Abstract
Nanophotonic devices, which control light in subwavelength volumes and enhance light–matter interactions, have opened up exciting prospects for biosensing. Numerous nanophotonic biosensors have emerged to address the limitations of the current bioanalytical methods in terms of sensitivity, throughput, ease-of-use and miniaturization. In this Review, we provide an overview of the recent developments of label-free nanophotonic biosensors using evanescent-field-based sensing with plasmon resonances in metals and Mie resonances in dielectrics. We highlight the prospects of achieving an improved sensor performance and added functionalities by leveraging nanostructures and on-chip and optoelectronic integration, as well as microfluidics, biochemistry and data science toolkits. We also discuss open challenges in nanophotonic biosensing, such as reducing the overall cost and handling of complex biological samples, and provide an outlook for future opportunities to improve these technologies and thereby increase their impact in terms of improving health and safety.
Similar content being viewed by others
Main
Biosensors have proved immensely useful in numerous vital areas, such as healthcare, food safety, environmental monitoring, security, pharmaceutics and forensics. One of the leading sectors in the biosensor market is diagnostic technologies, as these support about 70% of medical decisions1. Nowadays, diagnostic information is routinely obtained by hospitals and centralized laboratories that use analytical methods, such as labelled immunoassays, polymerase chain reaction, cell culturing and light microscopy. Labelled immunoassays employ enzymatic, fluorescent or chemiluminescent tags to enable the detection of target analytes. Although labelled immunoassays are sensitive and available for a broad range of analytes, they require expensive and bulky benchtop instrumentation and entail time-consuming multistep detection protocols. Some labelled immunoassays are available for non-clinical settings—most notably, the lateral-flow assay (LFA), with home pregnancy tests or COVID-19 tests being the best-known examples2,3. LFAs do not require equipment and enable measurements in a matter of minutes. However, they are typically semiquantitative and limited in terms of sensitivity and the number of analytes they can simultaneously measure.
Although conventional analytical methods will remain important in centralized laboratories when sample volumes are large and an immediate response is not necessary, easy-to-use and rapid point-of-care biosensors that can be used in virtually any location could be a game-changer for the management of diseases. Early diagnosis is crucial for a wide range of conditions, such as infectious diseases, cancer, cardiovascular diseases, autoimmune disorders and inflammatory diseases, for which timing is critical to maximize the efficacy of therapy. For example, the detection of cancer-associated biomarkers, such as proteins, antibodies, hormones, cytokines, prions, DNA or RNA, in biofluids (that is, a liquid biopsy) is being increasingly explored as a non-invasive alternative to surgical biopsies and could also potentially enable a faster detection of early-stage or residual disease after treatment4. This is an important, but also challenging, research direction, as most biomarkers are present at very low concentrations (often below pg ml–1 (ref. 5)). Important insights can also be gained by detecting structural changes in biomarker molecules6 related to neurodegenerative disorders, such as Alzheimer’s and Parkinson’s disease. Furthermore, continuous monitoring of biomarker or therapeutic drug levels at the bedside can provide valuable feedback to physicians and allow them to tailor the treatment options for individual patients7,8,9. In this regard, biosensors that can provide the molecular profile of a patient could become instrumental in paving the way towards precision medicine10,11,12. Low-cost, easy-to-use wearable biosensors are expected to find applications as personal monitoring devices13. Automated and autonomous biosensors integrated into public infrastructures (transportation hubs, schools, offices and so on) are envisaged to improve public safety by alerting the public about biological threats. These devices may also be connected through the ‘internet of things’ and generate large spatiotemporal datasets at the population scale14,15. Therefore, future biosensor technologies will inevitably need to harness artificial intelligence algorithms to handle the vast amount of information generated. Figure 1 illustrates this vision and gives a glimpse into the future of biosensors.
This review presents recent developments in nanophotonic biosensors based on plasmonic and dielectric nanostructures that could contribute to this vision. We focus on label-free optical biosensors based on biomolecular recognition and the measurement of refractive index changes, and also the measurement of infrared absorption or Raman scattering using surface-enhanced optical spectroscopies. We describe their features and benefits and discuss challenges for their further development and applications.
Evanescent-field-based nanophotonic biosensors
Optical affinity biosensors are devices that employ biomolecular receptors that interact with a particular analyte and an optical readout system that transforms information about such an interaction into a measurable output. Alternatively, the output can be obtained by directly exploiting the optical properties of the analyte, as in the case of spectroscopic (for example, Raman, infrared and chiral) biosensors. Evanescent-field-based photonic biosensors provide a convenient approach to probe analytes with light while separating the optoelectronic components from the liquid sample under study. The main characteristic of the evanescent field is that it decays exponentially along an axis perpendicular to the sensing surface, with a decay length on the order of hundreds of nanometres. This surface confinement is an important feature that increases the light–matter interaction and offers precise spatial control over the measurement.
The most well-known example of an evanescent-field-based affinity biosensor is the surface plasmon resonance (SPR) biosensor, which exploits surface plasmons that propagate at the interface between a noble metal layer and a dielectric16,17. Propagating surface plasmons (PSPs) are charge density waves that can be excited by an external light source at a momentum-matching condition, which depends on the refractive index of the dielectric medium at the surface of the metal layer. By measuring the changes in this resonance condition (for example, angle, wavelength, intensity or phase), the molecular interactions that occur at the sensor surface can be monitored directly in real time16. Other evanescent-field-based affinity biosensors utilize dielectric waveguides and resonators18,19,20. In both SPR and dielectric waveguide-based biosensors, the decay length of the evanescent field is on the order of ~200–400 nm, which is considerably larger than that of most categories of biomolecular analytes (Fig. 2). As a consequence, conventional evanescent-field-based sensors provide fundamentally limited interactions between the probing light and the analyte.
In contrast, nanophotonic structures can tightly confine light near their surface, down to distances of less than ten nanometres, and so create regions of strong electromagnetic fields, called ‘hot spots’. They can also enhance the intensities of the localized fields by more than 3–4 orders of magnitude relative to the incident light, acting as optical ‘nanoantennas’21. This strong field localization makes nanophotonic biosensors simultaneously more sensitive to analyte molecules and less susceptible to background interference (for example, due to variations in sample composition). The performance of nanophotonic biosensors depends on the resonance properties of the nanostructures, especially on the spatial overlap of the regions in which the analyte molecules are captured and the region in which the evanescent field intensity is highest. Figure 2 compares the size of various biological analytes and the evanescent field decay length of electromagnetic modes supported on various nanophotonic structures. Resonance properties can be engineered by nanostructure design parameters, such as materials, geometry and arrangement, as well as by the nature of the optical phenomenon that leads to the resonances (for example, gap plasmons, Fano-like modes or bound state in the continuum (BIC))22,23,24,25. Most nanophotonic biosensors exploit resonances supported by metallic and dielectric nanostructures. Below, we examine various nanophotonic biosensor design strategies based on affinity-based refractometric sensing and surface-enhanced spectroscopy.
Affinity biosensors based on plasmonic nanostructures
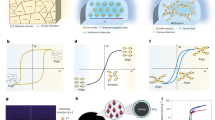
In the early 2000s, biosensors based on surface plasmons supported by metal nanostructures—commonly referred to as nanoplasmonic biosensors—emerged as an alternative to conventional SPR biosensors based on PSPs on thin films. The first generation of nanoplasmonic biosensors were based on localized surface plasmons (LSPs) excited on metal nanoparticles (NPs)26. The decay length of LSPs tends to increase with the size of the NP, but is typically ~5–25 nm (ref. 27)—more than an order of magnitude shorter than that of PSPs16. Plasmonic NPs absorb and scatter incident light and exhibit an extinction peak at the resonance wavelength that corresponds to the LSP. The sensitivity of the spectral peak position to the refractive index tends to increase with the NP size and resonance wavelength28,29. Spherical NPs have been shown to exhibit bulk sensitivity (S) and figure of merit (FOM) values of 40 to 180 nm RIU–1 (refractive index unit) and 1 to 2 RIU–1, respectively28,30 (see Box 1 for definitions). The refractive index sensitivity can be increased by changing the shape of the NP, and various shapes, such as nanorods, nanorings, nanoprisms and nanocubes, have been explored31. LSPs on individual NPs or small arrays of NPs are typically observed using total internal reflection microscopy32 and dark-field microscopy33. For instance, Ament et al. used total internal reflection microscopy with an intense white-light laser and an electron multiplier charge-coupled device camera-based spectrometer to detect the binding of individual biomolecules to a single gold NP34. Independently, Zijlstra et al. demonstrated the single-molecule detection of LSPs on a gold nanorod with photothermal microscopy (Fig. 3a)35.
