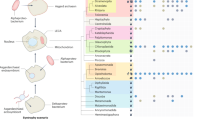
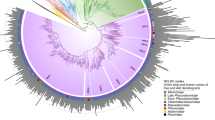
Abstract
The last universal cellular ancestor (LUCA) is the most recent population of organisms from which all cellular life on Earth descends. The reconstruction of the genome and phenotype of the LUCA is a major challenge in evolutionary biology. Given that all life forms are associated with viruses and/or other mobile genetic elements, there is no doubt that the LUCA was a host to viruses. Here, by projecting back in time using the extant distribution of viruses across the two primary domains of life, bacteria and archaea, and tracing the evolutionary histories of some key virus genes, we attempt a reconstruction of the LUCA virome. Even a conservative version of this reconstruction suggests a remarkably complex virome that already included the main groups of extant viruses of bacteria and archaea. We further present evidence of extensive virus evolution antedating the LUCA. The presence of a highly complex virome implies the substantial genomic and pan-genomic complexity of the LUCA itself.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Forterre, P. & Prangishvili, D. The great billion-year war between ribosome- and capsid-encoding organisms (cells and viruses) as the major source of evolutionary novelties. Ann. N. Y. Acad. Sci. 1178, 65–77 (2009).
Koonin, E. V., Senkevich, T. G. & Dolja, V. V. The ancient virus world and evolution of cells. Biol. Direct 1, 29 (2006).
Moreira, D. & Lopez-Garcia, P. Ten reasons to exclude viruses from the tree of life. Nat. Rev. Microbiol. 7, 306–311 (2009).
Koonin, E. V. & Dolja, V. V. A virocentric perspective on the evolution of life. Curr. Opin. Virol. 3, 546–557 (2013).
Raoult, D. & Forterre, P. Redefining viruses: lessons from Mimivirus. Nat. Rev. Microbiol. 6, 315–319 (2008).
Koonin, E. V., Wolf, Y. I. & Katsnelson, M. I. Inevitability of the emergence and persistence of genetic parasites caused by evolutionary instability of parasite-free states. Biol. Direct 12, 31 (2017).
Szathmary, E. & Demeter, L. Group selection of early replicators and the origin of life. J. Theor. Biol. 128, 463–486 (1987).
Takeuchi, N. & Hogeweg, P. The role of complex formation and deleterious mutations for the stability of RNA-like replicator systems. J. Mol. Evol. 65, 668–686 (2007).
Takeuchi, N. & Hogeweg, P. Evolutionary dynamics of RNA-like replicator systems: a bioinformatic approach to the origin of life. Phys. Life Rev. 9, 219–263 (2012).
Eigen, M. The origin of genetic information: viruses as models. Gene 135, 37–47 (1993).
Eigen, M. Selforganization of matter and the evolution of biological macromolecules. Naturwissenschaften 58, 465–523 (1971).
Baltimore, D. Expression of animal virus genomes. Bacteriol. Rev. 35, 235–241 (1971).
Koonin, E. V. et al. Global organization and proposed megataxonomy of the virus world. Microbiol. Mol. Biol. Rev. 84, e00061-19 (2020).
International Committee on Taxonomy of Viruses Executive Committee. The new scope of virus taxonomy: partitioning the virosphere into 15 hierarchical ranks. Nat. Microbiol. 5, 668–674 (2020).
Siddell, S. G. et al. Additional changes to taxonomy ratified in a special vote by the international committee on taxonomy of viruses (October 2018). Arch. Virol. 164, 943–946 (2019).
Prangishvili, D. et al. The enigmatic archaeal virosphere. Nat. Rev. Microbiol. 15, 724–739 (2017).
Munson-McGee, J. H., Snyder, J. C. & Young, M. J. Archaeal viruses from high-temperature environments. Genes 9, E128 (2018).
Iranzo, J., Koonin, E. V., Prangishvili, D. & Krupovic, M. Bipartite network analysis of the archaeal virosphere: evolutionary connections between viruses and capsidless mobile elements. J. Virol. 90, 11043–11055 (2016).
Krupovic, M., Dolja, V. V. & Koonin, E. V. Origin of viruses: primordial replicators recruiting capsids from hosts. Nat. Rev. Microbiol. 17, 449–458 (2019).
Koonin, E. V. Comparative genomics, minimal gene-sets and the last universal common ancestor. Nat. Rev. Microbiol. 1, 127–136 (2003).
Glansdorff, N., Xu, Y. & Labedan, B. The last universal common ancestor: emergence, constitution and genetic legacy of an elusive forerunner. Biol. Direct 3, 29 (2008).
Weiss, M. C. et al. The physiology and habitat of the last universal common ancestor. Nat. Microbiol. 1, 16116 (2016).
