Abstract
Mitochondria are believed to have originated ~2.5 billion years ago. As well as energy generation in cells, mitochondria have a role in defence against bacterial pathogens. Despite profound changes in mitochondrial morphology and functions following bacterial challenge, whether intracellular bacteria can hijack mitochondria to promote their survival remains elusive. We report that Listeria monocytogenes—an intracellular bacterial pathogen—suppresses LC3-associated phagocytosis (LAP) by modulation of mitochondrial Ca2+ (mtCa2+) signalling in order to survive inside cells. Invasion of macrophages by L. monocytogenes induced mtCa2+ uptake through the mtCa2+ uniporter (MCU), which in turn increased acetyl-coenzyme A (acetyl-CoA) production by pyruvate dehydrogenase. Acetylation of the LAP effector Rubicon with acetyl-CoA decreased LAP formation. Genetic ablation of MCU attenuated intracellular bacterial growth due to increased LAP formation. Our data show that modulation of mtCa2+ signalling can increase bacterial survival inside cells, and highlight the importance of mitochondrial metabolism in host–microbial interactions.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Source data are provided with this paper.
References
Mills, E. L., Kelly, B. & O’Neill, L. A. J. Mitochondria are the powerhouses of immunity. Nat. Immunol. 18, 488–498 (2017).
Zhang, Q. et al. Circulating mitochondrial DAMPs cause inflammatory responses to injury. Nature 464, 104–107 (2010).
Nakahira, K. et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat. Immunol. 12, 222–230 (2011).
West, A. P. & Shadel, G. S. Mitochondrial DNA in innate immune responses and inflammatory pathology. Nat. Rev. Immunol. 17, 363–375 (2017).
West, A. P., Shadel, G. S. & Ghosh, S. Mitochondria in innate immune responses. Nat. Rev. Immunol. 11, 389–402 (2011).
Mehta, M. M., Weinberg, S. E. & Chandel, N. S. Mitochondrial control of immunity: beyond ATP. Nat. Rev. Immunol. 17, 608–620 (2017).
Hamasaki, M. et al. Autophagosomes form at ER–mitochondria contact sites. Nature 495, 389–393 (2013).
Mills, E. L. et al. Succinate dehydrogenase supports metabolic repurposing of mitochondria to drive inflammatory macrophages. Cell 167, 457–470 (2016).
Naujoks, J. et al. IFNs modify the proteome of Legionella-containing vacuoles and restrict infection via IRG1-derived itaconic acid. PLoS Pathog. 12, e1005408 (2016).
Tannahill, G. M. et al. Succinate is an inflammatory signal that induces IL-1β through HIF-1α. Nature 496, 238–242 (2013).
Arts, R. J. et al. Glutaminolysis and fumarate accumulation integrate immunometabolic and epigenetic programs in trained immunity. Cell Metab. 24, 807–819 (2016).
Mills, E. L. et al. Itaconate is an anti-inflammatory metabolite that activates Nrf2 via alkylation of KEAP1. Nature 556, 113–117 (2018).
Bambouskova, M. et al. Electrophilic properties of itaconate and derivatives regulate the IκBζ–ATF3 inflammatory axis. Nature 556, 501–504 (2018).
Liu, P. S. et al. α-ketoglutarate orchestrates macrophage activation through metabolic and epigenetic reprogramming. Nat. Immunol. 18, 985–994 (2017).
West, A. P. et al. TLR signalling augments macrophage bactericidal activity through mitochondrial ROS. Nature 472, 476–480 (2011).
Abuaita, B. H., Schultz, T. L. & O'Riordan, M. X. Mitochondria-derived vesicles deliver antimicrobial reactive oxygen species to control phagosome-localized Staphylococcus aureus. Cell Host Microbe 24, 625–636 (2018).
McCormack, J. G., Halestrap, A. P. & Denton, R. M. Role of calcium ions in regulation of mammalian intramitochondrial metabolism. Physiol. Rev. 70, 391–425 (1990).
Pan, X. et al. The physiological role of mitochondrial calcium revealed by mice lacking the mitochondrial calcium uniporter. Nat. Cell Biol. 15, 1464–1472 (2013).