a, A gold nanorod functionalized at its tips with biotin (top) used to detect real-time binding of individual streptavidin–R-phycoerythrin conjugates (bottom). b, An array of submicrometre gold ‘mushrooms’ (left) exploited the coupling of LSP with Wood’s anomaly to achieve a high FOM for detecting alpha-fetoprotein (right). c, A multiplexed LSP-based sensor integrated with microfluidics (left) used for the quantitative detection of cytokine biomarkers in serum samples (right). d, Integration of silicon nanoresonator arrays with microfluidics (top) enabled the real-time detection of PSA (bottom); r and s indicate disk radius and separation, respectively. The inset is a diagram showing the sensor organization in the fluidic channels. e, High-Q silicon nanostructures that support BIC modes (top left) combined with hyperspectral imaging (top right) and an image-processing method to achieve the detection of just a few IgG molecules per μm2 (bottom). FP and TP stand for the ratio of false positives and the ratio of true positives, respectively. f, A nanogap antenna with a bow-tie-shaped resonator (left) coupled with a bottom reflector to detect a SEIRA signal from small numbers of molecules (right). g, Five pairs of plasmonic nanorod antennas with 30 nm gaps integrated onto a silicon waveguide as part of an on-chip SEIRA biosensor. h, A multiresonant antenna (left) used to support up to four resonances over a relatively broadband spectrum (right). Panels adapted with permission from: a, ref. 35, Springer Nature Limited; b, ref. 44, Springer Nature Limited; c, ref. 55, American Chemical Society; d, ref. 65, American Chemical Society; e, ref. 70, Springer Nature Limited; f, ref. 84, American Chemical Society. Panels reproduced with permission from: g, ref. 93, American Chemical Society; h, ref. 94, American Chemical Society. a.u., arbitrary units; INF-γ, interferon-gamma; PDMS, polydimethylsiloxane; TNF, tumour necrosis factor.
Nanostructures that comprise ordered groups of NPs (for example, arrays of nanodisks or nanoholes, binary nanodisks and double nanopillars) can support Fano resonances that originate from the interaction between a non-radiative (dark) mode and a continuum generated by a radiative (bright) mode, and exhibit dramatically reduced linewidths (and thus potentially higher FOMs)22,23,36. Arrays of nanoholes have attracted particular interest, as they exhibit the so-called ‘extraordinary optical transmission’ phenomenon associated with the interaction of the diffractively excited PSPs and the LSPs of the nanoholes37,38. Yanik et al. reported a suspended array of nanoholes in a thin gold layer that exhibited S and FOM values as high as ~700 nm RIU–1 and ~160 RIU–1, respectively, which enabled the naked-eye detection of protein monolayers39. Subsequently, a nanohole array was combined with high-resolution spectroscopic imaging and the simultaneous readout from a one-dimensional (1D) array of sensing spots for affinity measurements of cholera–toxin-B binding to membrane-bound receptors40. This system also enabled real-time analysis of secretions from single cells for cell therapy applications41. Arrayed nanostructures can also support plasmonic surface lattice resonances, which originate from the interaction between light diffracted by the periodic array of metal NPs and the LSPs of the NPs, and exhibit extremely narrow spectral features24. Recently, these resonances were observed with an array of gold NPs on a glass substrate using hyperspectral imaging, direct illumination and attenuated total reflection methods42,43. Shen et al. used such resonances on a periodic array of gold-capped mushroom-shaped structures on a gold film to achieve S and FOM values of up to ~400 nm RIU–1 and ~108 RIU–1, respectively44. The authors employed their sensor to detect cytochrome c and alpha-fetoprotein at concentrations of 200 pM and 15 ng ml–1, respectively (Fig. 3b).
The use of nanoplasmonic biosensors for the study of biomolecular interactions has been relatively limited, but bioanalytical applications were pursued in numerous studies45,46,47,48, most of which were devoted to medical applications and the detection of protein and nucleic acid disease biomarkers. Nanoplasmonic biosensors typically achieve limits of detection (LOD; see the definition in Box 1) that range from 1 pg ml–1 to 100 ng ml–1 for protein biomarkers and from 100 fM to 10 nM for nucleic acid biomarkers. These LODs are mostly based on analyte detection in simple solutions (for example, buffers), and only a few studies have pursued analyte detection in biofluid matrices. This is mainly due to three reasons. First is the lack of simple and robust functionalization strategies that can confer resistance to non-specific binding from interferents in complex matrices. Second is the lack of methods for the cost-effective fabrication of large batches of nanostructures that is necessary to optimize the surface functionalization and detection assays, and to execute the experiments necessary to characterize the biosensor performance. Finally, there has been a limited level of device integration to date, with most studies focused on a selected aspect or biosensor element (for example, plasmonic nanostructure) rather than on the system as a whole. Also, note that LOD is a system performance metric that results from the complex interplay of biosensor characteristics, which include nanostructure, optical reader, microfluidics, biofunctionalization layer and assay configuration. This makes it rather difficult to assess the contributions of different biosensor elements to the analytical performance; for example, improved sensing characteristics of plasmonic nanostructures can be overshadowed by the low affinity of immobilized receptors.
Studies that report side-by-side comparisons of different nanophotonic biosensors against each other or different types of biosensors are rather rare. One such comparison was conducted by Špačková et al., who investigated the performance of plasmonic biosensors based on randomly distributed gold nanorods with different fill fractions relative to the conventional SPR biosensor49. The authors demonstrated that biosensor performance depends both on the optical and mass transport characteristics of plasmonic nanostructures, and that optimized nanostructures allow for the detection of analytes at concentrations an order of magnitude lower than are detectable by a conventional SPR biosensor. Indirect comparisons of nanoplasmonic affinity biosensors should be made carefully, as such comparisons are often complicated by the diversity of the experimental conditions (for example, instrumentation, protocols and reagents), differences in methods for the determination of LOD and sometimes by the lack of relevant experimental details. Several review articles that feature critical analyses of the literature have, therefore, highlighted the need for caution in making such comparisons48,50,51. This demonstrates the necessity for common standards to characterize and document the performance of biosensors to enable critical assessments of their capabilities and performance limits.
Despite these challenges, the share of studies that deal with clinical applications of plasmonic biosensors has grown in recent years45,52,53. For example, Yuan et al. used an LSP biosensor based on silver NPs to detect human epididymis secretory protein 4 in blood samples from ovarian cancer patients with a LOD of 4 pM (ref. 54). Chen et al. reported an integrated LSP-based biosensor platform that combined a gold nanorod microarray with microfluidics (Fig. 3c), and demonstrated parallel multiplex immunoassays that achieved the detection of six cytokines in serum with LOD <20 pg ml (ref. 55). They observed a good correlation between the results generated by their biosensor relative to those of commercial enzyme-linked immunosorbent assays, and also showed that their device can measure elevated cytokine levels—particularly interleukin-6 (IL-6) and IL-10—in samples taken from neonates who had cardiopulmonary bypass surgery for congenital heart disease.
Affinity biosensors based on resonant dielectric nanostructures
Plasmonic nanostructures based on the coinage metals suffers from a limited spectral operation range that covers the visible and near-infrared spectrum for plasmon-resonant field enhancement, as well as issues related to loss56, instability at high temperatures and poor compatibility with CMOS (complementary metal oxide semiconductor) front-end processes. Transition metal nitrides, transparent conductive oxides, metal sulfides and doped oxides are currently being heavily investigated as complementary plasmonic materials57,58, and could potentially provide an alternative platform for both refractometric biosensing and surface-enhanced spectroscopy. An alternative approach dispenses with plasmon excitation altogether and instead employs high-index dielectric nanostructures taken from industry-relevant semiconductors and oxides as the transducing nanoresonator. Dielectric nanoresonators provide low intrinsic optical absorption losses and hence a high quality factor (Q) resonances as well as a substantial resonant enhancement of both electric and magnetic near fields25. Their low optical losses also reduce local heat generation on illumination at their resonance wavelengths59. The Q values can be orders of magnitude higher than those in plasmonics if higher-order resonances are employed. Remarkably, surface-enhanced Raman scattering (SERS) was observed for GaP particles as early as in 198860, although the field enhancement is lower than that with plasmonic approaches. In addition, the minimal quenching properties of dielectrics are appealing for surface-enhanced fluorescence methods61.
