Abstract
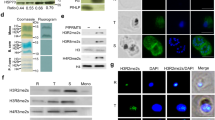
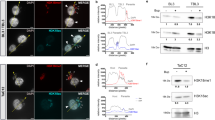
Malaria pathogenesis results from the asexual replication of Plasmodium falciparum within human red blood cells, which relies on a precisely timed cascade of gene expression over a 48-h life cycle. Although substantial post-transcriptional regulation of this hardwired program has been observed, it remains unclear how these processes are mediated on a transcriptome-wide level. To this end, we identified mRNA modifications in the P. falciparum transcriptome and performed a comprehensive characterization of N6-methyladenosine (m6A) over the course of blood-stage development. Using mass spectrometry and m6A RNA sequencing, we demonstrate that m6A is highly developmentally regulated, exceeding m6A levels known in any other eukaryote. We characterize a distinct m6A writer complex and show that knockdown of the putative m6A methyltransferase, PfMT-A70, by CRISPR interference leads to increased levels of transcripts that normally contain m6A. In accordance, we find an inverse correlation between m6A methylation and mRNA stability or translational efficiency. We further identify two putative m6A-binding YTH proteins that are likely to be involved in the regulation of these processes across the parasite’s life cycle. Our data demonstrate unique features of an extensive m6A mRNA methylation programme in malaria parasites and reveal its crucial role in dynamically fine-tuning the transcriptional cascade of a unicellular eukaryote.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
All sequencing data are accessible on the Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/geo) under study accession number GSE123839. Raw sequence data are accessible on the NCBI Sequence Read Archive (SRA) under BioProject accession number PRJNA473770. Proteomics and mRNA modification LC-MS/MS data are deposited at the Chorus database with accession number 1579.
References
World Malaria Report 2018 (World Health Organization, 2018).
Bozdech, Z. et al. The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum. PLoS Biol. 1, 85–100 (2003).
Scherf, A., Lopez-Rubio, J. J. & Riviere, L. Antigenic variation in Plasmodium falciparum. Annu. Rev. Microbiol. 62, 445–470 (2008).
Kafsack, B. F. C. et al. A transcriptional switch underlies commitment to sexual development in malaria parasites. Nature 507, 248–252 (2014).
Salcedo-Amaya, A. M. et al. Dynamic histone H3 epigenome marking during the intraerythrocytic cycle of Plasmodium falciparum. Proc. Natl Acad. Sci. USA 106, 9655–9660 (2009).
Kensche, P. R. et al. The nucleosome landscape of Plasmodium falciparum reveals chromatin architecture and dynamics of regulatory sequences. Nucleic Acids Res. 44, 2110–2124 (2016).
Toenhake, C. G. et al. Chromatin accessibility-based characterization of the gene regulatory network underlying Plasmodium falciparum blood-stage development. Cell Host Microbe 23, 557–569 (2018).
Ganesan, K. et al. A genetically hard-wired metabolic transcriptome in Plasmodium falciparum fails to mount protective responses to lethal antifolates. PLoS Pathog. 4, e1000214 (2008).
Llinás, M., Bozdech, Z., Wong, E. D., Adai, A. T. & DeRisi, J. L. Comparative whole genome transcriptome analysis of three Plasmodium falciparum strains. Nucleic Acids Res. 34, 1166–1173 (2006).
Painter, H. J. et al. Genome-wide real-time in vivo transcriptional dynamics during Plasmodium falciparum blood-stage development. Nat. Commun. 9, 2656 (2018).
Hughes, K. R., Philip, N., Starnes, G. L., Taylor, S. & Waters, A. P. From cradle to grave: RNA biology in malaria parasites. WIRES RNA 1, 287–303 (2010).
Vembar, S. S., Droll, D. & Scherf, A. Translational regulation in blood stages of the malaria parasite Plasmodium spp.: systems-wide studies pave the way. WIRES RNA 7, 772–792 (2016).