De Stefani, D., Raffaello, A., Teardo, E., Szabo, I. & Rizzuto, R. A forty-kilodalton protein of the inner membrane is the mitochondrial calcium uniporter. Nature 476, 336–340 (2011).
Baughman, J. M. et al. Integrative genomics identifies MCU as an essential component of the mitochondrial calcium uniporter. Nature 476, 341–345 (2011).
Rizzuto, R., De Stefani, D., Raffaello, A. & Mammucari, C. Mitochondria as sensors and regulators of calcium signalling. Nat. Rev. Mol. Cell Biol. 13, 566–578 (2012).
De Stefani, D., Rizzuto, R. & Pozzan, T. Enjoy the trip: calcium in mitochondria back and forth. Annu. Rev. Biochem. 85, 161–192 (2016).
Uhlen, P. et al. Alpha-haemolysin of uropathogenic E. coli induces Ca2+ oscillations in renal epithelial cells. Nature 405, 694–697 (2000).
TranVan Nhieu, G., Clair, C., Grompone, G. & Sansonetti, P. Calcium signalling during cell interactions with bacterial pathogens. Biol. Cell 96, 93–101 (2004).
Stavru, F., Bouillaud, F., Sartori, A., Ricquier, D. & Cossart, P. Listeria monocytogenes transiently alters mitochondrial dynamics during infection. Proc. Natl Acad. Sci. USA 108, 3612–3617 (2011).
Sanjuan, M. A. et al. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature 450, 1253–1257 (2007).
Mitchell, G. & Isberg, R. R. Innate immunity to intracellular pathogens: balancing microbial elimination and inflammation. Cell Host Microbe 22, 166–175 (2017).
Huang, J. & Brumell, J. H. Bacteria–autophagy interplay: a battle for survival. Nat. Rev. Microbiol. 12, 101–114 (2014).
Mehta, P., Henault, J., Kolbeck, R. & Sanjuan, M. A. Noncanonical autophagy: one small step for LC3, one giant leap for immunity. Curr. Opin. Immunol. 26, 69–75 (2014).
Florey, O., Kim, S. E., Sandoval, C. P., Haynes, C. M. & Overholtzer, M. Autophagy machinery mediates macroendocytic processing and entotic cell death by targeting single membranes. Nat. Cell Biol. 13, 1335–1343 (2011).
Martinez, J. et al. Noncanonical autophagy inhibits the autoinflammatory, lupus-like response to dying cells. Nature 533, 115–119 (2016).
Martinez, J. et al. Molecular characterization of LC3-associated phagocytosis reveals distinct roles for Rubicon, NOX2 and autophagy proteins. Nat. Cell Biol. 17, 893–906 (2015).
Yang, C. S. et al. Autophagy protein Rubicon mediates phagocytic NADPH oxidase activation in response to microbial infection or TLR stimulation. Cell Host Microbe 11, 264–276 (2012).
Mitchell, G. et al. Listeria monocytogenes triggers noncanonical autophagy upon phagocytosis, but avoids subsequent growth-restricting xenophagy. Proc. Natl Acad. Sci. USA 115, 210–217 (2018).
Gekara, N. O. et al. The multiple mechanisms of Ca2+ signalling by listeriolysin O, the cholesterol-dependent cytolysin of Listeria monocytogenes. Cell Microbiol. 9, 2008–2021 (2007).
Hamon, M., Bierne, H. & Cossart, P. Listeria monocytogenes: a multifaceted model. Nat. Rev. Microbiol. 4, 423–434 (2006).
Vadia, S. & Seveau, S. Fluxes of Ca2+ and K+ are required for the listeriolysin O-dependent internalization pathway of Listeria monocytogenes. Infect. Immun. 82, 1084–1091 (2014).
Vadia, S. et al. The pore-forming toxin listeriolysin O mediates a novel entry pathway of L. monocytogenes into human hepatocytes. PLoS Pathog. 7, e1002356 (2011).