Low-loss dielectric microstructures have been employed for refractometric label-free sensing for many decades. Prominent examples include higher-order Mie resonances, known as whispering-gallery modes62, which exhibit the hallmark of a very high Q—which often exceeds 106—for simple structures such as silica or silicon microspheres, disks or ring waveguides. Nanometric slots are a further route to additional field enhancement63. Turning from the micro- to the nanoscale, high-index contrast dielectric nanostructures show more modest Q values for their low-order electric and magnetic resonances; on the positive side, this allows more broadband operation under low-loss conditions. As in plasmonics, S and FOM are influenced by multiple resonator parameters, such as Q, near-field enhancement factors (EFs) and the spatial overlap of the near fields with the analytes. As these parameters tend to be coupled with each other, it is important to consider trade-offs. For example, an increase in Q can help to measure small resonance shifts, but if a higher Q comes at the expense of confining the field inside the resonator, this can reduce the field overlap with the analyte and thus lower the sensitivity. For simple dielectric resonator structures, wavelength shifts associated with analyte binding are generally smaller than those for plasmonic nanostructures. Nevertheless, a study in which biotin-functionalized silicon disks were used to detect streptavidin in buffer achieved low LODs—down to 5 ng ml–1—by measuring the spectral shift of the magnetic mode in the near-infrared spectrum64. Yavas et al. integrated arrays of antibody-functionalized silicon nanoresonators with microfluidics and demonstrated the detection of ~1 ng ml–1 prostate-specific antigen (PSA) in buffer (Fig. 3d)65. Although these results are encouraging, the applicability of such dielectric resonators to clinical samples has yet to be demonstrated.
One can increase the refractive index sensitivity of nanoscale dielectric resonators by employing strategies to suppress radiative coupling to the continuum, and thus narrow the resonance linewidth, although these gains come at the expense of a reduced operating bandwidth. For example, Yang et al. used Fano-type resonances for a low-loss silicon metasurface with a Q of ~500 (ref. 66). Although this is approximately one order of magnitude higher than Q values obtained with plasmonic Fano-type cavities, the S (~300 nm RIU–1) and FOM (100 RIU–1) values are comparable, which demonstrates the complex role of resonator parameters in the sensor performance. Here, the sharper Fano resonance was achieved due to a coherent coupling among a large number of unit cells (in essence, lattice resonances). A tighter spatial confinement for sensing was assessed by using a silicon oligomer approach to establish a lower Q (~35) Fano-type resonance, to achieve S and FOM values of ~400 nm RIU–1 and ~10 RIU–1, respectively67. In metasurfaces that consist of many unit cells, a high Q can be achieved due to coherent coupling and suppression of the scattering via lattice resonances, but at the expense of a reduced spatial resolution. A study of semirandom arrays of silicon nanoresonators revealed different contributions of lowest-order electric and magnetic Mie modes to both redshift and reducing extinction for the detection of PSA binding68. In the context of BIC in dielectric metasurfaces69, breaking the in-plane symmetry of the unit cell with a controllable asymmetry parameter can provide quasi-BIC modes with a very high Q. Recently, a BIC-type dielectric metasurface was used with hyperspectral imaging and data -science methods to optimally process spatially resolved spectra from millions of image pixels70. The metasurface was optimized to support resonance at ~850 nm with a high Q of ~145 and accessible, strong near fields, and exhibited an S of ~263 nm RIU–1 and a FOM of ~40 RIU–1 for measurements in aqueous media. This sensor achieved imaging at the diffraction limit, and enabled multiplexed detection in a single measurement at the level of less than three immunoglobulin G (IgG) molecules per μm2 (Fig. 3e). Another study utilized BICs in diatomic meta-units of dielectric metasurfaces, and realized a single-wavelength imaging-based optofluidic biosensor for the real-time detection of extracellular vesicle-binding events at concentrations as low as 200 fM (ref. 71).
Nanophotonic biosensors based on surface-enhanced spectroscopies
As discussed above, the specificity of affinity biosensors based on nanostructured metals and/dielectrics is solely determined by the characteristics of the receptors that may not be readily available for all the analytes of interest. Vibrational spectroscopies—in particular, infrared absorption and Raman scattering methods—can complement refractometric biosensors by providing selectivity without employing an analyte-specific receptor, and also generate insights into the analyte’s molecular structure. These methods excite molecular vibrations whose resonance frequencies depend on the chemical nature of the bonds and their conformations (Box 2). Therefore, they can be used to identify molecular ‘fingerprints’ and investigate molecular conformations without external labels. The key challenge for applying vibrational spectroscopies to small-volume biological samples is their relatively low sensitivity. Typical infrared absorption cross-sections (σIR) of molecules are ~10–20 cm2 per molecule, and non-resonant Raman scattering cross-sections (σRaman) are as small as about 10–31–10–29 cm2 per molecule. The corresponding low signal levels also limit the performance of vibrational spectroscopy techniques for accurate conformational analysis. The relatively strong absorption of mid-infrared radiation in water can obscure analyte signals and thus poses an additional challenge for biosensing in aqueous media.
Nanophotonic structures can boost analyte signals via surface-enhancement mechanisms with techniques such as SEIRA72,73 and surface-enhanced Raman spectroscopy (SERS)74,75,76 . For example, SERS can enhance Raman signals by up to ten orders of magnitude by plasmonic excitation, which has resulted in tremendous scientific interest in understanding the basis of this enhancement74. Interest is further fuelled by the observation of single-molecule SERS from plasmonic hot spots77,78. Using both top-down and bottom-up approaches, researchers have demonstrated a wide range of SERS substrates for biosensing applications79,80,81. Although the SERS community has mainly utilized noble metal structures, resonant dielectric nanostructures and metasurfaces represent promising alternatives in terms of lower absorption losses, CMOS compatibility and higher robustness. Caldarola et al. used an array of silicon disk dimers with a 20 nm gap to demonstrate SERS with a lower heat generation than that of metals59, and Cambiasso et al. used resonant silicon dimer structures for the SERS detection of a monolayer of β-carotenal molecules82. Resonant dielectric nanostructures can reduce unwanted heat generation—problematic for the degradation of adsorbed molecules and geometry deformation of the nanostructure itself—and enable facile surface functionalization (for example, through the use of silane chemistry and biomembranes). We envision these offering a valuable complement to metallic SERS substrates for affinity biosensing and bioanalytical applications.
One can likewise enhance SEIRA signals through the optimization of near fields using plasmonic antennas with nanometre-scale gaps, in which the field enhancement is caused by the ‘lightning rod effect’83. Dong et al. designed a bowtie-shaped gold structure with a <3 nm gap on a reflective substrate, which provided nearly seven orders of magnitude SEIRA signal enhancement (Fig. 3f)84. The authors quantitatively evaluated the performance of this 3D nanogap antenna and demonstrated the detection of SEIRA signals from as few as ∼500 4-nitrothiophenol molecules with a commercial Fourier transform infrared spectrometer. John-Herpin et al. used grating-order-coupled plasmonic nanogap antenna arrays to study the detection limits for protein sensing85. They demonstrated a chemically specific detection and secondary structure analysis of streptavidin in aqueous solution at concentrations as low as 100 pg ml–1 and 500 ng l–1, respectively85. In contrast, conventional infrared spectroscopy requires much higher concentrations (for example, 10 mg ml–1). Etezadi et al. presented the first application of SEIRA for the real-time secondary structure analysis of proteins86. The work showed that plasmonically enhanced amide-I fingerprints obtained by nanorod antennas can be utilized to measure random coil to cross-beta-sheet conformational changes that result from the pathological fibrillar aggregation of α-synuclein, a protein associated with plaque formation in Parkinson’s disease. Recent work expanded the palette of alternative SEIRA substrate materials, which include dielectric nanostructures, such as silicon and germanium, as well as van der Waals materials87,88,89,90 . Furthermore, continuous improvements in the development of mid-infrared laser sources91, detectors and low-loss waveguides92 show great promise towards building hybrid waveguide–integrated SEIRA platforms (Fig. 3g)93.
Vibrational spectroscopy is inherently broadband, because molecular fingerprint signatures cover a wide spectral range. However, the majority of nanophotonic substrates rely on single-resonant antennas that support a relatively narrow bandwidth. This spectral mismatch was recently addressed by engineering broadband metasurfaces based on multiresonant antennas. Aouani et al. used a log-periodic trapezoidal nanoantenna for multispectral SEIRA spectroscopy, and demonstrated EFs over 105 in a spectral window of 3 μm (Fig. 3h)94. Rodrigo et al. showed a self-similar multiperiodic nanorod antenna array that can support four independently controlled resonances covering an ultrawide spectral range from 1.5 to 10 μm (ref. 95). The dual-resonant version of this design was used to spectroscopically resolve the interactions of biomimetic lipid membranes with peptide toxins, as well as the dynamics of cargo release from synaptic vesicle mimics96. One of the next challenges is to simultaneously detect a large number of analytes from different molecular classes (for example, DNA, RNA, protein, small molecules and so on), which currently remains difficult because of their overlapping vibrational bands and the resulting complex spectral data. Recent papers highlighted that chemometric analysis—in particular, using machine learning and deep learning algorithms—will play an important role in exploiting the rich spectral information obtained with broadband metasurfaces for the effective discrimination of individual analytes in complex biological samples97,98,99,100.
