TRTE and TSCM from young donors (n = 20) (Fig. 5a–c); TCF-1 and SLAMF-6 have distinct dynamics during CD4 T-cell differentiation and aging. This observed age-associated decline in TCF-1 expression and decoupling with SLAMF-6 expression could reflect perturbations within the extended Wnt-dependent regulatory network that governs TSCM homeostasis. Such disturbances include an increased frequencies of autoantibodies—indirect predictors of signaling activity and immune response43,44—against several protein targets that act in the Wnt/β-catenin pathway with age (n = 60 and n = 93 for young and older donors, respectively), including CTTNB1 (p < 0.0001), GSK3B (p < 0.01), TCF-4 (p < 0.01), LEF1 (p < 0.01), IRF4 (p = 0.05), HDAC1 (p < 0.0001), and HDAC3 (p = 0.0064) (Fig. 5d; Supplementary Fig. 6A). Moreover, plasma concentrations of DKK-1, a natural inhibitor of the Wnt/β-catenin pathway, were increased during aging (Fig. 5e; p < 0.0001 and n = 78); while levels of SFRP1, a natural activator of Wnt/β-catenin pathway, were reduced (p < 0.0001 and n = 80). Both circulating levels of SFRP1 and DKK-1 correlated with CD4 TSCM frequencies (Fig. 5f).
a TCF-1 and SLAMF-6 expression in CD4 T cells. Representative zebra plots of SLAMF-6 and TCF-1 staining in CD4 T cells from a representative old individual. b TCF-1 and SLAMF-6 expression in TSCM CD4 cells during aging. Representative overlaid histograms plots of SLAMF-6 and TCF-1 expression in gated TSCM CD4 cells from young (n = 10) and older (n = 10) individuals. c Decreased expression of TCF-1 during CD4 T-cell differentiation and aging. The median fluorescence intensity of TCF-1 was measured in T-cell subsets. The statistical analysis was performed on paired (n = 20, Wilcoxon signed-rank test) or unpaired samples (n = 20, Mann–Whitney; *, **, ***, and **** for p < 0.05, p < 0.01, p < 0.001, and p < 0.0001, respectively). Source data are provided as a Source Data file. d Alternative activation of Wnt/β-catenin pathway by DKK-1 during aging. Cryopreserved plasma was used to measure autoantibodies directed against molecules involved in the Wnt/β-catenin pathway (n = 93 and n = 60 in young and old, respectively). The statistical analysis of immunone protein array data was performed on unpaired samples (U Mann–Whitney test, **** for p < 0.0001). Source data are provided as a Source Data file. e Modulation of the natural inhibitor and agonist of the Wnt/β-catenin pathway during aging. The plasmatic concentration of DKK-1 and SFRP1 was measured directly by ELISA (n = 43 and n = 37 in young and old donors, respectively). The statistical analysis was performed on unpaired samples (U Mann–Whitney test, **** for p < 0.0001). Source data are provided as a Source Data file. f Regulation of TSCM CD4 cells by DKK-1 and SFRP1. The frequency of TSCM CD4 cells correlated negatively or positively with the systemic concentration of DKK-1 and SFRP1, respectively (p = 0.0003 and p = 0.0118) (n = 77). The correlations were calculated with the Spearman’s rank-order test. Source data are provided as a Source Data file. g Inflammation and DKK-1 plasma levels. The concentration of DKK-1, sCD14, sCD163, and IL-26 was measured directly by ELISA. Plasma levels of tryptophan and L-kynurenine were measured by LC-MS/MS. The correlations were calculated with the Spearman’s rank-order test. Source data are provided as a Source Data file.
Concomitantly, age-associated differences in SFRP1 and DKK-1 activity may interact with the loss of TCF-1 expression to restrict signaling within the canonical Wnt/β-catenin pathway and limit the persistence/expansion of TSCM CD4 cells. We studied this phenomenon in HIV-infected patients, who are often used as a model of inflammatory aging. Higher concentrations of DKK-1 were detected in the plasma of older HIV donors (Supplementary Fig. 6B, p < 0.0001 and n = 97) and HAART concomitantly reduced inflammation and circulating DKK-1 (p = 0.0068 and n = 12). These improvements in inflammatory and DKK-1 profiles in treated donors suggested that antiretroviral therapy could promote TSCM differentiation via these mechanisms. We demonstrate further associations between DKK-1 and cellular markers of immune activation (sCD14, p = 0.0093, r = 0.2728, n = 90; IL-26, p = 0.0245, r = 0.2306, n = 95; IDO activity, p = 0.0008, r = 0.3362, n = 97; sCD163, p = 0.0056, r = 0.2795, n = 97) in Fig. 5g. Altogether, the latter strengthens our hypothesis that the age-related impairments of TSCM homeostasis could be result from meta-inflammation that affects Wnt/β-catenin signaling through DKK-1. Of note, our transcriptomic analysis of the Wnt/β-catenin pathway reveals that DKK-1 gene expression (as observed in tumors45) was only enriched in Cluster 1 TSCM from young donors (“PCP enriched”); CDH1 (another Wnt/β-catenin inhibitor), which is involved in noncanonical Wnt signaling for all donors (Supplementary Fig. 6C), was also dominantly expressed within the TSCM cluster. Among other genes related to Wnt/β-catenin inhibition in older donors, only SFRP5 and DACT1 were, respectively, upregulated in Clusters 0 (“PCP like”) and 1 (“Wnt/β-catenin like”). The increased DKK-1 activity, is therefore, unlikely to be due to an intrinsic expression of DKK-1 by TSCM of aged donors.
RTE CD4 T cells are fitter precursors of TSCM
The inhibition of glycogen synthase kinase-3β by TWS119 was shown to promote the in vitro activation of the Wnt/β-catenin pathway in naive T cells, which led to the generation of TSCM9. Borrowing this approach, we attempted to generate inducible CD4 TSCM (iTSCM), and observed that this process was significantly less efficient in older donors (p = 0.0098, n = 20) (Fig. 6a). Increasing the dosage of TWS119 (10 μM) ameliorated the loss of expansion capacity of CD4 TSCM (p = 0.007, n = 10)—suggesting that the reported dose of 5 μM may be insufficient to overcome the age/inflammatory-associated inhibition of this pathway. Congruent with the ex vivo phenotype of isolated TSCM, iTSCM had a CD62LhighCD45RA+CD45RO−/+CD95+ phenotype. We performed further phenotypic analysis to compare ex vivo and iTSCM in donors from different age groups (Fig. 6b). The increased expansion of iTSCM with higher concentrations of TWS119 was also associated with CD127 recovery (p = 0.0317, n = 5). Moreover, TWS119-induced CD4 TSCM presented with lower levels of activation markers: CD95 (p = 0.0013), CD26 (p = 0.0079), CCR5 (p = 0.0159), CXCR3 (p = 0.0391) and maintained high levels of CCR7 expression compared to control cells (p = 0.0079) (Supplementary Fig. 7A); altogether suggestive of a younger phenotype with greater proliferative, functional, and trafficking potential. The phenotypic similarities between iTSCM and ex vivo CD4 TSCM, in terms of surface marker expression (Supplementary Fig. 7B), can be corroborated by their compatible genetic signatures8,15,16,18.