Murakami, T. et al. Critical role for calcium mobilization in activation of the NLRP3 inflammasome. Proc. Natl Acad. Sci. USA 109, 11282–11287 (2012).
Mohapatra, N. P. et al. Combined deletion of four Francisella novicida acid phosphatases attenuates virulence and macrophage vacuolar escape. Infect. Immun. 76, 3690–3699 (2008).
Lam, G. Y., Cemma, M., Muise, A. M., Higgins, D. E. & Brumell, J. H. Host and bacterial factors that regulate LC3 recruitment to Listeria monocytogenes during the early stages of macrophage infection. Autophagy 9, 985–995 (2013).
Galluzzi, L., Pietrocola, F., Levine, B. & Kroemer, G. Metabolic control of autophagy. Cell 159, 1263–1276 (2014).
Shaughnessy, L. M., Hoppe, A. D., Christensen, K. A. & Swanson, J. A. Membrane perforations inhibit lysosome fusion by altering pH and calcium in Listeria monocytogenes vacuoles. Cell Microbiol. 8, 781–792 (2006).
Gluschko, A. et al. The β2 integrin Mac-1 induces protective LC3-associated phagocytosis of Listeria monocytogenes. Cell Host Microbe 23, 324–337 (2018).
Hornung, V. et al. Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat. Immunol. 9, 847–856 (2008).
Wong, S. W., Sil, P. & Martinez, J. Rubicon: LC3-associated phagocytosis and beyond. FEBS J. 285, 1379–1388 (2018).
Ryan, D. G. et al. Coupling Krebs cycle metabolites to signalling in immunity and cancer. Nat. Metab. 1, 16–33 (2019).
O’Neill, L. A. J. & Artyomov, M. N. Itaconate: the poster child of metabolic reprogramming in macrophage function. Nat. Rev. Immunol. 19, 273–281 (2019).
Li, T. et al. O-GlcNAc transferase links glucose metabolism to MAVS-mediated antiviral innate immunity. Cell Host Microbe 24, 791–803 (2018).
Li, X. et al. O-GlcNAc transferase suppresses inflammation and necroptosis by targeting receptor-interacting serine/threonine-protein kinase 3. Immunity 50, 576–590 (2019).
Li, X. et al. Myeloid-derived cullin 3 promotes STAT3 phosphorylation by inhibiting OGT expression and protects against intestinal inflammation. J. Exp. Med. 214, 1093–1109 (2017).
Alistar, A. et al. Safety and tolerability of the first-in-class agent CPI-613 in combination with modified FOLFIRINOX in patients with metastatic pancreatic cancer: a single-centre, open-label, dose-escalation, phase 1 trial. Lancet Oncol. 18, 770–778 (2017).
Pardee, T. S. et al. A phase I study of CPI-613 in combination with high-dose cytarabine and mitoxantrone for relapsed or refractory acute myeloid leukemia. Clin. Cancer Res. 24, 2060–2073 (2018).
Wellen, K. E. & Thompson, C. B. Cellular metabolic stress: considering how cells respond to nutrient excess. Mol. Cell 40, 323–332 (2010).
Liu, J., Qian, C. & Cao, X. Post-translational modification control of innate immunity. Immunity 45, 15–30 (2016).
Pietrocola, F., Galluzzi, L., Bravo-San Pedro, J. M., Madeo, F. & Kroemer, G. Acetyl coenzyme A: a central metabolite and second messenger. Cell Metab. 21, 805–821 (2015).
Cameron, A. M., Lawless, S. J. & Pearce, E. J. Metabolism and acetylation in innate immune cell function and fate. Semin. Immunol. 28, 408–416 (2016).
Schulthess, J. et al. The short chain fatty acid butyrate imprints an antimicrobial program in macrophages. Immunity 50, 432–445 (2019).
Wen, H., Ting, J. P. & O’Neill, L. A. A role for the NLRP3 inflammasome in metabolic diseases—did Warburg miss inflammation? Nat. Immunol. 13, 352–357 (2012).
Matsunaga, K. et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat. Cell Biol. 11, 385–396 (2009).
Zhong, Y. et al. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat. Cell Biol. 11, 468–476 (2009).