Doolittle, W. F. Uprooting the tree of life. Sci. Am. 282, 90–95 (2000).
Puigbo, P., Wolf, Y. I. & Koonin, E. V. Search for a ‘Tree of Life’ in the thicket of the phylogenetic forest. J. Biol. 8, 59 (2009).
Berkemer, S. J. & McGlynn, S. E. A new analysis of archaea-bacteria domain separation: variable phylogenetic distance and the tempo of early evolution. Mol. Biol. Evol. https://doi.org/10.1093/molbev/msaa089 (2020).
Fournier, G. P., Andam, C. P. & Gogarten, J. P. Ancient horizontal gene transfer and the last common ancestors. BMC Evol. Biol. 15, 70 (2015).
Ouzounis, C. A., Kunin, V., Darzentas, N. & Goldovsky, L. A minimal estimate for the gene content of the last universal common ancestor–exobiology from a terrestrial perspective. Res. Microbiol. 157, 57–68 (2006).
Gogarten, J. P. & Taiz, L. Evolution of proton pumping ATPases: rooting the tree of life. Photosynth. Res. 33, 137–146 (1992).
Mirkin, B. G., Fenner, T. I., Galperin, M. Y. & Koonin, E. V. Algorithms for computing parsimonious evolutionary scenarios for genome evolution, the last universal common ancestor and dominance of horizontal gene transfer in the evolution of prokaryotes. BMC Evol. Biol. 3, 2 (2003).
Brown, J. R. & Doolittle, W. F. Archaea and the prokaryote-to-eukaryote transition. Microbiol. Mol. Biol. Rev. 61, 456–502 (1997).
Leipe, D. D., Aravind, L. & Koonin, E. V. Did DNA replication evolve twice independently? Nucleic Acids Res. 27, 3389–3401 (1999).
Pereto, J., Lopez-Garcia, P. & Moreira, D. Ancestral lipid biosynthesis and early membrane evolution. Trends Biochem. Sci. 29, 469–477 (2004).
Lombard, J., Lopez-Garcia, P. & Moreira, D. The early evolution of lipid membranes and the three domains of life. Nat. Rev. Microbiol. 10, 507–515 (2012).
Mushegian, A. R. & Koonin, E. V. A minimal gene set for cellular life derived by comparison of complete bacterial genomes. Proc. Natl Acad. Sci. USA 93, 10268–10273 (1996).
Forterre, P. Three RNA cells for ribosomal lineages and three DNA viruses to replicate their genomes: a hypothesis for the origin of cellular domain. Proc. Natl Acad. Sci. USA 103, 3669–3674 (2006).
Raia, P. et al. Structure of the DP1-DP2 PolD complex bound with DNA and its implications for the evolutionary history of DNA and RNA polymerases. PLoS Biol. 17, e3000122 (2019).
Sauguet, L. The extended “two-barrel” polymerases superfamily: structure, function and evolution. J. Mol. Biol. 431, 4167–4183 (2019).
Koonin, E. V., Krupovic, M., Ishino, S. & Ishino, Y. The replication machinery of LUCA: common origin of DNA replication and transcription. BMC Biol. 18, 61 (2020).
Werner, F. & Grohmann, D. Evolution of multisubunit RNA polymerases in the three domains of life. Nat. Rev. Microbiol. 9, 85–98 (2011).
Martin, W. & Russell, M. J. On the origins of cells: a hypothesis for the evolutionary transitions from abiotic geochemistry to chemoautotrophic prokaryotes, and from prokaryotes to nucleated cells. Philos. Trans. R. Soc. Lond. B Biol. Sci. 358, 59–83 (2003).
Koonin, E. V. & Martin, W. On the origin of genomes and cells within inorganic compartments. Trends Genet. 21, 647–654 (2005).
Mulkidjanian, A. Y., Galperin, M. Y. & Koonin, E. V. Co-evolution of primordial membranes and membrane proteins. Trends Biochem. Sci. 34, 206–215 (2009).
Villanueva, L. et al. Bridging the divide: bacteria synthesizing archaeal membrane lipids. Preprint at bioRxiv https://doi.org/10.1101/448035 (2018).
Caforio, A. et al. Converting Escherichia coli into an archaebacterium with a hybrid heterochiral membrane. Proc. Natl Acad. Sci. USA 115, 3704–3709 (2018).
Rajagopal, M. & Walker, S. Envelope structures of gram-positive bacteria. Curr. Top. Microbiol. Immunol. 404, 1–44 (2017).
Auer, G. K. & Weibel, D. B. Bacterial cell mechanics. Biochemistry 56, 3710–3724 (2017).