a Hyporesponsiveness of the Wnt/β-catenin pathway during aging. The frequency of specifically TWS119-induced TSCM CD4 cells was evaluated by flow cytometry, and calculated after the subtraction of nonspecific DMSO-induced TSCM CD4 cells frequencies. The statistical analysis was performed on unpaired samples (U Mann–Whitney test, ** for p < 0.01). Source data are provided as a Source Data file. b CD127 expression characterizes induced-TSCM CD4 cells. The phenotype of induced-TSCM CD4 T cells was performed at day 7. The statistical analysis was performed on paired samples (Wilcoxon signed-rank test) (* and ** for p < 0.05 and p < 0.01, respectively). Source data are provided as a Source Data file. c Identification of recent thymic emigrants in cord blood or in peripheral blood of young and old donors by flow cytometry. TRTE were defined as PTK7+CD31+CD4 T cells. d Induction of TSCM CD4 cells from TRTE cells. The correlations between ex vivo TRTE and in vitro TWS119-induced TSCM CD4 cells at day 7 were calculated with the Spearman’s rank-order test (n = 15). Source data are provided as a Source Data file. e TSCM CD4 cells induction depends on CD31 expression in naive CD4 T cells. Flow cytometry staining of TWS119 dose-dependent induced-TSCM CD4 T cells derived from CD31− or CD31high naive CD4 T cells. f Increased potential of CD31high naive to TSCM CD4 cells differentiation. Frequencies of TSCM CD4 cells derived from CD31− or CD31high naive CD4 T cells in response to the stimulation of the Wnt/β-catenin pathway with low dose of TWS119 (5 μM). Young and old donors are represented with black and gray symbols, respectively. The statistical analysis was performed on paired samples (Wilcoxon signed-rank test) (***for p < 0.001). Source data are provided as a Source Data file. g Phenotype of induced TSCM from TRTE and CD31low naive CD4 T cells. Flow cytometry staining of induced-TSCM CD4 cells derived from CD31− or CD31high (TRTE) naive CD4 T cells. Histograms represented overlaid expression of individual markers after 7 days of culture in presence of vehicle alone (DMSO) or Wnt/β-catenin stimulating drug (TWS119) at 5 or 10 μM.
The parallel fates—and ontogenetic proximity—of TNAIVE and TSCM cells suggested that the age-related decline of thymic function is likely to contribute to the loss of peripheral TSCM. Furthermore, a decay in the expression of CD31 and PTK7 expression in CD4 T cells from cord blood to young followed by older donors (Fig. 6c), supports a hypothesis that lifelong immune stimulation contributes to CD31 shedding46. We also observed a positive correlation between the frequencies of TRTE cells (defined as PTK7+CD31+) at day 0 and the frequencies of induced TSCM at day 7 (Fig. 6d), supporting the idea that TSCM could be derived from TRTE cells. This hypothesis was evaluated in silico by STEM analysis of naive CD4 T-cell subsets (Supplementary Fig. 7C, D). The dynamics of gene expression between TRTE and TSCM CD4 cells was a more plausible outcome than comparisons of other putative ontogenic pathways. Next, Monocle was used to generate pseudotime from single-cell RNA-seq data and determine CD4 T-cell differentiation trajectory- our transcriptome supports a model where TRTE could differentiate into TSCM and CD31neg naive CD4 T cells (Supplementary Fig. 7E).
Due to the overlapping features of TRTE and TSCM with age, particularly the loss of Wnt/β-catenin pathway activity with age—we speculated that high CD31 expression could reduce the threshold of naive CD4 T cells to differentiate into TSCM via the Wnt/β-catenin activation (Fig. 6e). We observed that, independent of donor age, 5 μM of TWS119 concentration could efficiently mediate the induction of FACS-sorted CD31high naive CD4 T cells into TSCM CD4 cells (iTSCM) (Fig. 6f, p = 0.0008 and n = 9). Conversely, CD31neg naive CD4 T cells were resilient to conversion at 5 μM concentration of TWS119, but responded when 10 μM was used (p = 0.0007 and n = 9). In monitoring the expression of CD45RA, CCR7, CD127, and CD27 on CD31neg naive CD4 T cells, we also obtained evidence that TWS119 could promote a more “naive” phenotype in CD31neg naive CD4 T cells at higher dosage (Fig. 6g).
Metabolism of TSCM CD4 cells during aging
Several groups have described the age-related hypermethylation of genes (IFNG, CCR7, CD27, etc.) that result in functional changes in naive T-cell behavior22,47,48. Guided by these principles, we studied whether groups of genes were simultaneously modulated (grouped by behavioral profile) with progressive T-cell differentiation (i.e., from TRTE to TTMNP) using STEM analysis (Supplementary Fig. 7C). We identified the top canonical pathways that were associated with T-cell differentiation in different age groups, as well as the respective upstream regulators that were responsible for the observed phenotype. T-cell differentiation was associated with different metabolic profiles between the young and old, suggesting differences in energy management with age. For example, in young donors, anabolic pathways such as diacylglycerol and phosphatidylglycerol biosynthesis were modulated with T-cell differentiation (Cluster 11), while genes involved in catabolic processes such as oxidative phosphorylation were highlighted in older donors. We also observed differences in Cluster 5, where major shifts in the regulation of acid biosynthesis (glutamine, serine, and glycine) and glycogen biosynthesis were observed in young and elderly donors, respectively (Cluster 5; Supplementary Fig. 7D). In examining the signaling targets that are altered with progressive naive CD4 T-cell differentiation, we observed possible alterations in the activation of specific signaling and metabolic pathways (RhoA, Sirtuin, mTOR, and MYC). These canonical pathways are regulated by upstream regulators, which were distinct for each age group within the same clusters of concordantly regulated genes. We detected the naive T-cell differentiation could be differentially guided by the influence of homeostatic cytokines (STAT5A) as well as by the environment through the alternate engagement of viral sensors (IRF3, IFNB1, and IL12B) within the two age groups. For example, the energetic requirements for the development (TSC22D3, POU2F2), differentiation, or acquisition of effector functions (TSC22D3, IRF3, and LEPR for Th17 cells) are specific to each CD4 T-cell subset. The priming and differentiation of naive CD4 T cells are thus coupled with specific changes in gene expression and metabolic gene signature during aging.
Polarization of TSCM CD4 cells during aging
In addition to phenotypic and molecular dissimilarities, we endeavored to identify morphological and structural changes that may develop in TSCM with age as a possible response to the differential engagement of Wnt signaling pathways (PCP in particular and possibly due to DKK-1) with age—as any visible differences in their surface architecture could also help to explain differences in TSCM behavior. We investigated on the potential implication of the Wnt pathway in the CD4 TSCM polarization. The atypical expression of CDC42 in Wnt/β-catenin cluster in TSCM from old donors (Supplementary Fig. 3B) led us to propose that the orchestration of cytoskeletal events, including the distribution of proteins associated with polarity, might be impaired in the elderly. However, TCR-mediated stimulation led to the expected unipolar recruitment of Cdc42 in CD4 T cells from young donors, but such polarization was infrequent in aged donors (Supplementary Fig. 8A, B). The latter was particularly the case for CD31− naive CD4 T cells, but this trend was also observed for TCM and TSCM cells, albeit absent in CD31high naive CD4 T cells (TRTE).
Due to the distinct polarization profiles of naive CD4 T-cell subsets, we sought to determine whether the main regulator and source of chemical energy, i.e., the mitochondria, behaved differently in CD4 TSCM cells during aging49,50 (Supplementary Fig. 8C). We observed a reduction in the average mitochondrial volume (but not of mitochondria numbers, Supplementary Fig. 8D) in TSCM CD4 cells in the elderly as compared with young donors (p < 0.05) (Supplementary Fig. 6D).
Overall, these multidimensional changes in the patterns of TSCM gene and protein expression advocate strongly for the argument that systemic changes in the frequency and function of TSCM cells in the elderly could to a large extent, be explained by disturbances to the cellular environment (summarized in Fig. 7).