Garaude, J. et al. Mitochondrial respiratory-chain adaptations in macrophages contribute to antibacterial host defense. Nat. Immunol. 17, 1037–1045 (2016).
Osborne, S. E. & Brumell, J. H. Listeriolysin O: from bazooka to Swiss army knife. Phil. Trans. R. Soc. Lond. B Biol. Sci. 372, 20160222 (2017).
Seveau, S. Multifaceted activity of listeriolysin O, the cholesterol-dependent cytolysin of Listeria monocytogenes. Subcell. Biochem. 80, 161–195 (2014).
Lam, G. Y. et al. Listeriolysin O suppresses phospholipase C-mediated activation of the microbicidal NADPH oxidase to promote Listeria monocytogenes infection. Cell Host Microbe 10, 627–634 (2011).
Wellen, K. E. et al. ATP–citrate lyase links cellular metabolism to histone acetylation. Science 324, 1076–1080 (2009).
Covarrubias, A. J. et al. Akt–mTORC1 signaling regulates Acly to integrate metabolic input to control of macrophage activation. eLife 5, e11612 (2016).
Murphy, E. et al. Unresolved questions from the analysis of mice lacking MCU expression. Biochem. Biophys. Res. Commun. 449, 384–385 (2014).
Wen, H. et al. Fatty acid-induced NLRP3–ASC inflammasome activation interferes with insulin signaling. Nat. Immunol. 12, 408–415 (2011).
Wiederkehr, A. et al. Mitochondrial matrix calcium is an activating signal for hormone secretion. Cell Metab. 13, 601–611 (2011).
Luhrmann, A. & Haas, A. A method to purify bacteria-containing phagosomes from infected macrophages. Methods Cell Sci. 22, 329–341 (2000).
Lu, Y. et al. Palmitoylation of NOD1 and NOD2 is required for bacterial sensing. Science 366, 460–467 (2019).
Gunda, V., Yu, F. & Singh, P. K. Validation of metabolic alterations in microscale cell culture lysates using hydrophilic interaction liquid chromatography (HILIC)-tandem mass spectrometry-based metabolomics. PLoS ONE 11, e0154416 (2016).
Noubade, R. et al. NRROS negatively regulates reactive oxygen species during host defence and autoimmunity. Nature 509, 235–239 (2014).
Simons, K. T., Kooperberg, C., Huang, E. & Baker, D. Assembly of protein tertiary structures from fragments with similar local sequences using simulated annealing and Bayesian scoring functions. J. Mol. Biol. 268, 209–225 (1997).
Barth, P., Wallner, B. & Baker, D. Prediction of membrane protein structures with complex topologies using limited constraints. Proc. Natl Acad. Sci. USA 106, 1409–1414 (2009).
Yang, Y. & Zhou, Y. Ab initio folding of terminal segments with secondary structures reveals the fine difference between two closely related all-atom statistical energy functions. Protein Sci. 17, 1212–1219 (2008).
Laskowski, R. A., Rullmannn, J. A., MacArthur, M. W., Kaptein, R. & Thornton, J. M. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 8, 477–486 (1996).
Chen, R., Li, L. & Weng, Z. ZDOCK: an initial-stage protein-docking algorithm. Proteins 52, 80–87 (2003).
Jo, S., Kim, T., Iyer, V. G. & Im, W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 29, 1859–1865 (2008).
Van Der Spoel, D. et al. GROMACS: fast, flexible, and free. J. Comput. Chem. 26, 1701–1718 (2005).
Huang, J. & MacKerell, A. D. Jr CHARMM36 all-atom additive protein force field: validation based on comparison to NMR data. J. Comput. Chem. 34, 2135–2145 (2013).
Acknowledgements
We thank members of the Wen laboratory for discussions, Y. Shi from the UNC Genomics Core Facility for the microarray experiment, and M. Yuan for the metabolomics assay. This work was supported by National Institutes of Health grants R01GM120496 and R01GM135234 (to H. Wen), 5P01CA120964 and 5P30CA006516 (to J.M.A.), P30CA016086 (to the UNC Lineberger Comprehensive Cancer Center), R01CA163649, R01CA210439 and R01CA216853 (to P.K.S.), R01DE026728 (to Y.L.), R37AI044828 and R35CA231620 (to D.R.G.) and R01AI107250 (to S.S.).