Future perspectives and challenges
Further advancement of nanophotonic biosensors for clinical applications will require a broad multidisciplinary effort on a number of different fronts. We discuss some of the technical hurdles and opportunities for platform development below.
Optoelectronic integration and miniaturization
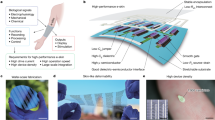
Progress in integrated photonic circuits is enabling the development of nanophotonic biosensors that achieve a greatly improved miniaturization, portability and throughput. Integration can be classified into vertical and planar schemes (Fig. 4). In the vertical integration scheme, nanostructures can simplify light-coupling requirements compared with those of conventional SPR, which needs external optical couplers, such as prisms, gratings or waveguides. Excitation of the resonant nanostructures with normally incident light in a collinear optical path is helpful for miniaturization and multiplexing, as recently demonstrated with intensity, phase, lens-free or hyperspectral imaging strategies101,102. Figure 4a illustrates a vertical integration scheme that operates in transmission mode. Here, each nanostructure with its subwavelength sensing region can act as a transducer, which expands the throughput capabilities to nanoscale dimensions. In principle, if we consider only optics and assume that each nanostructure can be functionalized selectively, this scenario could lead to massive multiplexing, limited only by the diffraction limit of light. In practice, however, individual sensing elements typically utilize nanostructure arrays to produce a stronger output signal than that of individual particles. These elements can be arranged in 2D arrays and functionalized for multiplexed detection. But, as vertical integration involves light passing through the sample, one needs to consider possible issues when analysing highly absorptive or turbid solutions due to the effects of optical loss, interference and scattering.
a, Examples of vertical and planar integration schemes for the implementation of nanophotonic biosensors with different device layers and components. b, Nanophotonic building blocks and commonly employed surface biofunctionalization approaches are shown. The vertical configuration is advantageous for high-throughput screening applications with large-area imaging and the implementation of disposable biochips. In contrast, a higher level of miniaturization can be achieved by the planar integration of optoelectronic components and electronics on a single sensor chip.
In planar integration schemes, the sensor utilizes optical waveguides that can be arranged in a 1D array and functionalized separately for multiplexed detection (Fig. 4b). A higher level of miniaturization can be achieved in this configuration relative to that of the vertical scheme by integrating active and passive optoelectronic components as well as the electronics layer onto a single sensor chip, either monolithically or by hybrid bonding. Most planar sensors still rely on external light sources and detectors that are coupled to the waveguides with gratings, at the expense of a reduced optical bandwidth. There are successful research and commercial examples of silicon-based integrated sensors that use conventional waveguides and optical microresonators, such as microrings and photonic crystal cavities18,19,103,104,105 . The inclusion of subwavelength nanostructures and nanopatterns on low-loss waveguides is a promising route to both increase the sensor performance and also benefit from the light-guiding function of the waveguides. For instance, nanoplasmonic structures were recently fabricated onto waveguides to control their spectra and increase their sensitivity for refractometric methods, as well as SERS and SEIRA, through subwavelength confinement of the sensing volumes93,106,107,108 .
Cost-effectiveness
Inexpensive disposable biosensor chips are desirable to avoid cross-contamination issues and complicated cleaning procedures while handling biological samples. In this regard, integration schemes that allow single-use cartridges and a stand-alone reader are most practical. For example, the nanophotonic biochip in Fig. 4a can be housed in a cartridge and separated from the light source and detector; such disposable cartridges can be customized as consumables for detecting different analytes with the same reader. This scenario can optimize reader costs by enabling the use of off-the-shelf optoelectronic components, and has other advantages relative to multiuse biosensor formats, in which procedures associated with the regeneration of surface functionalization typically reduce sensor performance over time and increase the overall cost. However, this approach requires caution in terms of the cost of biomaterials (for example, reagents) to produce inexpensive single-use cartridges109. There is a trend to merge nanophotonic biosensors with smartphones, because their light sources, cameras, image processing and communication capabilities can reduce costs and facilitate large-scale distribution13,101,102 Such sensors can be used to measure signals from patient samples, analyse data with personalized apps and then send the results wirelessly to clinicians for interpretation. Although biosensors with planar integration can lead to a smaller footprint and portability, their cost is likely to be higher for single-use scenarios because of the elaborate fabrication process associated with the multiple device layers and packaging steps.
Nanophotonic biochips depend on repeated patterning of engineered nanostructures, and researchers have typically employed electron-beam or focused-ion-beam lithography to date due to the flexibility in patterning different nanostructures. However, the low throughput and high cost of these serial patterning methods has created demand for low-cost alternative manufacturing methods for commercialization. One promising route is to leverage Moore’s law—that is, utilize silicon-compatible manufacturing methods and infrastructure. Despite some early developments in this area, there are still only limited examples of work using semiconductor foundries in the literature. Materials pose an important challenge here; for example, gold and silver are not compatible with front-end CMOS processing. We expect that large-scale and low-cost top-down lithography approaches, such as nanoimprinting, nanostencils, interference lithography and deep and extreme ultraviolet lithography, will gain more attention as alternative manufacturing strategies110,111,112,113 .
Sample handling
Microfluidic systems are useful in biosensor integration because they enable functions, such as sample treatment, sample concentration and analyte delivery, while minimizing the required sample volume and the consumption of expensive reagents114. Although nanophotonic biosensors benefit from field confinement in subwavelength hot spots for high sensitivity, the slow transport of analytes from bulk samples to the hot spots due to mass transport limitations can lead to longer detection times115. Approaches that address this challenge are now being pursued, such as microfluidic components that enable analyte concentration and improved transportation to the sensor surface, or the inclusion of external lasers and electrical fields116,117,118,119. The elimination or on-chip integration of the bulky and expensive external microfluidic components (for example, pumps and valves) poses an additional challenge. Capillary microfluidics is an attractive alternative, as it can enable liquid sample delivery without peripheral equipment by utilizing surface-tension effects120. Another emerging method is digital microfluidics, which uses electric forces to manipulate microdroplets121. Here, metallic nanostructures can simultaneously function as electrodes, and thereby facilitate the integration of digital microfluidics to enable the manipulation of even picolitre droplets without the need for pumps or valves.
Sample collection and processing is a key consideration for on-site biosensing. The large diversity in analytes and the matrix composition (for example, bodily fluids in medical diagnostics versus food samples in food safety) presents unique challenges. Although blood collection is minimally invasive and blood is the most commonly used biofluid for biomarker detection, it requires the separation of blood components and the suppression of matrix effects due to interferents (for example, proteins, cells and immunoglobulins)122. Sample dilution with a buffer can help to reduce the matrix effects and enable detection when analyte concentrations are high, but sample concentration might, instead, be necessary to detect scarce analytes122,123. Biofluids, such as saliva, sweat, tears or urine, that are simpler in composition than blood and that can be collected in a non-invasive manner are now being widely explored for bioanalysis123,124.
Surface functionalization
In highly complex clinical samples (for example, blood plasma and serum), the non-specific binding of non-target molecules from the matrix to the biosensor surface is a major hurdle. For example, the lowest analyte concentrations that can be detected in the absence of interferents (for example, in buffer) are typically much lower than those in clinical samples. Therefore, there is an urgent need for functionalization methods that confer resistance to non-specific binding in complex samples while enabling a robust immobilization of the receptors125. Traditional approaches for non-specific binding suppression include surface treatment with protein blockers (for example, bovine serum albumin or casein) or detergents (for example, Tween-20), or the use of poly(ethylene glycol). However, these confer limited resistance to non-specific binding in clinical samples. Alternative approaches that employ polymers and copolymers, such as zwitterionic polymers, are now being explored126,127,128, but there is currently no universal strategy to prevent fouling from complex biological media. An important consideration for antifouling coatings in nanophotonic affinity biosensors is that the coatings typically require a minimum thickness (~10–20 nm) to function properly, which can situate the immobilized receptor outside the hot spot. The size of the receptor is another consideration. Small receptors, such as aptamers129 and nanobodies130, are likely to be increasingly used to ensure that analyte–receptor binding takes place in these hot spots (Fig. 2). Immobilization is commonly achieved through covalent binding—via thiol, epoxy, carboxyl or aldehyde functional groups present on the receptors—or streptavidin-biotin binding. Although high-affinity receptors are generally desirable for stronger interaction, Visser et al. and Lubken et al. have shown that molecular switches that enable reversible interactions can be used for continuous biosensing7,131.