Graphical summary of key molecules involved in the specific signature of individual TSCM cluster identified by scRNAseq gene expression analysis. Gray, blue, and green boxes represented highlighted molecules in Wnt/calcium, Wnt/β-catenin, and Wnt/PCP, respectively. Red arrows and characters summarized the alterations of Wnt signaling observed in aged donors and/or chronic inflammation described in this study.
Discussion
Naive CD4 T cells are a heterogeneous population in terms of gene expression, phenotype, and function, and are divided into subclasses that respond differently to external signals—such as chronic infection, vaccination, and inflammaging. The various inflammatory contexts examined in this study demonstrate that CD4 TSCM and their progenitors are sensitive to the external environment. Immune activation induced by persistent infections such as HIV and CMV may imprint specific behavior to CD4 TSCM cells. The clonal expansion of differentiated virus-specific T cells may also indirectly shape T-cell repertoire and therefore limit the responsiveness to future challenges. In this study, we demonstrate a quantitative and qualitative (proliferation, effector function) defect in CD4 TSCM cells during aging and chronic infections. We also provide multiple evidence to show that persistent inflammation could indeed interfere with the functioning of these subsets at the single-cell level—these changes were accompanied by changes to Wnt/β-catenin gene expression, and associated with specific proteomic and metabolic signatures. Essentially, while all naive T cell can differentiate, the most likely precursors of CD4 TSCM cells appear to reside in the TRTE compartment, which is itself severely compromised in the contexts of aging (reduced thymopoeisis, inflammation) and chronic infections (clonal expansion of memory T cells, which may compete for space and resources). Immune activation, TLR stimulation, and the binding of innate viral sensors may also activate putative upstream TFs that act to orchestrate biased T-cell differentiation in the elderly, possibly via DKK-1 modulation51. Inflammation could thus affect CD4 TSCM cells directly and indirectly even at the RTE precursor stage.
In describing the extent of CD4 TSCM depletion that accompanies aging and chronic inflammation induced by HIV infection, and linking these phenomena to immune activation and the Wnt/β-catenin pathway in this phenomenon—we propose that modulation in the gene expression of TSCM cells, which manifest most strikingly in their impact on metabolic and signaling pathways—could be significantly explained by alterations in the inflammatory environment (Fig. 7). This age-dependent signature of TSCM could contribute to sub-optimal TSCM differentiation and increased susceptibility to cellular senescence via a mechanism that is independent of antigenic source and linked to the nature of the inflammatory environment. Thus, we demonstrate that the sub-optimal immune response that is observed during aging and HIV infection may evolve partly from the loss of CD4 TSCM heterogeneity through altered Wnt signaling engagement.
Our conclusions are further substantiated by observations that CD8 TSCM depletion is been associated with disease progression, in the contexts of HIV52,53 or symptomatic Trypanosoma cruzi infections54; the generation of antigen-specific CD4 TSCM cell with more diverse effector molecules (Granzyme A, K) and chemokines (CCR5, CCR6) profiles during acute Mycobacterium tuberculosis infection also contributes to a better prognosis55. The importance of having adequate CD4 TSCM heterogeneity is also supported by the results from the RV144 prime-boost HIV-vaccine trial56. Similarly, a selective depletion of CCR5+ TSCM CD4 cells was observed in pathogenic but not in non-pathogenic simian immunodeficiency virus infections57. The loss of these memory precursors could be due to HIV replication in CD4 TSCM, which in turn affects the ability of the host to repopulate the entire library of T cells and to control current and further infections58.
Whether the alternate engagement of canonical or noncanonical Wnt/β-catenin signaling contributes to heterogeneous clusters of CD4 TSCM warrants further investigation. The ambiguous role of the Wnt pathway activation in HIV replication59 or in the establishment of HIV latency could result from differences in how TSCM are distributed within the clusters described in this study. Alternatively, HIV prognosis may be affected by the availability of DKK-1 or virus- derived molecules like HIV TAT and YAP that could interact with Wnt signaling pathways. The dynamic flux of TSCM has been recently demonstrated by tracking genetically modified TSCM60, and using stable isotopes and heavy water61, which supports a model where the TSCM reservoir is continuously replenished and maintained by continuous TSCM turnover in the absence of any telomeric erosion due to the active expression of telomerase. However, the kinetics of human TSCM also follows a dichotomous distribution, with two sub-populations described as either short- or long-lived (5 months versus 9 years, respectively)11. These segmentation contributes to three speculations: (i) the alternate Wnt pathway engagement may affect the longevity of these different sub-populations, (ii) these short- and long-lived-TSCM may arise from TNAIVE and CD31high naive T cells, respectively, and (iii) TSCM heterogeneity may be driven by the type of antigen. The integration of environmental signals by TSCM and their proliferative capacity could trigger homeostatic adjustments that preserve T-cell compartmentalization57. The age-associated decline of TSCM cells may therefore compromise this plasticity and the entire memory immune capacity.
Next, the diversity and clonal availability of TSCM were also found to be crucial for generating recall responses62 and for the long-term maintenance of adult T-cell leukemia63. However, CDR3 diversity in TSCM is reduced during aging and, in the absence of cognate epitopes, TSCM persistence depends heavily on homeostatic proliferation (mediated through IL-7, IL-15, or IL-21) and the differentiation of scarce TRTE11. An increase in IL-7 levels was further correlated to the loss of naive64 and CD4 TRTE cells65, which are both predictive of poor HIV prognosis64,65,66. Furthermore, co-infection with HCV presents an additional burden that contributes to the loss of TRTE67. The reduced survivability and persistence of TRTE and TSCM CD4 cells in the elderly and during HIV infection are likely to be related to both the increased presence of inflammatory cytokines and homeostatic IL-7 levels. In support of this hypothesis, we observed differences in CD127 expression within TSCM clusters that strongly express molecules associated with canonical Wnt/β-catenin signaling. CD127 is considered as a marker of long-term memory68. Here, we show this role could evolve through the promotion of Wnt/β-catenin signaling, which leads to enhanced CD4 TSCM proliferative capacity. The loss of CD127 has been further described as a hallmark effect of inflammation, and we confirm this through the context of HIV-associated immune aging. Moreover, some studies have described an increased infectivity of CD4 TSCM58,69 and TRTE by HIV (or SIV in the case of the Rhesus Macaque) in progressors57. A preserved TRTE compartment is also associated with higher CD4 nadir66. That HIV could gain an evolutionary advantage by undermining CD4 TSCM and TRTE function suggests their importance in the control of viral replication. Moreover, the reconstitution of CD4 TSCM accompanies successful HAART administration—whether this is a cause or effect of successful HIV control warrants further investigation.
The other striking observation in this study was the increased hyporesponsiveness of the Wnt/β-catenin pathway and the concomitant loss of active Wnt/β-catenin genetic signature at the single-cell level during aging and HIV infection. In addition to driving the TSCM differentiation via the route of CD31high CD4 T cells, stimulation of the Wnt/β-catenin pathway with high dosage agonist promoted the acquisition of a CD4 TSCM phenotype even in CD31− naive CD4 T cells, which often possess a homeostatic proliferation history. While the resistance of total naive T cells to iTSCM differentiation in aged donors was linked to lower TRTE frequencies, we observed that TRTE were most pliant to the Wnt/β-catenin pathway stimulation, since they responded regardless of the donor’s age, and with minimum agonist dosage. The preservation (or acquisition) of CD127 on naive CD4 T cells was also found to be a reliable indicator of the ease of iTSCM induction. Thus, data from these experiments suggest that the pliability of CD4 TRTE to TSCM differentiation erodes progressively from the time when CD4 TRTE egress from the thymus and that such a phenomenon may be due to alterations in Wnt/β-catenin signaling. Importantly, our results show that there is clinical potential in targeting Wnt/β-catenin signaling to promote the in vivo genesis of CD4 TSCM.