Author information
Authors and Affiliations
Contributions
H. Wen and H. Wang designed the experiments, supervised the study and interpreted the data. T.L., L.K., X.L., Y.L., W.G., B.Z., L.L. and L.X. performed the experiments and provided intellectual input. L.E.H. performed the key mass spectrometry experiment. J.M.A., K.S.A. and P.K.S. performed the key metabolomics experiments and provided intellectual input. S.W. and X.C. performed computational modelling of Rubicon–p22phox interaction and provided intellectual input. Q.M. performed biostatistical analyses and provided intellectual input. X.L., Y.L.L., S.S., J.S.G. and D.R.G. contributed intellectual input and generated critical reagents. H.W. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 MCU Is required for L. monocytogenes-induced mitochondrial Ca2+ uptake in macrophages.
a−d, THP-1 cells (a and b) or BMMs (c and d) were incubated with rhod-2 (a and c) or Fluo-4 (b and d), followed by either WT or Δhly L. monocytogenes (MOI, 10) challenge. Fluorescence signal was read using a microplate reader. e−f, Fluorescence signal-based Ca2+ measurement to monitor mtCa2+ (e) and cytosolic Ca2+ (f) in Mcufl/fl and Mcu∆mye BMMs upon L. monocytogenes challenge. g-h, mtCa2+ (g) and cytosolic Ca2+ (h) in Mcufl/fl or Mcu∆mye BMMs were measured upon ATP stimulation (100 μM). i-j, mtCa2+ (i) and cytosolic Ca2+ (j) in Mcufl/fl or Mcu∆mye BMMs were measured upon purified protein listeriolysin O (LLO) (0.5 nM) challenge. k, mtCa2+ in BMMs pretreated with Xestospongin C (5 μM) contained in normal DMEM or Ca2+ free DMEM for 2 hours, were measured upon 0.5 nM LLO protein challenge. l, mtCa2+ in BMMs pretreated with cytochalasin D (10 μM) were measured upon L. monocytogenes challenge. The results presented are presentative of three independent experiments.
Extended Data Fig. 2 McuΔmye mice show no change in global immune cell populations or activation phenotype at naïve status.
Supplementary Figure 2. McuΔmye mice show no change in global immune cell populations or activation phenotype at naïve status. a, Cartoon of the strategy to generate myeloid-specific Mcu deletion mice and the primers used for genotyping. b, Genotyping result for indicated mice. c, Immunoblotting of MCU in Mcufl/fl and McuΔmye mice. d, Gating strategy to determine the percentage of differentiated macrophages (CD11b+F4/80+) and B cells (CD19+) from peritoneal cavity of Mcufl/fl and McuΔmye mice. e, The percentage of differentiated macrophages and B cells was shown. f, Gating strategy to determine the percentage of activated macrophage (CD80+, CD86+, MHCII+ and CD206+) from peritoneal cavity of Mcufl/fl and McuΔmye mice. g, Quantification of the mean fluorescence intensity (MFI) of the activated macrophages. h, Gating strategy to determine the percentage of macrophages (CD11b+F4/80+), neutrophils (CD11b+Ly6G+), monocytes (CD11b+Ly6C+) and conventional dendritic cells (CD11b+CD11c+) from spleen of Mcufl/fl and McuΔmye mice. i, The percentage of macrophages, neutrophils, monocytes and dendritic cells was shown. j, Gating strategy to determine the percentage of B cells (CD19+) and T cells (CD3+CD4+ and CD3+CD8+) from spleen of Mcufl/fl and McuΔmye mice. k and l, The percentage of B cells (k), and T cells (l) was shown. The averages of n = 3 biologically independent samples are shown. The error bars represent the SEM. Statistical significance was determined using t test (and nonparametric tests).