Unlike conventional biochips with flat surfaces, nanostructured biochips typically include multiple materials and exhibit uneven sensitivity along the sensor surface. These aspects make the functionalization process more demanding and call for new approaches, such as material132,133 or site-specific functionalization methods. For example, Zijlstra et al. reported a site-specific procedure that enables preferential functionalization of the tips of gold nanorods (Fig. 3a)35. A light-assisted functionalization approach using a three-photon absorption process was employed by Galloway et al. to immobilize proteins in the hotspot of a plasmonic dimer134. Tijunelyte et al. described a localized click reaction in the vicinity of gold NPs, with plasmonic tuning of the click chemistry135.
Emerging directions
Progress in materials science and fundamental optics will continue to advance nanophotonic biosensors. 2D quantum materials, hybrid optical materials and phase-change materials can enable active, tunable or reconfigurable biochips. For instance, the unique optoelectronic properties of graphene can provide a dynamic control of plasmonic resonances by electrostatic gating. Rodrigo et al. used this feature to realize electrically tunable mid-infrared plasmonic biochemical sensors89. The atomic-layer thickness of 2D materials136 and acoustic graphene plasmons137 were exploited to achieve tight near-field confinements, which are useful for small-molecule detection. Likewise, light generation from graphene, metals or hybrid nanomaterials that consist of active components, such as quantum dots, could eliminate the need for an external light source and lead to ultracompact electrically or optically addressable biosensors. Concurrently, the combination of different optical and non-optical (for example, electrochemical, impedance or mechanical) detection techniques on a single platform could enable multifunctional biosensors to extract more information from a given sample138. Nanophotonic biochips that operate over a broad electromagnetic spectrum can be engineered to perform refractometric sensing, SERS and SEIRA on the same platform. Another technique that can be included is chiral sensing, and metasurfaces that can generate strong electric and magnetic dipole moments are currently attracting considerable interest as a means to generate superchiral near fields for the detection and separation of chiral biomolecules139,140,141,142,143. Even though we have focused on label-free approaches in this review, surface-enhanced luminescence mechanisms (for example, fluorescence, chemiluminescence or upconversion) with plasmonic and dielectric nanostructures also present promising avenues61,144. Overall, gathering more data through the spectral, temporal, spatial and polarization degrees of freedom in optics will be instrumental in harnessing artificial intelligence145,146. In combination with autonomous, connected device operation, intelligent biosensors can be embedded in the internet of things of the future digital healthcare systems, as envisioned in Fig. 1.
In conclusion, nanophotonic biosensors have made substantial progress and demonstrated sufficient sensitivity for the observation of even individual molecular binding events147,148. However, translating these exciting scientific advances into biosensor devices for everyday life requires further efforts on multiple fronts. Research into the phenomena-rich realm of nanoscale optical physics is expected to lead to the development of new sensing mechanisms. The established fields of integrated photonics and optoelectronics as well as connections with commercial foundries will provide gateways to miniaturized and inexpensive nanophotonic biosensors and cost-effective nanofabrication methods. Marrying nanophotonic structures with advanced functional coatings that can efficiently capture target analytes while remaining inert to other molecules is a prerequisite for the reliable analysis of complex samples and operation in constantly changing biological milieux. Likewise, the need to couple nanophotonic biochips with microfluidics for sample collection and treatment in bioanalytical applications will continue to grow. These developments are expected to lead to mobile and affordable biosensors that can respond to the needs of modern healthcare, food safety and environmental monitoring, and thereby improve our well-being and overall quality of life.
References
Ngo, A., Gandhi, P. & Miller, W. G. Frequency that laboratory tests influence medical decisions. J. Appl. Lab. Med. 1, 410–414 (2017).
Mak, W. C., Beni, V. & Turner, A. P. F. Lateral-flow technology: from visual to instrumental. Trends Anal. Chem. 79, 297–305 (2016).
Kevadiya, B. D. et al. Diagnostics for SARS-CoV-2 infections. Nat. Mater. 20, 593–605 (2021).
Soda, N., Rehm, B. H. A., Sonar, P., Nguyen, N.-T. & Shiddiky, M. J. A. Advanced liquid biopsy technologies for circulating biomarker detection. J. Mater. Chem. B 7, 6670–6704 (2019).
Byrnes, S. A. & Weigl, B. H. Selecting analytical biomarkers for diagnostic applications: a first principles approach. Expert Rev. Mol. Diagn. 18, 19–26 (2018).
Toyama, B. H. & Weissman, J. S. Amyloid structure: conformational diversity and consequences. Annu. Rev. Biochem. 80, 557–585 (2011).
Visser, E. W. A., Yan, J., van IJzendoorn, L. J. & Prins, M. W. J. Continuous biomarker monitoring by particle mobility sensing with single molecule resolution. Nat. Commun. 9, 2541 (2018).
Heikenfeld, J. et al. Accessing analytes in biofluids for peripheral biochemical monitoring. Nat. Biotechnol. 37, 407–419 (2019).
Mage, P. L. et al. Closed-loop control of circulating drug levels in live animals. Nat. Biomed. Eng. 1, 0070 (2017).
Ho, D. et al. Enabling technologies for personalized and precision medicine. Trends Biotechnol. 38, 497–518 (2020).
Ginsburg, G. S. & Phillips, K. A. Precision medicine: from science to value. Health Aff. 37, 694–701 (2018).
Ahmed, M. U., Saaem, I., Wu, P. C. & Brown, A. S. Personalized diagnostics and biosensors: a review of the biology and technology needed for personalized medicine. Crit. Rev. Biotechnol. 34, 180–196 (2014).
Shrivastava, S., Trung, T. Q. & Lee, N.-E. Recent progress, challenges, and prospects of fully integrated mobile and wearable point-of-care testing systems for self-testing. Chem. Soc. Rev. 49, 1812–1866 (2020).
Pateraki, M. et al. in Wearable and Implantable Medical Devices (eds Dey, N., Ashour, A. S., Fong, S, J. & Bhatt, C.) 25–53 (Elsevier, 2020).
Jain, S. et al. Internet of medical things (IoMT)—integrated biosensors for point-of-care testing of infectious diseases. Biosens. Bioelectron. 179, 113074 (2021).
Homola, J. Surface Plasmon Resonance Based Sensors Vol. 4 (Springer, 2006).
Liedberg, B., Nylander, C. & Lundström, I. Biosensing with surface plasmon resonance—how it all started. Biosens. Bioelectron. 10, i–ix (1995).
Luan, E., Shoman, H., Ratner, D., Cheung, K. & Chrostowski, L. Silicon photonic biosensors using label-free detection. Sensors 18, 3519 (2018).
Wang, J. et al. Silicon‐based integrated label‐free optofluidic biosensors: latest advances and roadmap. Adv. Mater. Technol. 5, 1901138 (2020).
Chen, Y.-F. et al. Optofluidic opportunities in global health, food, water and energy. Nanoscale 4, 4839–4857 (2012).
Novotny, L. & van Hulst, N. Antennas for light. Nat. Photon. 5, 83–90 (2011).
Luk’yanchuk, B. et al. The Fano resonance in plasmonic nanostructures and metamaterials. Nat. Mater. 9, 707–715 (2010).
Limonov, M. F., Rybin, M. V., Poddubny, A. N. & Kivshar, Y. S. Fano resonances in photonics. Nat. Photon. 11, 543–554 (2017).
Kravets, V. G., Kabashin, A. V., Barnes, W. L. & Grigorenko, A. N. Plasmonic surface lattice resonances: a review of properties and applications. Chem. Rev. 118, 5912–5951 (2018).
Kuznetsov, A. I., Miroshnichenko, A. E., Brongersma, M. L., Kivshar, Y. S. & Luk’yanchuk, B. Optically resonant dielectric nanostructures. Science 354, aag2472 (2016).
Haes, A. J. & Van Duyne, R. P. A nanoscale optical biosensor: sensitivity and selectivity of an approach based on the localized surface plasmon resonance spectroscopy of triangular silver nanoparticles. J. Am. Chem. Soc. 124, 10596–10604 (2002).
Kedem, O., Tesler, A. B., Vaskevich, A. & Rubinstein, I. Sensitivity and optimization of localized surface plasmon resonance transducers. ACS Nano 5, 748–760 (2011).