Our data consistently reveal an age- or inflammation-dependent dysregulation in the balance of natural agonists and antagonists (DKK-1/SFRP1) of the Wnt/β-catenin pathway and an increased prevalence of autoantibodies against members of canonical Wnt/β-catenin pathway signaling (CTTNB1, GSK3B, IRF4, and HDAC1). All these factors could further contribute to the hyporesponsiveness of this pathway in CD4 TSCM from elderly donors, which dampens downstream T-cell functions. That loss of CD4 TSCM integrity can be mediated through compromised Wnt/β-catenin signaling can be supported by the observed overexpression of DKK-1 in various cancers45,70 and by Tregs in the contexts of autoimmune disease and colitis71. It was suggested that the inhibitory nature of DKK-1 can be directly influenced by a tumor suppressor gene or indirectly via the induction of myeloid-derived suppressor cells. Accordingly, DKK-1 has been proposed as a target for immune therapy and anti-DKK-1 vaccination was shown to strengthen antitumor immunity72.
Finally, the aberrant morphological changes in CD4 naive, TSCM, and TCM subsets during TCR engagement are indicative that elderly T cells are unable to orchestrate appropriate physiological responses to critical signaling events. Among immature CD4 T-cell subsets from the elderly, spatial organization following TCR engagement was only preserved in TRTE. A similar characteristic was observed in HSCs, and this phenomenon was closely related to the noncanonical engagement of the Wnt/β-catenin pathway73. Cdc42, a Rho GTPase, is involved in the immunological synapse formation and involved in signal transduction, acto-myosin organization, cell proliferation, and Wnt/PCP and Wnt/Ca2+ noncanonical pathways74,75. Defective Cdc42 expression, the observed loss of polarity and mitochondrial regulation may therefore contribute to the reduced fitness and metabolic activity of CD4 TSCM cells during aging.
The paucity of TRTE in elderly—driven both by thymic involution and immune activation—results in a ticking time-bomb scenario where responses to new and repeat infections are increasingly crippled by the loss of memory T-cell precursors and naive T-cell heterogeneity. Nevertheless, our data suggest that modulation of the Wnt/β-catenin pathway by introducing high levels of agonist (or better agonists) may be helpful in delaying or preventing the latter scenario; this is an attractive proposal as alternative methods (such as thymic rejuvenation) have been shown to result in adverse effects. The importance of CD4 TSCM in promoting survival and immune competency remains largely unexplored, since most studies have focused on CD8 TSCM. In the CD8 context, TSCM have been shown to be important for preventing GVHD, promoting antitumor immunity8,13 and is required for mounting adequate responses to a plethora of pathogens. The stemness of TSCM CD8 T cells also correlates with the long-term persistence of yellow fever -specific CD8 TSCM during successful vaccination76. With regard to CD4 function, our data support the idea that the preservation of CD4 TSCM frequencies is also important for immune competency—particularly in the context of viral control. Thus, promoting CD4 TSCM differentiation could, likewise, be relevant to enhancing vaccine efficacy; this is logical as TSCM constitute an important reservoir for the development of memory T-cell responses. Altogether, our data suggest that strategies that reduce the exposure of CD4 T cells to systemic inflammation may be instrumental to preserving TSCM immunocompetency.
Methods
Donors and sample preparation
Blood was collected from participants of the Singapore Longitudinal Aging Study (SLAS) cohort. Characteristics of the SLAS cohort are detailed in the Supplementary Table 2 and in our previous publication, related to physical frailty77. Blood was collected into BD Vacutainer CPT Cell Preparation tubes with Sodium Citrate (BD Biosciences, San Jose, CA, USA). After centrifugation at 300 rcf for 20 min at room temperature, plasma and peripheral blood mononuclear cells (PBMCs) were isolated. Plasma was stored at −80 °C before use. PBMCs were frozen in 90% fetal bovine serum (FBS) containing 10% DMSO and stored in liquid nitrogen.
The study has been approved by the National University of Singapore-Institutional Review Board 04–140 and all participants gave informed consent. Young donors were recruited at the National University of Singapore. The study has been approved by the Ethics Committee of the NUS-IRB 09-256. All study participants provided informed written consent.
Blood was collected from HIV-infected individuals attending the University Malaya Medical Centre (UMMC), Malaysia. Data on HIV-specific characteristics including HIV RNA, CD4 T-cell counts, antiretroviral drug history, and history of co-infections were obtained from patient medical records. The study was approved by the hospital institutional review board for Malaysian HIV-infected patients (MEC 975.6).
All experiments using human buffy coats were approved by the Humanitas Clinical and Research Institute IRB (approval 28/01/2016).
Animal studies
All experiments using mice were conducted upon the approval of the Italian Ministry of Health (protocols 256/2015-PR). The permission to perform animal experiments was granted by the Italian Ministry of Health. NOD.Cg-Prkdcscid IL2rgtm1Wjl/SzJ (NSG) mice (Jackson Laboratories) were bred in specific-pathogens-free (SPF) conditions.
Screening of serum
All serum samples of the entire SLAS cohort were first tested for IgG antibodies against H. pylori, EBV, VZV, HSV1, HSV2, CMV, by anti-enzyme-linked immunosorbent assay (ELISA) using commercial test kits (Virion\Serion, Germany) according to the manufacturer’s recommended procedure. IgG antibodies against CHIKV and DENV were detected by ELISA from Abcam (USA) and Panbio (USA), respectively.
Phenotyping of PBMCs
Cell phenotyping was performed by flow cytometry on 23 fresh PBMC samples from young donors and 78 fresh PBMC samples from older donors. For each staining, 1 × 106 PBMCs were used. Lymphocytes were gated based on FSC/SSC profile and doublets/dead cell exclusion.
Absolute cell count was performed by flow cytometry on freshly collected blood of ten healthy donors and six HAART HIV-infected patients. For each staining, 100 μL of blood was used in Trucount tubes. After doublets/dead cell exclusion, lymphocytes were gated based on FSC/SSC profile and CD45 expression.
The antibodies are listed in Supplementary Table 3. Flow cytometry was performed on an LSR Fortessa Cell Analyzer (BD Biosciences), and automatic compensation was applied.
We used BDSymphony A5 (BD Biosciences) to perform high-dimensional single cells immunophenotyping and characterize the heterogeneity of naive and TSCM CD4 cells from 2 million frozen PBMCs.
Flow cytometry of HIV participants was performed on a BD FACS Celesta (BD Biosciences) at University of Malaya and automatic compensation was applied.
Flow cytometry functional assay
Proliferation: CFSE-stained sorted CD4 T-cell subsets were stimulated with anti-CD3/CD28 microbeads or IL-7 during 5 and 7 days, respectively. Proliferation was measured through the dilution of CFSE. Proliferation/ICS: after 5 days of TCR stimulation, CFSE-stained sorted CD4 T-cell subsets were re-stimulated with PMA/Ionomycin (1 µg/ml and 100 ng/ml, respectively) during 4 h to detect the secretion of cytokines by proliferating CD4 T cells. Brefeldin A and Monensin (eBioscience) were added during the final 2 h of incubation. For the list of antibodies used, refer to Supplementary Table 3. Flow cytometry was performed on an LSR Fortessa Cell Analyzer (BD Biosciences).