Shock, J. L., Fischer, K. F. & DeRisi, J. L. Whole-genome analysis of mRNA decay in Plasmodium falciparum reveals a global lengthening of mRNA half-life during the intra-erythrocytic development cycle. Genome Biol. 8, R134 (2007).
Foth, B. J., Zhang, N., Mok, S., Preiser, P. R. & Bozdech, Z. Quantitative protein expression profiling reveals extensive post-transcriptional regulation and post-translational modifications in schizont-stage malaria parasites. Genome Biol. 9, R177 (2008).
Caro, F., Ahyong, V., Betegon, M. & DeRisi, J. L. Genome-wide regulatory dynamics of translation in the Plasmodium falciparum asexual blood stages. eLife 3, e04106 (2014).
Roundtree, I. A., Evans, M. E., Pan, T. & He, C. Dynamic RNA modifications in gene expression regulation. Cell 169, 1187–1200 (2017).
Meyer, K. D. & Jaffrey, S. R. Rethinking m6A readers, writers, and erasers. Annu. Rev. Cell Dev. Biol. 33, 319–342 (2017).
Śledź, P. & Jinek, M. Structural insights into the molecular mechanism of the m(6)A writer complex. eLife 5, e18434 (2016).
Wang, Y. et al. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat. Cell Biol. 16, 191–198 (2014).
Ping, X.-L. et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 24, 177–189 (2014).
Liu, N. et al. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 518, 560–564 (2015).
Meyer, K. D. et al. 5’ UTR m(6)A promotes cap-independent translation. Cell 163, 999–1010 (2015).
Wang, X. et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120 (2014).
Patil, D. P., Pickering, B. F. & Jaffrey, S. R. Reading m6A in the transcriptome: m6A-binding proteins. Trends Cell Biol. 28, 113–127 (2018).
Vu, L. P. et al. The N6-methyladenosine (m6A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat. Med. 23, 1369–1376 (2017).
Lence, T. et al. m6A modulates neuronal functions and sex determination in Drosophila. Nature 540, 242–247 (2016).
Schwartz, S. et al. High-resolution mapping reveals a conserved, widespread, dynamic mRNA methylation program in yeast meiosis. Cell 155, 1409–1421 (2013).
Gardner, M. J. et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419, 498–511 (2002).
Larson, M. H. et al. CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat. Protoc. 8, 2180–2196 (2013).
Xiao, B. et al. Epigenetic editing by CRISPR/dCas9 in Plasmodium falciparum. Proc. Natl Acad. Sci. USA 116, 255–260 (2019).
Fujita, T. & Fujii, H. Efficient isolation of specific genomic regions and identification of associated proteins by engineered DNA-binding molecule-mediated chromatin immunoprecipitation (enChIP) using CRISPR. Biochem. Biophys. Res. Commun. 439, 132–136 (2013).
Ponts, N. et al. Nucleosome landscape and control of transcription in the human malaria parasite. Genome Res. 20, 228–238 (2010).
Bártfai, R. et al. H2A.Z demarcates intergenic regions of the Plasmodium falciparum epigenome that are dynamically marked by H3K9ac and H3K4me3. PLoS Pathog. 6, e1001223 (2010).
Adjalley, S. H., Chabbert, C. D., Klaus, B., Pelechano, V. & Steinmetz, L. M. Landscape and dynamics of transcription Initiation in the malaria parasite Plasmodium falciparum. Cell Rep. 14, 2463–2475 (2016).
Linder, B. et al. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods 12, 767–772 (2015).
Stevens, A. T., Howe, D. K. & Hunt, A. G. Characterization of mRNA polyadenylation in the apicomplexa. PLoS One 13, e0203317 (2018).
Engel, M. et al. The role of m6A/m-RNA methylation in stress response regulation. Neuron 99, 389–403 (2018).
Iyer, L. M., Zhang, D. & Aravind, L. Adenine methylation in eukaryotes: apprehending the complex evolutionary history and functional potential of an epigenetic modification. Bioessays 38, 27–40 (2016).