Chen, H., Kou, X., Yang, Z., Ni, W. & Wang, J. Shape- and size-dependent refractive index sensitivity of gold nanoparticles. Langmuir 24, 5233–5237 (2008).
Zalyubovskiy, S. J. et al. Theoretical limit of localized surface plasmon resonance sensitivity to local refractive index change and its comparison to conventional surface plasmon resonance sensor. J. Opt. Soc. Am. A 29, 994–1002 (2012).
Martinsson, E. et al. Optimizing the refractive index sensitivity of plasmonically coupled gold nanoparticles. Plasmonics 9, 773–780 (2014).
Mayer, K. M. & Hafner, J. H. Localized surface plasmon resonance sensors. Chem. Rev. 111, 3828–3857 (2011).
Sönnichsen, C. et al. Spectroscopy of single metallic nanoparticles using total internal reflection microscopy. Appl. Phys. Lett. 77, 2949–2951 (2000).
McFarland, A. D. & Van Duyne, R. P. Single silver nanoparticles as real-time optical sensors with zeptomole sensitivity. Nano Lett. 3, 1057–1062 (2003).
Ament, I., Prasad, J., Henkel, A., Schmachtel, S. & Sönnichsen, C. Single unlabeled protein detection on individual plasmonic nanoparticles. Nano Lett. 12, 1092–1095 (2012).
Zijlstra, P., Paulo, P. M. R. & Orrit, M. Optical detection of single non-absorbing molecules using the surface plasmon resonance of a gold nanorod. Nat. Nanotechnol. 7, 379–382 (2012).
Hao, F. et al. Symmetry breaking in plasmonic nanocavities: subradiant LSPR sensing and a tunable Fano resonance. Nano Lett. 8, 3983–3988 (2008).
Brolo, A. G., Gordon, R., Leathem, B. & Kavanagh, K. L. Surface plasmon sensor based on the enhanced light transmission through arrays of nanoholes in gold films. Langmuir 20, 4813–4815 (2004).
Dahlin, A. et al. Localized surface plasmon resonance sensing of lipid-membrane-mediated biorecognition events. J. Am. Chem. Soc. 127, 5043–5048 (2005).
Yanik, A. A. et al. Seeing protein monolayers with naked eye through plasmonic Fano resonances. Proc. Natl Acad. Sci. USA 108, 11784–11789 (2011).
Lee, S. H., Lindquist, N. C., Wittenberg, N. J., Jordan, L. R. & Oh, S.-H. Real-time full-spectral imaging and affinity measurements from 50 microfluidic channels using nanohole surface plasmon resonance. Lab Chip 12, 3882–3890 (2012).
Li, X. et al. Label-free optofluidic nanobiosensor enables real-time analysis of single-cell cytokine secretion. Small 14, 1800698 (2018).
Zopf, D. et al. Plasmonic nanosensor array for multiplexed DNA-based pathogen detection. ACS Sens. 4, 335–343 (2019).
Danilov, A. et al. Ultra-narrow surface lattice resonances in plasmonic metamaterial arrays for biosensing applications. Biosens. Bioelectron. 104, 102–112 (2018).
Shen, Y. et al. Plasmonic gold mushroom arrays with refractive index sensing figures of merit approaching the theoretical limit. Nat. Commun. 4, 2381 (2013).
Mauriz, E., Dey, P. & Lechuga, L. M. Advances in nanoplasmonic biosensors for clinical applications. Analyst 144, 7105–7129 (2019).
Chocarro-Ruiz, B., Fernández-Gavela, A., Herranz, S. & Lechuga, L. M. Nanophotonic label-free biosensors for environmental monitoring. Curr. Opin. Biotechnol. 45, 175–183 (2017).
Jackman, J. A., Rahim Ferhan, A. & Cho, N.-J. Nanoplasmonic sensors for biointerfacial science. Chem. Soc. Rev. 46, 3615–3660 (2017).
Bonyár, A. Label-free nucleic acid biosensing using nanomaterial-based localized surface plasmon resonance imaging: a review. ACS Appl. Nano Mater. 3, 8506–8521 (2020).
Špačková, B., Lynn, N. S., Slabý, J., Šípová, H. & Homola, J. A route to superior performance of a nanoplasmonic biosensor: consideration of both photonic and mass transport aspects. ACS Photon. 5, 1019–1025 (2018).
Yang, T., Chen, S., He, X., Guo, H. & Sun, X. How to convincingly measure low concentration samples with optical label-free biosensors. Sens. Actuators B 306, 127568 (2020).
Dahlin, A. Biochemical sensing with nanoplasmonic architectures: we know how but do we know why? Annu. Rev. Anal. Chem. 14, 281–297 (2021).
Špačková, B., Wrobel, P., Bockova, M. & Homola, J. Optical biosensors based on plasmonic nanostructures: a review. Proc. IEEE 104, 2380–2408 (2016).
Masson, J.-F. Surface plasmon resonance clinical biosensors for medical diagnostics. ACS Sens. 2, 16–30 (2017).
Yuan, J., Duan, R., Yang, H., Luo, X. & Xi, M. Detection of serum human epididymis secretory protein 4 in patients with ovarian cancer using a label-free biosensor based on localized surface plasmon resonance. Int. J. Nanomed. 7, 2921–2928 (2012).
Chen, P. et al. Multiplex serum cytokine immunoassay using nanoplasmonic biosensor microarrays. ACS Nano 9, 4173–4181 (2015).
Khurgin, J. B. How to deal with the loss in plasmonics and metamaterials. Nat. Nanotechnol. 10, 2–6 (2015).
Doiron, B. et al. Quantifying figures of merit for localized surface plasmon resonance applications: a materials survey. ACS Photon. 6, 240–259 (2019).
Naik, G. V., Shalaev, V. M. & Boltasseva, A. Alternative plasmonic materials: beyond gold and silver. Adv. Mater. 25, 3264–3294 (2013).
Caldarola, M. et al. Non-plasmonic nanoantennas for surface enhanced spectroscopies with ultra-low heat conversion. Nat. Commun. 6, 7915 (2015).
Hayashi, S., Koh, R., Ichiyama, Y. & Yamamoto, K. Evidence for surface-enhanced Raman scattering on nonmetallic surfaces: copper phthalocyanine molecules on GaP small particles. Phys. Rev. Lett. 60, 1085–1088 (1988).
Romano, S. et al. Surface-enhanced Raman and fluorescence spectroscopy with an all-dielectric metasurface. J. Phys. Chem. C 122, 19738–19745 (2018).
Foreman, M. R., Swaim, J. D. & Vollmer, F. Whispering gallery mode sensors. Adv. Opt. Photon. 7, 168–240 (2015).
Robinson, J. T., Chen, L. & Lipson, M. On-chip gas detection in silicon optical microcavities. Opt. Express 16, 4296–4301 (2008).
Bontempi, N. et al. Highly sensitive biosensors based on all-dielectric nanoresonators. Nanoscale 9, 4972–4980 (2017).
Yavas, O., Svedendahl, M., Dobosz, P., Sanz, V. & Quidant, R. On-a-chip biosensing based on all-dielectric nanoresonators. Nano Lett. 17, 4421–4426 (2017).
Yang, Y., Kravchenko, I. I., Briggs, D. P. & Valentine, J. All-dielectric metasurface analogue of electromagnetically induced transparency. Nat. Commun. 5, 5753 (2014).
Chong, K. E. et al. Refractive index sensing with Fano resonances in silicon oligomers. Philos. Trans. R. Soc. A 375, 20160070 (2017).
Yavas, O., Svedendahl, M. & Quidant, R. Unravelling the role of electric and magnetic dipoles in biosensing with Si nanoresonators. ACS Nano 13, 4582–4588 (2019).
Hsu, C. W., Zhen, B., Stone, A. D., Joannopoulos, J. D. & Soljačić, M. Bound states in the continuum. Nat. Rev. Mater. 1, 16048 (2016).
Yesilkoy, F. et al. Ultrasensitive hyperspectral imaging and biodetection enabled by dielectric metasurfaces. Nat. Photon. 13, 390–396 (2019).
Jahani, Y. et al. Imaging-based spectrometer-less optofluidic biosensors based on dielectric metasurfaces for detecting extracellular vesicles. Nat. Commun. 12, 3246 (2021).
Ataka, K. & Heberle, J. Biochemical applications of surface-enhanced infrared absorption spectroscopy. Anal. Bioanal. Chem. 388, 47–54 (2007).
Neubrech, F., Huck, C., Weber, K., Pucci, A. & Giessen, H. Surface-enhanced infrared spectroscopy using resonant nanoantennas. Chem. Rev. 117, 5110–5145 (2017).