Multiplex analytes screening
Cell sorting was performed with a FACS Aria III (BD Biosciences) on 12 samples according to CCR7, CD27, CD31, CD45RO, CD49d, CXCR3, and CD95 expression in CD4 T cells. For the list of antibodies used for sorting, refer to Supplementary Table 3. After 18-h incubation with PMA/Ionomycin, supernatants were collected and tested by Luminex assay. The Milliplex HTH17MAG-14K (Millipore) was used according to the manufacturer’s instructions and signal detected by Flexmap.
The Milliplex HCYTOMAG-60K (Millipore) was used according to the manufacturer’s instructions to measure cytokines in the plasma of young and old donors.
ELISA
Soluble CD14 and Galectin-9 (R&D Systems), soluble CD163 and IL-21 (eBioscience), IL-26, DKK-1, and SFRP1 (USCN) were measured according to the manufacturer's instructions. Plasma samples of HIV-infected patients and corresponding controls were Triton-inactivated and diluted in the appropriate buffer and assayed in duplicate.
Autoantibody profiling using the ImmunomeTM protein arrays
Plasma samples were assayed using the ImmunomeTM protein array (Sengenics Corporation, Singapore)78. The array contains quadruplicate spots of 1627 full-length, correctly folded, and fully functional immobilized self- and cancer proteins. These include cancer antigens (mainly cancer–testis antigens (CTAs)), transcription factors, kinases, signaling proteins, and others. Raw data were processed and normalized using a robust customized pipeline78.
In vivo transfer into NSG mice of induced TSCM CD4 cells
Seven days before the transfer, CD4 naive T cells were FACS sorted from aged (n = 2) and young (n = 2) healthy control’s PBMC as CD45RO–CCR7+CD27+CD95– and activated with aCD3/aCD28 magnetic beads (Invitrogen) (1:2 bead:cells ratio) in the presence of IL-7 and IL-15 (10 ng/ml each, Peprotech). Purity of sorted naive CD4+ T cells was >97% (not shown). At day 0, magnetic beads were detached and in vitro generated CD4 TSCM-enriched cells (8 × 106/mouse) were co-transfer with (5–10 × 106) CD4-depleted autologous PBMCs obtained by negative magnetic separation with MACS beads (Miltenyi). Mice were weighed every week. Three (day 21; Exp#1) or 4 (day 28; Exp#2) weeks after the transfer, mice were killed, spleens and lungs were collected, weighed, dissociated into single-cell suspension, stained with fluorochrome-conjugated antibodies and analyzed by flow cytometry (LSR Fortessa, BD).
In vitro induction of TSCM CD4 cells
CD4 naive T cells were FACS sorted from aged (n = 15) and young (n = 25) healthy donor’s PBMC as CD45RO–CCR7+CD27+CD95– and activated with aCD3/aCD28 magnetic beads (Invitrogen) (1:2 bead:cells ratio) in the presence of DMSO or TWS119 (5 and 10 μM). At day 7, magnetic beads were detached, and in vitro-induced CD4 TSCM were studied for their phenotype and gene expression.
Quantitative real-time PCR
Sorted CD4 T-cell subsets were immediately lysed. RNA extraction was performed using an RNeasy Plus Micro kit (Qiagen) and reverse transcribed into cDNA using the SuperScript First Strand kit (Invitrogen). cDNA was analyzed by real-time PCR with the KAPA SYBR qPCR Master Mix kit (KAPA Biosystems) or TAQMAN. The following primers were provided by Qiagen: BATF (QT00078449), IRF4 (QT00065716), HDAC1 (QT00015239), PCNA (QT00024633), or by TAQMAN: LEF1 (Hs01547250_m1), TCF7 (Hs01556515_m1), and Notch1 (Hs01062014_m1).
nCounter Human Inflammation v2
Direct mRNA expression levels of the samples were measured using the NanoString nCounter gene expression system. In all, 18,125–20,714 sorted CD4 T-cell subsets in 5 μL of RLT buffer from Qiagen RNeasy Mini kit (Qiagen, Hilden, Germany) were hybridized with probes from the nCounter Human Inflammation v2 panel (Nanostring, Seattle, USA) at 65 °C for 16–19 h according to the nCounter™ Gene Expression Assay Manual. Excess probes were washed away using a two-step magnetic bead-based purification on the nCounter™ Prep Station (GEN1). The nCounter™ Digital Analyzer (GEN1) was used to count individual fluorescent barcodes and quantify target molecules present in each sample. For each assay, a high-density scan (600 fields of view) was performed.
RNA-seq
The total RNA was extracted following the double-extraction protocol: RNA isolation by acid guanidinium thiocyanate–phenol–chloroform extraction (TRIzol, Thermo Fisher Scientific, Waltham, MA, USA) followed by a Qiagen RNeasy Micro clean-up procedure (Qiagen, Hilden, Germany). All human RNAs were analyzed on the Agilent Bioanalyzer for quality assessment with RNA integrity number (RIN) range from 6.2 to 9.6 and median RIN 8.9 (Agilent, Santa Clara, CA, USA). cDNA libraries were prepared using 1 ng of the total RNA and 0.5 µl of a 1:50,000 dilution of ERCC RNA Spike in Controls (Ambion Thermo Fisher Scientific, Waltham, MA, USA) using SMARTSeq v2 protocol79, except for the following modifications: (1) use of 20 µM TSO, (2) use of 250 pg of cDNA with 1/5 reaction of Illumina Nextera XT kit (Illumina, San Diego, CA, USA). The length distribution of the cDNA libraries was monitored using DNA High Sensitivity Reagent Kit on the Perkin Elmer Labchip (Perkin Elmer, Waltham, MA, USA). All samples were subjected to an indexed PE sequencing run of 2 × 51 cycles on an Illumina HiSeq 2000 (16 samples/lane). The paired-end reads were mapped to Human GRCh38 reference genome using the STAR alignment tool. The number of reads mapped to each gene was counted using feature Counts (part of Subread package) and GENCODE gene annotation version V25.
Single cells RNA-seq
Single-cell cDNA libraries were using the SMARTSeq v2 protocol79 with the following modifications: (1) 1 mg/ml BSA lysis buffer (Ambion Thermo Fisher Scientific, Waltham, MA, USA); (2) use of 250 pg of cDNA with 1/5 reaction of Illumina Nextera XT kit (Illumina, San Diego, CA, USA). The length distribution of the cDNA libraries was monitored using a DNA High Sensitivity Reagent Kit on the Perkin Elmer Labchip (Perkin Elmer, Waltham, MA, USA). All samples were subjected to an indexed paired-end sequencing run of 2 × 51 cycles on an Illumina HiSeq 2000 system (Illumina, San Diego, CA, USA) (192 samples/lane). Pair-endraw reads were aligned to human reference genome using RSEM version 1.3.0. Human reference genome version 25 released by Gencode was used (https://www.gencodegenes.org/human/release_25.html). Transcript Per Million read(TPM) values were calculated using RSEM version 1.3.0 and used for downstream analysis.
Confocal microscopy
Mitochondria contents: Sorted cells were stained with 100 nm MitoTracker Green (Life Technologies) and 1 µg/ml Hoechst 33258 for 1 h in a humidified incubator. Prior to imaging, cells were washed and resuspended in R-10 before plating in an eight-well glass-bottom µ-plate (ibidi, Germany). Images were taken using the FV-1000 confocal microscope system (Olympus) under controlled temperature conditions using a 60× oil objective. Mitochondrial staining was analyzed using Imaris (Bitplane, Switzerland).