Sleytr, U. B., Schuster, B., Egelseer, E. M. & Pum, D. S-layers: principles and applications. FEMS Microbiol. Rev. 38, 823–864 (2014).
Vernikos, G., Medini, D., Riley, D. R. & Tettelin, H. Ten years of pan-genome analyses. Curr. Opin. Microbiol. 23, 148–154 (2015).
Medini, D., Donati, C., Tettelin, H., Masignani, V. & Rappuoli, R. The microbial pan-genome. Curr. Opin. Genet. Dev. 15, 589–594 (2005).
Puigbo, P., Lobkovsky, A. E., Kristensen, D. M., Wolf, Y. I. & Koonin, E. V. Genomes in turmoil: quantification of genome dynamics in prokaryote supergenomes. BMC Biol. 12, 66 (2014).
Parks, D. H. et al. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat. Biotechnol. 36, 996–1004 (2018).
Embley, T. M. & Martin, W. Eukaryotic evolution, changes and challenges. Nature 440, 623–630 (2006).
Kristensen, D. M., Cai, X. & Mushegian, A. Evolutionarily conserved orthologous families in phages are relatively rare in their prokaryotic hosts. J. Bacteriol. 193, 1806–1814 (2011).
Iranzo, J., Krupovic, M. & Koonin, E. V. The double-stranded DNA virosphere as a modular hierarchical network of gene sharing. mBio 7, e00978-16 (2016).
Duda, R. L. & Teschke, C. M. The amazing HK97 fold: versatile results of modest differences. Curr. Opin. Virol. 36, 9–16 (2019).
Krupovic, M., Forterre, P. & Bamford, D. H. Comparative analysis of the mosaic genomes of tailed archaeal viruses and proviruses suggests common themes for virion architecture and assembly with tailed viruses of bacteria. J. Mol. Biol. 397, 144–160 (2010).
Sencilo, A. et al. Snapshot of haloarchaeal tailed virus genomes. RNA Biol. 10, 803–816 (2013).
Cheng, H., Shen, N., Pei, J. & Grishin, N. V. Double-stranded DNA bacteriophage prohead protease is homologous to herpesvirus protease. Protein Sci. 13, 2260–2269 (2004).
Low, S. J., Dzunkova, M., Chaumeil, P. A., Parks, D. H. & Hugenholtz, P. Evaluation of a concatenated protein phylogeny for classification of tailed double-stranded DNA viruses belonging to the order Caudovirales. Nat. Microbiol. 4, 1306–1315 (2019).
Dion, M. B., Oechslin, F. & Moineau, S. Phage diversity, genomics and phylogeny. Nat. Rev. Microbiol. 18, 125–138 (2020).
Philosof, A. et al. Novel abundant oceanic viruses of uncultured marine group II euryarchaeota. Curr. Biol. 27, 1362–1368 (2017).
Kazlauskas, D., Krupovic, M. & Venclovas, C. The logic of DNA replication in double-stranded DNA viruses: insights from global analysis of viral genomes. Nucleic Acids Res. 44, 4551–4564 (2016).
Yutin, N., Bäckström, D., Ettema, T. J. G., Krupovic, M. & Koonin, E. V. Vast diversity of prokaryotic virus genomes encoding double jelly-roll major capsid proteins uncovered by genomic and metagenomic sequence analysis. Virol. J. 15, 67 (2018).
Kauffman, K. M. et al. A major lineage of non-tailed dsDNA viruses as unrecognized killers of marine bacteria. Nature 554, 118–122 (2018).
Krupovic, M. & Bamford, D. H. Archaeal proviruses TKV4 and MVV extend the PRD1-adenovirus lineage to the phylum euryarchaeota. Virology 375, 292–300 (2008).
Krupovic, M. et al. Integrated mobile genetic elements in Thaumarchaeota. Env. Microbiol. 21, 2056–2078 (2019).
Jalasvuori, M. & Koskinen, K. Extending the hosts of Tectiviridae into four additional genera of Gram-positive bacteria and more diverse Bacillus species. Virology 518, 136–142 (2018).
Roux, S. et al. Cryptic inoviruses revealed as pervasive in bacteria and archaea across earth’s biomes. Nat. Microbiol. 4, 1895–1906 (2019).
Roux, S., Krupovic, M., Poulet, A., Debroas, D. & Enault, F. Evolution and diversity of the microviridae viral family through a collection of 81 new complete genomes assembled from virome reads. PLoS One 7, e40418 (2012).
Creasy, A., Rosario, K., Leigh, B. A., Dishaw, L. J. & Breitbart, M. Unprecedented diversity of ssDNA phages from the family Microviridae detected within the gut of a protochordate model organism (Ciona robusta). Viruses 10, 404 (2018).