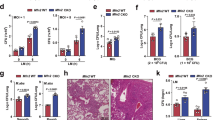
Extended Data Fig. 3 MCU deficiency in myeloid cells attenuates bacterial growth without affecting cytokine production.
a, Colony-forming units in BMMs generated from Mcufl/fl and McuΔmye mice challenged with L. monocytogenes (MOI, 10) for 1 hour followed by gentamicin treatment for indicated time before cell lysis. Intracellular bacteria were plated on brain-heart-infusion plates. b, GFP-L. monocytogenes containing cells in BMMs generated from Mcufl/fl and McuΔmye mice challenged with L. monocytogenes (MOI, 10) for indicated periods were measured by FACS analysis. c-f, BMMs (c-d) or peritoneal macrophages (e-f) from Mcufl/fl and McuΔmye mice were left untreated or challenged with L. monocytogenes (MOI, 10) for indicated periods. Gene transcripts of Il6 and Tnfa in the cells (c and e), IL-6 and TNF-α proteins in the supernatants (d and f) were measured with RT-PCR and ELISA, respectively. g, Gene transcripts of IL6 and TNFA in THP-1 cells left untreated or stimulated with L. monocytogenes (MOI, 10) for indicated periods were measured with RT-PCR. h-i, NF-κB signaling molecules including IKKα, p65 and IκBα, and MAPK signaling molecules including ERK1/2, JNK1/2 and p38, in Mcufl/fl and McuΔmye BMMs (h) or peritoneal macrophages (i) left untreated or stimulated with L. monocytogenes (MOI, 10) for indicated periods were analyzed by immunoblotting. j, Immunoblotting of NF-κB signaling molecules and MAPK signaling molecules in THP-1 cells left untreated or stimulated with L. monocytogenes (MOI, 10) for indicated periods. The averages of n = 3 biologically independent samples are shown. The error bars represent the SEM. Statistical significance was determined using t test (and nonparametric tests).
Extended Data Fig. 4 MCU deficiency does not affect canonical autophagy.
a-e, Confocal imaging (a and d) and quantification (b and e) of the colocalization of Zymosan (red) with either LC3B puncta (green) (a and b) or LysoTracker (green) (d-e), immunoblotting of LAP-associated molecules in isolated phagosomes or total cell lysates from Mcufl/fl and Mcu∆mye BMMs in the presence of LLO (5 nM) (c). Scale bar, 2 µm. f-i, Immunofluorescence staining of LC3B in Mcufl/fl and Mcu∆mye BMMs (f and g), or THP-1 MCU-WT and MCU-KO cells (h and i) left untreated or challenged with EBSS for 2 h, or rapamycin (100 nM) for 16 h. LC3B puncta per cell (g and i) were shown. Scale bar, 2 μm. j-l, Immunoblotting of LC3B and p62 in Mcufl/fl and Mcu∆mye BMMs (j), or THP-1 MCU-WT and MCU-KO cells (k) left untreated or pretreated with bafilomycin (50 nM) for 1 h, followed by EBSS or rapamycin (100 nM) incubation for another 2 or 16 h, respectively. Immunoblotting of AMPK and mTOR signaling molecules AKT, S6K and S6 in Mcufl/fl and Mcu∆mye BMMs challenged with L. monocytogenes (MOI, 10) for indicated periods (l). The averages of n = 3 biologically independent samples are shown. The error bars represent the SEM. Statistical significance was determined using t test (and nonparametric tests).
Extended Data Fig. 5 L. monocytogenes-induced LAP in McuΔmye macrophages depends on NOX2.
a, Confocal imaging of DQ-ovalbumin in Mcufl/fl, Mcu∆mye, Cybb−/− and McuΔmyeCybb−/− BMMs challenged with L. monocytogenes (MOI, 10) for 2 h Scale bar, 10 μm. b-c, Confocal imaging (b) and quantification (c) of the colocalization of LysoTracker (red) and L. monocytogenes (green) in Mcufl/fl, Mcu∆mye, Cybb−/− and McuΔmyeCybb−/− BMMs challenged with GFP-L. monocytogenes for indicated periods. Scale bar, 2 μm. The averages of n = 3 biologically independent samples are shown. The error bars represent the SEM. Statistical significance was determined using t test (and nonparametric tests).