Cdc42 polarization: Frozen sorted naive CD4 T cells subsets were thawed and activated during 2 h at 37 °C with reversible anti-CD3/CD28 Streptamers (IBA Lifesciences, Germany). T cells were seeded on fibronectin-coated glass coverslips in PBS + 10% FBS. After 2 h of incubation at 37 °C (5% CO2) in RPMI-20% FCS with 1% antibiotics, cells were fixed with 4% PFA. After fixation cells were gently washed with PBS, permeabilized with 0.2% Triton X-100 (Sigma) in PBS for 20 min, and blocked with 10% donkey serum (Sigma) for 30 min. Primary and secondary antibody incubations were performed overnight at 4 °C and for 1 h at room temperature, respectively. Coverslips were mounted with ProLong Gold Antifade Reagent with or without DAPI (Invitrogen, Molecular Probes). The cells were coimmunostained with an anti-alpha tubulin antibody (Abcam, rat monoclonal ab6160) detected with an anti-rat AMCA-conjugated secondary antibody or an anti-rat DyLight488-conjugated antibody (Jackson ImmunoResearch), an anti-Cdc42 antibody (Millipore, rabbit polyclonal). Samples were imaged with an AxioObserver Z1 microscope (Zeiss) equipped with a 63 × PH objective. Images were analyzed with AxioVision 4.6 software. Alternatively, samples were analyzed with an LSM710 confocal microscope (Zeiss) equipped with a 63× objective. Primary raw data were imported into the Volocity Software package (Version 6.0, Perkin Elmer) for further processing and conversion into 3D images. As for polarity scoring, the localization of each single-stained protein was considered polarized when a clear asymmetric distribution was visible by drawing a line across the middle of the cell. A total of 50–100 naive CD4 T cells were singularly analyzed per sample. The data are plotted as percentage of the total number of cells scored per sample.
Scanning electron microscopy: For imaging by scanning electron microscopy, sorted cells were fixed in 2.5% glutaraldehyde in 0.1 M phosphate buffer for 1 h (pH 7.4) at room temperature, treated post fixation with 1% osmium tetroxide (Ted Pella Inc) at room temperature for 1 h, and then dehydrated through a graded ethanol series from 25 to 100% and critical point dried using a CPD 030 critical point dryer (Bal-Tec AG, Liechtenstein). The cell surfaces on which the cells were grown coated with, and the adhesive surface was coated with 15 nm of gold by sputter coating using a SCD005 high-vacuum sputter coater (Bal-Tec AG). The coated samples were examined with a field emission JSM-6701F Scanning Electron Microscope (JEOL Ltd., USA) at an acceleration voltage of 8 kV using the in-lens secondary electron detector.
Data analysis
Flow cytometry data were analyzed using FlowJo (Treestar) and FACSDiva (BD Biosciences). Samples were compared using GraphPad Prism software (v.8.0c). Unbiased t-SNE analysis of flow cytometry data: unbiased representations of multiparameter flow cytometry data were obtained using the t-distributed stochastic neighbor-embedding (t-SNE) algorithm. t-SNE is a nonlinear dimensionality reduction method that optimally locates cells with similar expression levels near to each other and cells with dissimilar expression levels further apart. t-SNE and UMAP analysis and were performed using FlowJo, custom R scripts, and Cytofkit software. Gene expression: Metascape, STEM (Short Time-series Expression Matrix) analysis were performed on the differentially expressed genes between naive T-cell subsets and during aging. For differentially expressed gene (DEG) analysis of RNA-seq data, comparisons between each condition of young and old donors were performed using edgeR, and DEGs were chosen with Benjamini–Hochberg adjusted p-values < 0.05. RPKM (reads per kilobase of transcript per million mapped reads) values were computed using edgeR R package version 3.3.2 by filtering the genes that have zero total count and by normalizing the count with TMM algorithm. The averages of RPKM values by condition have been used for STEM analysis. TCR repertoire analysis was performed with MiXCR. Similar amount of mRNA from sorted naive T-cell subsets and identical sequence reads were analyzed to normalize the results. scRNAseq: Young and older donors have been analyzed separately. For both, the donor effect was erased by removing some genes differentially expressed between donors, 183 genes for young donors and 216 genes for old donors, in order to match the density plots of each donor onto the first two PCs. These removed genes were not involved in the population differentiation. We filtered out low-quality cells from our data set based on a threshold for the number of genes detected (a minimum of 200 unique genes per cell). All genes that were not detected in at least 0.4% of all our single cells were discarded, leaving 22,116 genes for young donors and 21,786 for old donors for all further analyses. Normalized data were log-transformed—log(expression+1)—for all downstream analyses. t-SNE and clustering of cells were performed using Seurat R package (https://github.com/satijalab/seurat). FACS data were transformed with a logicle transformation. To summarize the expression of FACS data into each cluster found with the single-cell RNA-sequencing data, a “cluster score” was calculated as the average protein expression in each cluster and row scaled before generating a heatmap using the pheatmap R package. In order to determine the enrichment score for each cluster of TSCM CD4 cells population regarding the Wnt signaling pathway (GO:0016055) and the inflammatory response pathway (GO:0006954), we performed a gene set enrichment analysis (GSEA v3.0). The pathways were retrieved from AMIGO2 website. Monocle analysis was performed using the monocle R package version 2.1080.
Statistical analysis
Groups of young and elderly donors were analyzed by Mann–Whitney U test to compare values. The Wilcoxon matched-pairs signed-rank test was used for paired testing of median values of different subsets from the same donor. Analysis with p < 0.05 (*), p < 0.01 (**), p < 0.001 (***), and p < 0.0001 (****) were considered significantly different between the groups.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The authors declare that the data supporting the findings of this study are available within the paper (and its supplementary information files). The source data underlying Figs. 2a–g, 4b–c, 5c–g, and 6a–f and Supplementary Figs. 1, 2, 3, 5, 6, and 7 are provided as a Source Data file. The RNA-Seq/scRNAseq have been deposited in in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE143215 (including sub-series GSE143214 and GSE143213). The data that support the other findings of this study are available from the corresponding author upon reasonable request.
References
Fearon, D. T., Manders, P. & Wagner, S. D. Arrested differentiation, the self-renewing memory lymphocyte, and vaccination. Science 293, 248–250 (2001).
Graef, P. et al. Serial transfer of single-cell-derived immunocompetence reveals stemness of CD8(+) central memory T cells. Immunity 41, 116–126 (2014).
Demkowicz, W. E. Jr., Littaua, R. A., Wang, J. & Ennis, F. A. Human cytotoxic T-cell memory: long-lived responses to vaccinia virus. J. Virol. 70, 2627–2631 (1996).
Hammarlund, E. et al. Duration of antiviral immunity after smallpox vaccination. Nat. Med. 9, 1131–1137 (2003).
Capece, T. et al. A novel intracellular pool of LFA-1 is critical for asymmetric CD8(+) T cell activation and differentiation. J. Cell Biol. 216, 3817–3829 (2017).
Nish, S. A. et al. CD4+ T cell effector commitment coupled to self-renewal by asymmetric cell divisions. J. Exp. Med. 214, 39–47 (2017).
Paley, M. A. et al. Progenitor and terminal subsets of CD8+ T cells cooperate to contain chronic viral infection. Science 338, 1220–1225 (2012).
Gattinoni, L. et al. A human memory T cell subset with stem cell-like properties. Nat. Med. 17, 1290–1297 (2011).
Gattinoni, L. et al. Wnt signaling arrests effector T cell differentiation and generates CD8+ memory stem cells. Nat. Med. 15, 808–813 (2009).
Lugli, E. et al. Identification, isolation and in vitro expansion of human and nonhuman primate T stem cell memory cells. Nat. Protoc. 8, 33–42 (2013).
Del Amo, P. C. et al. Human TSCM cell dynamics in vivo are compatible with long-lived immunological memory and stemness. PLoS Biol. 16, e2005523 (2018).