Schwartz, S. et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5’ sites. Cell Rep. 8, 284–296 (2014).
Wen, J. et al. Zc3h13 regulates nuclear RNA m6A methylation and mouse embryonic stem cell self-renewal. Mol. Cell 69, 1028–1038 (2018).
Růžička, K. et al. Identification of factors required for m6A mRNA methylation in Arabidopsis reveals a role for the conserved E3 ubiquitin ligase HAKAI. New Phytol. 215, 157–172 (2017).
Oehring, S. C. et al. Organellar proteomics reveals hundreds of novel nuclear proteins in the malaria parasite Plasmodium falciparum. Genome Biol. 13, R108 (2012).
Yue, Y. et al. VIRMA mediates preferential m6A mRNA methylation in 3’UTR and near stop codon and associates with alternative polyadenylation. Cell Disco. 4, 10 (2018).
Garcia-Campos, M. A. et al. Deciphering the ‘m6A code’ via quantitative profiling of m6A at single-nucleotide resolution. Preprint at https://doi.org/10.1101/571679 (2019).
Ke, S. et al. m6A mRNA modifications are deposited in nascent pre-mRNA and are not required for splicing but do specify cytoplasmic turnover. Genes Dev. 31, 990–1006 (2017).
Patil, D. P. et al. M6A RNA methylation promotes XIST-mediated transcriptional repression. Nature 537, 369–373 (2016).
Lasonder, E. et al. Integrated transcriptomic and proteomic analyses of P. falciparum gametocytes: molecular insight into sex-specific processes and translational repression. Nucleic Acids Res. 44, 6087–6101 (2016).
Lindner, S. E. et al. Extensive transcriptional and translational regulation occur during the maturation of malaria parasite sporozoites. Preprint at https://doi.org/10.1101/642298 (2019).
Cui, L., Lindner, S. & Miao, J. Translational regulation during stage transitions in malaria parasites. Ann. New York Acad. Sci. 1342, 1–9 (2015).
Lopez-Rubio, J. J., Mancio-Silva, L. & Scherf, A. Genome-wide analysis of heterochromatin associates clonally variant gene regulation with perinuclear repressive centers in malaria parasites. Cell Host Microbe 5, 179–190 (2009).
Vembar, S. S., Macpherson, C. R., Sismeiro, O., Coppée, J.-Y. & Scherf, A. The PfAlba1 RNA-binding protein is an important regulator of translational timing in Plasmodium falciparum blood stages. Genome Biol. 16, 212 (2015).
Ng, C. S. et al. tRNA epitranscriptomics and biased codon are linked to proteome expression in Plasmodium falciparum. Mol. Syst. Biol. 14, e8009 (2018).
Crabb, B. S. et al. Transfection of the human malaria parasite Plasmodium falciparum. Methods Mol. Biol. 270, 263–276 (2004).
Ghorbal, M. et al. Genome editing in the human malaria parasite Plasmodium falciparum using the CRISPR-Cas9 system. Nat. Biotechnol. 32, 819–821 (2014).
Mesén-Ramírez, P. et al. Stable translocation intermediates jam global protein export in Plasmodium falciparum parasites and link the PTEX component EXP2 with translocation activity. PLoS Pathog. 12, e1005618 (2016).
Perkins, D. N., Pappin, D. J., Creasy, D. M. & Cottrell, J. S. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 (1999).
Qi, L. S. et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152, 1173–1183 (2013).
MacPherson, C. R. & Scherf, A. Flexible guide-RNA design for CRISPR applications using Protospacer Workbench. Nat. Biotechnol. 33, 805–806 (2015).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013).
Kearse, M. et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649 (2012).
Capella-Gutierrez, S., Silla-Martinez, J. M. & Gabaldon, T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973 (2009).
Darriba, D., Taboada, G. L., Doallo, R. & Posada, D. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27, 1164–1165 (2011).