Langer, J. et al. Present and future of surface-enhanced Raman scattering. ACS Nano 14, 28–117 (2020).
Guerrini, L. & Graham, D. Molecularly-mediated assemblies of plasmonic nanoparticles for surface-enhanced Raman spectroscopy applications. Chem. Soc. Rev. 41, 7085 (2012).
Fan, M., Andrade, G. F. S. & Brolo, A. G. A review on the fabrication of substrates for surface enhanced Raman spectroscopy and their applications in analytical chemistry. Anal. Chim. Acta 693, 7–25 (2011).
Le Ru, E. C. & Etchegoin, P. G. Single-molecule surface-enhanced Raman spectroscopy. Annu. Rev. Phys. Chem. 63, 65–87 (2012).
Xu, H., Aizpurua, J., Käll, M. & Apell, P. Electromagnetic contributions to single-molecule sensitivity in surface-enhanced Raman scattering. Phys. Rev. E 62, 4318–4324 (2000).
Stöckel, S., Kirchhoff, J., Neugebauer, U., Rösch, P. & Popp, J. The application of Raman spectroscopy for the detection and identification of microorganisms. J. Raman Spectrosc. 47, 89–109 (2016).
Ngo, H. T., Wang, H.-N., Fales, A. M. & Vo-Dinh, T. Plasmonic SERS biosensing nanochips for DNA detection. Anal. Bioanal. Chem. 408, 1773–1781 (2016).
Galarreta, B. C., Tabatabaei, M., Guieu, V., Peyrin, E. & Lagugné-Labarthet, F. Microfluidic channel with embedded SERS 2D platform for the aptamer detection of ochratoxin A. Anal. Bioanal. Chem. 405, 1613–1621 (2013).
Cambiasso, J., König, M., Cortés, E., Schlücker, S. & Maier, S. A. Surface-enhanced spectroscopies of a molecular monolayer in an all-dielectric nanoantenna. ACS Photon. 5, 1546–1557 (2018).
Huck, C. et al. Surface-enhanced infrared spectroscopy using nanometer-sized gaps. ACS Nano 8, 4908–4914 (2014).
Dong, L. et al. Nanogapped Au antennas for ultrasensitive surface-enhanced infrared absorption spectroscopy. Nano Lett. 17, 5768–5774 (2017).
John-Herpin, A., Tittl, A. & Altug, H. Quantifying the limits of detection of surface-enhanced infrared spectroscopy with grating order-coupled nanogap antennas. ACS Photon. 5, 4117–4124 (2018).
Etezadi, D., Warner, J. B., Lashuel, H. A. & Altug, H. Real-time in situ secondary structure analysis of protein monolayer with mid-infrared plasmonic nanoantennas. ACS Sens. 3, 1109–1117 (2018).
Tittl, A. et al. Imaging-based molecular barcoding with pixelated dielectric metasurfaces. Science 360, 1105–1109 (2018).
Tseng, M. L., Jahani, Y., Leitis, A. & Altug, H. Dielectric metasurfaces enabling advanced optical biosensors. ACS Photon. 8, 47–60 (2021).
Rodrigo, D. et al. Mid-infrared plasmonic biosensing with graphene. Science 349, 165–168 (2015).
Oh, S.-H. et al. Nanophotonic biosensors harnessing van der Waals materials. Nat. Commun. 12, 3824 (2021).
Schwaighofer, A., Brandstetter, M. & Lendl, B. Quantum cascade lasers (QCLs) in biomedical spectroscopy. Chem. Soc. Rev. 46, 5903–5924 (2017).
Mizaikoff, B. Waveguide-enhanced mid-infrared chem/bio sensors. Chem. Soc. Rev. 42, 8683–8699 (2013).
Chen, C. et al. Waveguide-integrated compact plasmonic resonators for on-chip mid-infrared laser spectroscopy. Nano Lett. 18, 7601–7608 (2018).
Aouani, H. et al. Ultrasensitive broadband probing of molecular vibrational modes with multifrequency optical antennas. ACS Nano 7, 669–675 (2013).
Rodrigo, D., Tittl, A., John-Herpin, A., Limaj, O. & Altug, H. Self-similar multiresonant nanoantenna arrays for sensing from near- to mid-infrared. ACS Photon. 5, 4903–4911 (2018).
Rodrigo, D. et al. Resolving molecule-specific information in dynamic lipid membrane processes with multi-resonant infrared metasurfaces. Nat. Commun. 9, 2160 (2018).
Tittl, A., John-Herpin, A., Leitis, A., Arvelo, E. R. & Altug, H. Metasurface-based molecular biosensing aided by artificial intelligence. Angew. Chem. Int. Ed. 58, 14810–14822 (2019).
Morais, C. L. M., Lima, K. M. G., Singh, M. & Martin, F. L. Tutorial: multivariate classification for vibrational spectroscopy in biological samples. Nat. Protocols 15, 2143–2162 (2020).
Lansford, J. L. & Vlachos, D. G. Infrared spectroscopy data- and physics-driven machine learning for characterizing surface microstructure of complex materials. Nat. Commun. 11, 1513 (2020).
John-Herpin, A., Kavungal, D., Mücke, L. & Altug, H. Infrared metasurface augmented by deep learning for monitoring dynamics between all major classes of biomolecules. Adv. Mater. 33, 2006054 (2021).
Lopez, G. A., Estevez, M.-C., Soler, M. & Lechuga, L. M. Recent advances in nanoplasmonic biosensors: applications and lab-on-a-chip integration. Nanophotonics 6, 123–136 (2017).
Zanchetta, G., Lanfranco, R., Giavazzi, F., Bellini, T. & Buscaglia, M. Emerging applications of label-free optical biosensors. Nanophotonics 6, 627–645 (2017).
Chen, C. & Wang, J. Optical biosensors: an exhaustive and comprehensive review. Analyst 145, 1605–1628 (2020).
Angelopoulou, M., Kakabakos, S. & Petrou, P. Label-free biosensors based onto monolithically integrated onto silicon optical transducers. Chemosensors 6, 52 (2018).
Mudumba, S. et al. Photonic ring resonance is a versatile platform for performing multiplex immunoassays in real time. J. Immunol. Methods 448, 34–43 (2017).
Chamanzar, M., Xia, Z., Yegnanarayanan, S. & Adibi, A. Hybrid integrated plasmonic–photonic waveguides for on-chip localized surface plasmon resonance (LSPR) sensing and spectroscopy. Opt. Express 21, 32086 (2013).
Agnarsson, B. et al. Evanescent light-scattering microscopy for label-free interfacial imaging: from single sub-100 nm vesicles to live cells. ACS Nano 9, 11849–11862 (2015).
Peyskens, F., Dhakal, A., Van Dorpe, P., Le Thomas, N. & Baets, R. Surface enhanced Raman spectroscopy using a single mode nanophotonic–plasmonic platform. ACS Photon. 3, 102–108 (2016).
Oliverio, M., Perotto, S., Messina, G. C., Lovato, L. & De Angelis, F. Chemical functionalization of plasmonic surface biosensors: a tutorial review on issues, strategies, and costs. ACS Appl. Mater. Interfaces 9, 29394–29411 (2017).
Aksu, S. et al. High-Throughput nanofabrication of infrared plasmonic nanoantenna arrays for vibrational nanospectroscopy. Nano Lett. 10, 2511–2518 (2010).
Henzie, J., Lee, M. H. & Odom, T. W. Multiscale patterning of plasmonic metamaterials. Nat. Nanotechnol. 2, 549–554 (2007).
Lindquist, N. C., Nagpal, P., McPeak, K. M., Norris, D. J. & Oh, S.-H. Engineering metallic nanostructures for plasmonics and nanophotonics. Rep. Prog. Phys. 75, 036501 (2012).
Yesilkoy, F. et al. Phase-sensitive plasmonic biosensor using a portable and large field-of-view interferometric microarray imager. Light.: Sci. Appl. 7, 17152–17152 (2018).
Luka, G. et al. Microfluidics integrated biosensors: a leading technology towards lab-on-a-chip and sensing applications. Sensors 15, 30011–30031 (2015).
Squires, T. M., Messinger, R. J. & Manalis, S. R. Making it stick: convection, reaction and diffusion in surface-based biosensors. Nat. Biotechnol. 26, 417–426 (2008).
Huang, M., Galarreta, B. C., Cetin, A. E. & Altug, H. Actively transporting virus like analytes with optofluidics for rapid and ultrasensitive biodetection. Lab Chip 13, 4841–4847 (2013).