Lugli, E. et al. Superior T memory stem cell persistence supports long-lived T cell memory. J. Clin. Investig. 123, 594–599 (2013).
Gattinoni, L., Speiser, D. E., Lichterfeld, M. & Bonini, C. T memory stem cells in health and disease. Nat. Med. 23, 18–27 (2017).
Cieri, N. et al. IL-7 and IL-15 instruct the generation of human memory stem T cells from naive precursors. Blood 121, 573–584 (2013).
Sabatino, M. et al. Generation of clinical-grade CD19-specific CAR-modified CD8+ memory stem cells for the treatment of human B-cell malignancies. Blood 128, 519–528 (2016).
Kondo, T. et al. Notch-mediated conversion of activated T cells into stem cell memory-like T cells for adoptive immunotherapy. Nat. Commun. 8, 15338 (2017).
Scholz, G. et al. Modulation of mTOR signalling triggers the formation of stem cell-like memory T cells. EBioMedicine 4, 50–61 (2016).
Pulko, V. et al. Human memory T cells with a naive phenotype accumulate with aging and respond to persistent viruses. Nat. Immunol. 17, 966–975 (2016).
Quinn, K. M. et al. Age-related decline in primary CD8(+) T cell responses is associated with the development of senescence in virtual memory CD8(+) T cells. Cell Rep. 23, 3512–3524 (2018).
van den Broek, T., Borghans, J. A. M. & van Wijk, F. The full spectrum of human naive T cells. Nat. Rev. Immunol. 18, 363–373 (2018).
White, J. T. et al. Virtual memory T cells develop and mediate bystander protective immunity in an IL-15-dependent manner. Nat. Commun. 7, 11291 (2016).
Mitchell, C. J. et al. A multi-omic analysis of human naive CD4+ T cells. BMC Syst. Biol. 9, 75 (2015).
den Braber, I. et al. Maintenance of peripheral naive T cells is sustained by thymus output in mice but not humans. Immunity 36, 288–297 (2012).
Haines, C. J. et al. Human CD4+ T cell recent thymic emigrants are identified by protein tyrosine kinase 7 and have reduced immune function. J. Exp. Med. 206, 275–285 (2009).
Kimmig, S. et al. Two subsets of naive T helper cells with distinct T cell receptor excision circle content in human adult peripheral blood. J. Exp. Med. 195, 789–794 (2002).
Kohler, S. & Thiel, A. Life after the thymus: CD31+ and CD31- human naive CD4+ T-cell subsets. Blood 113, 769–774 (2009).
van den Broek, T. et al. Neonatal thymectomy reveals differentiation and plasticity within human naive T cells. J. Clin. Investig. 126, 1126–1136 (2016).
Kilpatrick, R. D. et al. Homeostasis of the naive CD4+ T cell compartment during aging. J. Immunol. 180, 1499–1507 (2008).
Kared, H. et al. Adaptive NKG2C(+)CD57(+) natural killer cell and Tim-3 expression during viral infections. Front. Immunol. 9, 686 (2018).
Chahroudi, A., Silvestri, G. & Lichterfeld, M. T memory stem cells and HIV: a long-term relationship. Curr. HIV/AIDS Rep. 12, 33–40 (2015).
Douek, D. C., Koup, R. A., McFarland, R. D., Sullivan, J. L. & Luzuriaga, K. Effect of HIV on thymic function before and after antiretroviral therapy in children. J. Infect. Dis. 181, 1479–1482 (2000).
Napolitano, L. A. et al. Increased production of IL-7 accompanies HIV-1-mediated T-cell depletion: implications for T-cell homeostasis. Nat. Med. 7, 73–79 (2001).
Younes, S. A. et al. IL-15 promotes activation and expansion of CD8+ T cells in HIV-1 infection. J. Clin. Investig. 126, 2745–2756 (2016).
Pallikkuth, S., Parmigiani, A. & Pahwa, S. The role of interleukin-21 in HIV infection. Cytokine Growth Factor Rev. 23, 173–180 (2012).
Phan, A. T., Goldrath, A. W. & Glass, C. K. Metabolic and epigenetic coordination of T cell and macrophage immunity. Immunity 46, 714–729 (2017).
Britanova, O. V. et al. Age-related decrease in TCR repertoire diversity measured with deep and normalized sequence profiling. J. Immunol. 192, 2689–2698 (2014).
Qi, Q. et al. Diversity and clonal selection in the human T-cell repertoire. Proc. Natl Acad. Sci. USA 111, 13139–13144 (2014).
Brummelman, J. et al. High-dimensional single cell analysis identifies stem-like cytotoxic CD8(+) T cells infiltrating human tumors. J. Exp. Med. 215, 2520–2535 (2018).
Jeannet, G. et al. Essential role of the Wnt pathway effector Tcf-1 for the establishment of functional CD8 T cell memory. Proc. Natl Acad. Sci. USA 107, 9777–9782 (2010).
Kratchmarov, R., Magun, A. M. & Reiner, S. L. TCF1 expression marks self-renewing human CD8(+) T cells. Blood Adv. 2, 1685–1690 (2018).
Miller, B. C. et al. Subsets of exhausted CD8(+) T cells differentially mediate tumor control and respond to checkpoint blockade. Nat. Immunol. 20, 326–336 (2019).
Utzschneider, D. T. et al. T cell factor 1-expressing memory-like CD8(+) T cells sustain the immune response to chronic viral infections. Immunity 45, 415–427 (2016).
Haller-Kikkatalo, K. et al. Demographic associations for autoantibodies in disease-free individuals of a European population. Sci. Rep. 7, 44846 (2017).
Leslie, D., Lipsky, P. & Notkins, A. L. Autoantibodies as predictors of disease. J. Clin. Investig. 108, 1417–1422 (2001).
Kagey, M. H. & He, X. Rationale for targeting the Wnt signalling modulator Dickkopf-1 for oncology. Br. J. Pharmacol. 174, 4637–4650 (2017).
Fornasa, G. et al. TCR stimulation drives cleavage and shedding of the ITIM receptor CD31. J. Immunol. 184, 5485–5492 (2010).
Abdelsamed, H. A. et al. Human memory CD8 T cell effector potential is epigenetically preserved during in vivo homeostasis. J. Exp. Med. 214, 1593–1606 (2017).
Berkley, A. M., Hendricks, D. W., Simmons, K. B. & Fink, P. J. Recent thymic emigrants and mature naive T cells exhibit differential DNA methylation at key cytokine loci. J. Immunol. 190, 6180–6186 (2013).
Buck, M. D. et al. Mitochondrial dynamics controls T cell fate through metabolic programming. Cell 166, 63–76 (2016).
Sukumar, M. et al. Mitochondrial membrane potential identifies cells with enhanced stemness for cellular therapy. Cell Metab. 23, 63–76 (2016).
Osada, M. et al. DKK1 mediated inhibition of Wnt signaling in postnatal mice leads to loss of TEC progenitors and thymic degeneration. PLoS ONE 5, e9062 (2010).
Ribeiro, S. P. et al. The CD8(+) memory stem T cell (T(SCM)) subset is associated with improved prognosis in chronic HIV-1 infection. J. Virol. 88, 13836–13844 (2014).
Vigano, S. et al. Prolonged antiretroviral therapy preserves HIV-1-specific CD8 T cells with stem cell-like properties. J. Virol. 89, 7829–7840 (2015).
Mateus, J. et al. Low frequency of circulating CD8+ T stem cell memory cells in chronic chagasic patients with severe forms of the disease. PLoS Negl. Trop. Dis. 9, e3432 (2015).