Kumar, S., Stecher, G. & Tamura, K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874 (2016).
Oates, M. E. et al. D2P2: database of disordered protein predictions. Nucleic Acids Res. 41, D508–D516 (2013).
Aurrecoechea, C. et al. EuPathDB: the eukaryotic pathogen genomics database resource. Nucleic Acids Res. 45, D581–D591 (2017).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Li, H. et al. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Ramírez, F. et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165 (2016).
Krzywinski, M. et al. Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645 (2009).
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010).
Marçais, G. & Kingsford, C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 27, 764–770 (2011).
Crooks, G. E., Hon, G., Chandonia, J.-M. & Brenner, S. E. WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190 (2004).
Meyer, K. D. et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 149, 1635–1646 (2012).
Sims, D. et al. CGAT: computational genomics analysis toolkit. Bioinformatics 30, 1290–1291 (2014).
Anders, S., Pyl, P. T. & Huber, W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
R: A language and environment for statistical computing (R Development Core Team, 2012).
Broadbent, K. M. et al. Strand-specific RNA sequencing in Plasmodium falciparum malaria identifies developmentally regulated long non-coding RNA and circular RNA. BMC Genom. 16, 454 (2015).
Acknowledgements
We thank A. Claës, C. Scheidig-Benatar and P. Chen for help with parasite culture. Protein mass-spectrometry was performed at the Biopolymers and Proteomics core of The Koch Institute Swanson Biotechnology Center. This work was supported by a European Research Council Advanced Grant (PlasmoSilencing 670301) and the French Parasitology consortium ParaFrap (ANR-11-LABX0024) to A.Scherf. Work in the labs of P.R.P. and P.C.D. was funded by the National Research Foundation Singapore under its Singapore-MIT Alliance for Research and Technology (SMART) Centre, Infectious Disease and Antimicrobial Resistance IRGs. S.B. and J.M.B. were supported by an EMBO fellowship (S.B.: ALTF 1444-2016; J.M.B.: ALTF 180-2015). A.Si. acknowledges financial support from the Singapore-MIT Alliance (SMA) Graduate Fellowships.
Author information
Authors and Affiliations
Contributions
P.R.P, P.C.D. and A.Sc. conceptualized the project. S.B., J.M.B. and A.Sc. conceived experiments. J.M.B. developed and performed CRISPR interference and dCas9 ChIP-seq experiments. S.B. performed m6A-seq and RT-qPCR experiments. A.Si. performed and analysed LC-MS/MS and protein co-IP experiments. S.B., J.M.B. and T.R. generated constructs, transfectants and parasite material. S.B. performed bioinformatic analyses. P.R.P., P.C.D. and A.Sc. supervised and helped interpret analyses. All authors discussed and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Table 8, Supplementary Figures 1–5 and Supplementary References.
Supplementary Tables
Supplementary Tables 1–7 and Supplementary Tables 9–12.
Rights and permissions
About this article
Cite this article
Baumgarten, S., Bryant, J.M., Sinha, A. et al. Transcriptome-wide dynamics of extensive m6A mRNA methylation during Plasmodium falciparum blood-stage development. Nat Microbiol 4, 2246–2259 (2019). https://doi.org/10.1038/s41564-019-0521-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-019-0521-7
This article is cited by
-
Epigenetic regulation as a therapeutic target in the malaria parasite Plasmodium falciparum
Malaria Journal (2024)
-
Extracellular vesicles could be a putative posttranscriptional regulatory mechanism that shapes intracellular RNA levels in Plasmodium falciparum
Nature Communications (2023)
-
A single-cell atlas of Plasmodium falciparum transmission through the mosquito
Nature Communications (2021)
-
Linking the YTH domain to cancer: the importance of YTH family proteins in epigenetics
Cell Death & Disease (2021)
-
N6-Adenosine methylation on mRNA is recognized by YTH2 domain protein of human malaria parasite Plasmodium falciparum
Epigenetics & Chromatin (2020)