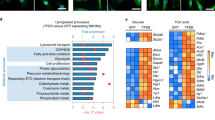
Extended Data Fig. 6 MCU promotes acetyl-CoA production.
a-d, Fold changes in intermediate metabolites of the glycolysis (a), PPP (b), HBP (c) or TCA cycle (d) in Mcufl/fl and Mcu∆mye BMMs left untreated or treated with L. monocytogenes (MOI, 10) for 2 h. The averages of n = 3 biologically independent samples are shown. The error bars represent the SEM. Statistical significance was determined using t test (and nonparametric tests).
Extended Data Fig. 7 Acetylation of Rubicon on K549 inhibits macrophage bactericidal effect.
a-b, Venn diagram analysis across four groups of genes. c, Immunoblotting of Ac-H3, H3K4me3, H3K9me2, H3K27me3 and H3 in Mcufl/fl and Mcu∆mye BMMs left untreated or stimulated with L. monocytogenes (MOI, 10) for indicated periods. d, Colocalization of L. monocytogenes (green) and LysoTracker (red) in RUBCN-KO THP-1 cells reconstituted with either empty vector or Flag-tagged Rubicon WT or K549R mutant upon GFP-L. monocytogenes challenge. Scale bar, 2 µm. The averages of n = 3 biologically independent samples are shown. The error bars represent the SEM. Statistical significance was determined using t test (and nonparametric tests).
Extended Data Fig. 8 Acetylation of Rubicon on K549 inhibits its interaction with p22phox/NOX2 complex.
a-b, Model showing the structural difference of RUBCNWT and Ac-RUBCN complex after 100 ns MD simulations. RUBCN, cyan cartoon; p22, yellow cartoon; Glu5 in p22, blue stick; Lys549 and Ac-Lys549 in RUBCN, red stick. c, Schematic representation of the role of MCU in LC3-associated phagocytosis and bactericidal effect.
Supplementary information
Supplementary Information
Supplementary Tables 1–6.
Source data
Source Data Fig. 1
Unprocessed Western Blots
Source Data Fig. 1
Statistical Source Data
Source Data Fig. 2
Unprocessed Western Blots
Source Data Fig. 2
Statistical Source Data
Source Data Fig. 3
Unprocessed Western Blots
Source Data Fig. 3
Statistical Source Data
Source Data Fig. 4
Unprocessed Western Blots
Source Data Fig. 4
Statistical Source Data
Source Data Fig. 5
Unprocessed Western Blots
Source Data Fig. 5
Statistical Source Data
Source Data Fig. 6
Unprocessed Western Blots
Source Data Extended Data Fig. 2
Unprocessed Western Blots
Source Data Extended Data Fig. 2
Statistical Source Data
Source Data Extended Data Fig. 3
Unprocessed Western Blots
Source Data Extended Data Fig. 3
Statistical Source Data
Source Data Extended Data Fig. 4
Unprocessed Western Blots
Source Data Extended Data Fig. 4
Statistical Source Data
Source Data Extended Data Fig. 5
Statistical Source Data
Source Data Extended Data Fig. 6
Statistical Source Data
Source Data Extended Data Fig. 7
Unprocessed Western Blots
Source Data Extended Data Fig. 7
Statistical Source Data
Rights and permissions
About this article
Cite this article
Li, T., Kong, L., Li, X. et al. Listeria monocytogenes upregulates mitochondrial calcium signalling to inhibit LC3-associated phagocytosis as a survival strategy. Nat Microbiol 6, 366–379 (2021). https://doi.org/10.1038/s41564-020-00843-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-020-00843-2
This article is cited by
-
Reduced mitochondrial calcium uptake in macrophages is a major driver of inflammaging
Nature Aging (2023)
-
Immunometabolic crosstalk during bacterial infection
Nature Microbiology (2022)
-
Rubicon-deficiency sensitizes mice to mixed lineage kinase domain-like (MLKL)-mediated kidney ischemia-reperfusion injury
Cell Death & Disease (2022)