Mpande, C. A. M. et al. Functional, antigen-specific stem cell memory (TSCM) CD4(+) T cells are induced by human mycobacterium tuberculosis. Infect. Front. Immunol. 9, 324 (2018).
Wang, Y. et al. A novel mechanism linking memory stem cells with innate immunity in protection against HIV-1. Infect. Sci. Rep. 7, 1057 (2017).
Cartwright, E. K. et al. Divergent CD4+ T memory stem cell dynamics in pathogenic and nonpathogenic simian immunodeficiency virus infections. J. Immunol. 192, 4666–4673 (2014).
Klatt, N. R. et al. Limited HIV infection of central memory and stem cell memory CD4+ T cells is associated with lack of progression in viremic individuals. PLoS Pathog. 10, e1004345 (2014).
Kumar, A. et al. Active beta-catenin signaling is an inhibitory pathway for human immunodeficiency virus replication in peripheral blood mononuclear cells. J. Virol. 82, 2813–2820 (2008).
Biasco, L. et al. In vivo tracking of T cells in humans unveils decade-long survival and activity of genetically modified T memory stem cells. Sci. Transl. Med. 7, 273ra213 (2015).
Ahmed, R. et al. Human stem cell-like memory T cells are maintained in a state of dynamic flux. Cell Rep. 17, 2811–2818 (2016).
Miyama, T. et al. Highly functional T-cell receptor repertoires are abundant in stem memory T cells and highly shared among individuals. Sci. Rep. 7, 3663 (2017).
Nagai, Y. et al. T memory stem cells are the hierarchical apex of adult T-cell leukemia. Blood 125, 3527–3535 (2015).
Aguilera-Sandoval, C. R. et al. Supranormal thymic output up to 2 decades after HIV-1 infection. AIDS 30, 701–711 (2016).
Fabre-Mersseman, V. et al. CD4(+) recent thymic emigrants are infected by HIV in vivo, implication for pathogenesis. AIDS 25, 1153–1162 (2011).
Zakhour, R. et al. Recent thymus emigrant CD4+ T cells predict HIV disease progression in patients with perinatally acquired HIV. Clin. Infect. Dis. 62, 1029–1035 (2016).
Shmagel, K. V. et al. Influence of hepatitis C virus coinfection on CD4(+) T cells of HIV-infected patients receiving HAART. AIDS 28, 2381–2388 (2014).
Kaech, S. M. et al. Selective expression of the interleukin 7 receptor identifies effector CD8 T cells that give rise to long-lived memory cells. Nat. Immunol. 4, 1191–1198 (2003).
Buzon, M. J. et al. HIV-1 persistence in CD4+ T cells with stem cell-like properties. Nat. Med. 20, 139–142 (2014).
Qian, J. et al. Dickkopf-1 (DKK1) is a widely expressed and potent tumor-associated antigen in multiple myeloma. Blood 110, 1587–1594 (2007).
Chae, W. J. et al. Membrane-bound Dickkopf-1 in Foxp3(+) regulatory T cells suppresses T-cell-mediated autoimmune colitis. Immunology 152, 265–275 (2017).
Qian, J. et al. Active vaccination with Dickkopf-1 induces protective and therapeutic antitumor immunity in murine multiple myeloma. Blood 119, 161–169 (2012).
Florian, M. C. et al. A canonical to non-canonical Wnt signalling switch in haematopoietic stem-cell ageing. Nature 503, 392–396 (2013).
Chemin, K. et al. Cytokine secretion by CD4+ T cells at the immunological synapse requires Cdc42-dependent local actin remodeling but not microtubule organizing center polarity. J. Immunol. 189, 2159–2168 (2012).
Guo, F. et al. Coordination of IL-7 receptor and T-cell receptor signaling by cell-division cycle 42 in T-cell homeostasis. Proc. Natl Acad. Sci. USA 107, 18505–18510 (2010).
Fuertes Marraco, S. A. et al. Long-lasting stem cell-like memory CD8+ T cells with a naive-like profile upon yellow fever vaccination. Sci. Transl. Med. 7, 282ra248 (2015).
Lu, Y. et al. Inflammatory and immune markers associated with physical frailty syndrome: findings from Singapore longitudinal aging studies. Oncotarget 7, 28783–28795 (2016).
Suwarnalata, G. et al. Augmentation of autoantibodies by Helicobacter pylori in parkinson’s disease patients may be linked to greater severity. PLoS ONE 11, e0153725 (2016).
Picelli, S. et al. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9, 171–181 (2014).
Trapnell, C. et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 32, 381–386 (2014).
Acknowledgements
We thank Ivy Low (Flow Cytometry Platform), Esther Mok (Multiplex Analysis Platform), Alicia Seok Wei Tay, and Foo Shihui (Genomic Platform) from SIgN Immunomonitoring Platform (BMRC IAF 311006 grant and BMRC transition funds #H16/99/b0/011). We thank BD Biosciences and BioMarketing Services (BMS) for the generous loan of a BD FACS celesta at University of Malaya. We would like to acknowledge Dr. Robert Balderas (BD Biosciences) for his input in antibody panel design and high-dimensional flow cytometry experiments. We thank Dr. Etienne Becht and Dr. Evan W. Newell for the customized t-SNE and UMAP scripts for flow cytometry unbiased analysis. We thank Dr. Petronela Ancuta (University of Montreal) and Dr. Laurent Reina (SIgN) for the critical reading of the paper. The study is supported by a research grant from the Agency for Science, Technology and Research (No. 10-036), by the Singapore Immunology Network and by a Starting Grant from the European Research Council (ERC-StG-2014 PERSYST 640511 to E.L.). A.L. is a scholar of International Society for Advancement of Cytometry (ISAC). R.R. and A.K. are funded by the High Impact Research/Ministry of Higher Education Research Grant, Malaysia (HIR/MOHE; H-20001-E000001) and the RU grant (UMRG RP029-14HTM). E.R-M was supported by Consejería de Salud y Bienestar Social of Junta de Andalucía through the Nicolás Monardes Program (C-0032/17) and Fondo de Investigación Sanitaria, Instituto de Salud Carlos III, Fondos Europeos para el Desarrollo Regional, FEDER, grants PI16/00684, PI19/01127, RETICS, Red de Investigación en SIDA (RD16/0025/0020).
Author information
Authors and Affiliations
Contributions
Conceptualization, H.K. and A.L.; experimental work, H.K., S.W.T., C.T., M.S., B.M., K.P., V.Z. and J.L.; samples acquisition, R.R., A.K., N.M.G., F.G., E.L. and T.P.N.; data analysis, H.K., M.C.L., M.C., W.H., B.L., A.A., M.C.F., J.M.C., B.M., E.R.M., T.F., H.G. and E.L.; writing—original the draft, H.K.; writing—review and editing, A.L. and G.W.; supervision, H.K. and A.L.; funding acquisition, A.L., T.P.N. and R.R.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kared, H., Tan, S.W., Lau, M.C. et al. Immunological history governs human stem cell memory CD4 heterogeneity via the Wnt signaling pathway. Nat Commun 11, 821 (2020). https://doi.org/10.1038/s41467-020-14442-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-020-14442-6
This article is cited by
-
Homeostatic cytokines tune naivety and stemness of cord blood-derived transgenic T cells
Cancer Gene Therapy (2022)
-
Hallmarks of T cell aging
Nature Immunology (2021)
-
The GSK3β-β-catenin-TCF1 pathway improves naive T cell activation in old adults by upregulating miR-181a
npj Aging and Mechanisms of Disease (2021)
-
Immunosenescence: a key player in cancer development
Journal of Hematology & Oncology (2020)
-
The untwining of immunosenescence and aging
Seminars in Immunopathology (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.