Escobedo, C., Brolo, A. G., Gordon, R. & Sinton, D. Optofluidic concentration: plasmonic nanostructure as concentrator and sensor. Nano Lett. 12, 1592–1596 (2012).
Barik, A. et al. Dielectrophoresis-enhanced plasmonic sensing with gold nanohole arrays. Nano Lett. 14, 2006–2012 (2014).
Ndukaife, J. C. et al. Long-range and rapid transport of individual nano-objects by a hybrid electrothermoplasmonic nanotweezer. Nat. Nanotechnol. 11, 53–59 (2016).
Olanrewaju, A., Beaugrand, M., Yafia, M. & Juncker, D. Capillary microfluidics in microchannels: from microfluidic networks to capillaric circuits. Lab Chip 18, 2323–2347 (2018).
Samiei, E., Tabrizian, M. & Hoorfar, M. A review of digital microfluidics as portable platforms for lab-on a-chip applications. Lab Chip 16, 2376–2396 (2016).
Sin, M. L., Mach, K. E., Wong, P. K. & Liao, J. C. Advances and challenges in biosensor-based diagnosis of infectious diseases. Expert Rev. Mol. Diagn. 14, 225–244 (2014).
Sonker, M., Sahore, V. & Woolley, A. T. Recent advances in microfluidic sample preparation and separation techniques for molecular biomarker analysis: a critical review. Anal. Chim. Acta 986, 1–11 (2017).
Senf, B., Yeo, W.-H. & Kim, J.-H. Recent advances in portable biosensors for biomarker detection in body fluids. Biosensors 10, 127 (2020).
Vaisocherová, H., Brynda, E. & Homola, J. Functionalizable low-fouling coatings for label-free biosensing in complex biological media: advances and applications. Anal. Bioanal. Chem. 407, 3927–3953 (2015).
Maan, A. M. C., Hofman, A. H., Vos, W. M. & Kamperman, M. Recent developments and practical feasibility of polymer‐based antifouling coatings. Adv. Funct. Mater. 30, 2000936 (2020).
Hinman, S. S., McKeating, K. S. & Cheng, Q. Surface plasmon resonance: material and interface design for universal accessibility. Anal. Chem. 90, 19–39 (2018).
Lísalová, H. et al. Ultralow-fouling behavior of biorecognition coatings based on carboxy-functional brushes of zwitterionic homo- and copolymers in blood plasma: functionalization matters. Anal. Chem. 89, 3524–3531 (2017).
Yoo, S. M., Kim, D.-K. & Lee, S. Y. Aptamer-functionalized localized surface plasmon resonance sensor for the multiplexed detection of different bacterial species. Talanta 132, 112–117 (2015).
Muyldermans, S. Nanobodies: natural single-domain antibodies. Annu. Rev. Biochem. 82, 775–797 (2013).
Lubken, R. M., de Jong, A. M. & Prins, M. W. J. Multiplexed continuous biosensing by single-molecule encoded nanoswitches. Nano Lett. 20, 2296–2302 (2020).
Ferreira, J. et al. Attomolar protein detection using in-hole surface plasmon resonance. J. Am. Chem. Soc. 131, 436–437 (2009).
Feuz, L., Jönsson, P., Jonsson, M. P. & Höök, F. Improving the limit of detection of nanoscale sensors by directed binding to high-sensitivity areas. ACS Nano 4, 2167–2177 (2010).
Galloway, C. M. et al. Plasmon-assisted delivery of single nano-objects in an optical hot spot. Nano Lett. 13, 4299–4304 (2013).
Tijunelyte, I. et al. Nanoplasmonics tuned ‘click chemistry’. Nanoscale 8, 7105–7112 (2016).
Hu, H. et al. Gas identification with graphene plasmons. Nat. Commun. 10, 1131 (2019).
Lee, I.-H., Yoo, D., Avouris, P., Low, T. & Oh, S.-H. Graphene acoustic plasmon resonator for ultrasensitive infrared spectroscopy. Nat. Nanotechnol. 14, 313–319 (2019).
Dahlin, A. B. et al. Electrochemical plasmonic sensors. Anal. Bioanal. Chem. 402, 1773–1784 (2012).
Hentschel, M., Schäferling, M., Duan, X., Giessen, H. & Liu, N. Chiral plasmonics. Sci. Adv. 3, e1602735 (2017).
Mohammadi, E. et al. Nanophotonic platforms for enhanced chiral sensing. ACS Photon. 5, 2669–2675 (2018).
Solomon, M. L., Hu, J., Lawrence, M., García-Etxarri, A. & Dionne, J. A. Enantiospecific optical enhancement of chiral sensing and separation with dielectric metasurfaces. ACS Photon. 6, 43–49 (2019).
Zhang, Q. et al. Unraveling the origin of chirality from plasmonic nanoparticle-protein complexes. Science 365, 1475–1478 (2019).
García-Guirado, J., Svedendahl, M., Puigdollers, J. & Quidant, R. Enhanced chiral sensing with dielectric nanoresonators. Nano Lett. 20, 585–591 (2020).
Bauch, M., Toma, K., Toma, M., Zhang, Q. & Dostalek, J. Plasmon-enhanced fluorescence biosensors: a review. Plasmonics 9, 781–799 (2014).
Cui, F., Yue, Y., Zhang, Y., Zhang, Z. & Zhou, H. S. Advancing biosensors with machine learning. ACS Sens. 5, 3346–3364 (2020).
Wu, Y. et al. Deep learning enables high-throughput analysis of particle-aggregation-based bio-sensors imaged using holography. ACS Photon. 6, 294–301 (2018).
Akkilic, N., Geschwindner, S. & Höök, F. Single-molecule biosensors: recent advances and applications. Biosens. Bioelectron. 151, 111944 (2020).
Huang, Q. et al. Critical review: digital resolution biomolecular sensing for diagnostics and life science research. Lab Chip 20, 2816–2840 (2020).
Jung, L. S., Campbell, C. T., Chinowsky, T. M., Mar, M. N. & Yee, S. S. Quantitative interpretation of the response of surface plasmon resonance sensors to adsorbed films. Langmuir 14, 5636–5648 (1998).
Wu, Y., Tilley, R. D. & Gooding, J. J. Challenges and solutions in developing ultrasensitive biosensors. J. Am. Chem. Soc. 141, 1162–1170 (2019).
Lindquist, N. C., de Albuquerque, C. D. L., Sobral-Filho, R. G., Paci, I. & Brolo, A. G. High-speed imaging of surface-enhanced Raman scattering fluctuations from individual nanoparticles. Nat. Nanotechnol. 14, 981–987 (2019).
Willets, K. A. & Van Duyne, R. P. Localized surface plasmon resonance spectroscopy and sensing. Annu. Rev. Phys. Chem. 58, 267–297 (2007).
Limaj, O. et al. Infrared plasmonic biosensor for real-time and label-free monitoring of lipid membranes. Nano Lett. 16, 1502–1508 (2016).
Acknowledgements
We acknowledge funding from the European Research Council under contract no. 682167 and no. 875672, and the European Union Horizon 2020 Framework Programme for Research and Innovation under contract no. FETOPEN-737071 and no. 777714 (H.A.), Samsung Global Research Outreach program, the Sanford P. Bordeau Chair in Electrical Engineering, and the Minnesota Environment and Natural Resources Trust Fund as recommended by the Legislative-Citizen Commission on Minnesota Resources (S.-H.O.), Deutsche Forschungsgemeinschaft, Engineering and Physical Sciences Research Council, and the Lee-Lucas Chair in Physics (S.A.M.) and the Czech Science Foundation under contract no. 20-23787X (J.H.). We also acknowledge A. John-Herpin, A. Belushkin and C. T. Ertsgaard for their help with the figure preparation.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Nanotechnology thanks Thomas Haertling, Mikael Käll and Jean-Francois Masson for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Altug, H., Oh, SH., Maier, S.A. et al. Advances and applications of nanophotonic biosensors. Nat. Nanotechnol. 17, 5–16 (2022). https://doi.org/10.1038/s41565-021-01045-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41565-021-01045-5
This article is cited by
-
Synthesized complex-frequency excitation for ultrasensitive molecular sensing
eLight (2024)
-
An optofluidic antenna for enhancing the sensitivity of single-emitter measurements
Nature Communications (2024)
-
Quantum-mechanical effects in photoluminescence from thin crystalline gold films
Light: Science & Applications (2024)
-
Phase-driven progress in nanophotonic biosensing
Light: Science & Applications (2024)
-
Recent Advances in Electrochemical and Nanophotonic Biosensors for SARS-CoV-2 Detection
BioChip Journal (2024)