Tisza, M. J. et al. Discovery of several thousand highly diverse circular DNA viruses. eLife 9, e51971 (2020).
Koonin, E. V., Dolja, V. V. & Krupovic, M. Origins and evolution of viruses of eukaryotes: the ultimate modularity. Virology 479-480, 2–25 (2015).
Wolf, Y. I. et al. Origins and evolution of the global RNA virome. mBio 9, e02329-18 (2018).
Krishnamurthy, S. R., Janowski, A. B., Zhao, G., Barouch, D. & Wang, D. Hyperexpansion of RNA bacteriophage diversity. PLoS Biol. 14, e1002409 (2016).
Callanan, J. et al. Expansion of known ssRNA phage genomes: from tens to over a thousand. Sci. Adv. 6, eaay5981 (2020).
Gladyshev, E. A. & Arkhipova, I. R. A widespread class of reverse transcriptase-related cellular genes. Proc. Natl Acad. Sci. USA 108, 20311–20316 (2011).
Zimmerly, S. & Wu, L. An unexplored diversity of reverse transcriptases in bacteria. Microbiol. Spectr. 3, MDNA3-0058-2014 https://doi.org/10.1128/microbiolspec.MDNA3-0058-2014 (2015).
Gilbert, W. The origin of life: the RNA world. Nature 319, 618 (1986).
Joyce, G. F. The antiquity of RNA-based evolution. Nature 418, 214–221 (2002).
Bernhardt, H. S. The RNA world hypothesis: the worst theory of the early evolution of life (except for all the others)(a). Biol. Direct 7, 23 (2012).
Catchpole, R. J. & Forterre, P. The evolution of reverse gyrase suggests a nonhyperthermophilic last universal common ancestor. Mol. Biol. Evol. 36, 2737–2747 (2019).
Cantine, M. D. & Fournier, G. P. Environmental adaptation from the origin of life to the last universal common ancestor. Orig. Life Evol. Biosph. 48, 35–54 (2018).
Boussau, B., Blanquart, S., Necsulea, A., Lartillot, N. & Gouy, M. Parallel adaptations to high temperatures in the Archaean eon. Nature 456, 942–945 (2008).
Groussin, M. & Gouy, M. Adaptation to environmental temperature is a major determinant of molecular evolutionary rates in archaea. Mol. Biol. Evol. 28, 2661–2674 (2011).
Forterre, P. The common ancestor of archaea and eukarya was not an archaeon. Archaea 2013, 372396 (2013).
Krupovic, M., Quemin, E. R., Bamford, D. H., Forterre, P. & Prangishvili, D. Unification of the globally distributed spindle-shaped viruses of the Archaea. J. Virol. 88, 2354–2358 (2014).
Krupovic, M. & Koonin, E. V. Multiple origins of viral capsid proteins from cellular ancestors. Proc. Natl Acad. Sci. USA 114, E2401–E2410 (2017).
Santos-Perez, I. et al. Structural basis for assembly of vertical single β-barrel viruses. Nat. Commun. 10, 1184 (2019).
De Colibus, L. et al. Assembly of complex viruses exemplified by a halophilic euryarchaeal virus. Nat. Commun. 10, 1456 (2019).
Rissanen, I. et al. Bacteriophage P23-77 capsid protein structures reveal the archetype of an ancient branch from a major virus lineage. Structure 21, 718–726 (2013).
Liu, Y. et al. A novel type of polyhedral viruses infecting hyperthermophilic archaea. J. Virol. 91, e00589-17 (2017).
Wang, F. et al. A packing for A-form DNA in an icosahedral virus. Proc. Natl Acad. Sci. USA 116, 22591–22597 (2019).
Kelley, L. L. et al. Structure of the hypothetical protein PF0899 from pyrococcus furiosus at 1.85 a resolution. Acta Crystallogr. Sect. F. Struct. Biol. Cryst. Commun. 63, 549–552 (2007).
Holm, L. DALI and the persistence of protein shape. Protein Sci. 29, 128–140 (2020).
Grininger, M., Zeth, K. & Oesterhelt, D. Dodecins: a family of lumichrome binding proteins. J. Mol. Biol. 357, 842–857 (2006).
Krupovic, M. & Bamford, D. H. Order to the viral universe. J. Virol. 84, 12476–12479 (2010).
Ilyina, T. V. & Koonin, E. V. Conserved sequence motifs in the initiator proteins for rolling circle DNA replication encoded by diverse replicons from eubacteria, eucaryotes and archaebacteria. Nucleic Acids Res. 20, 3279–3285 (1992).
Kazlauskas, D., Varsani, A., Koonin, E. V. & Krupovic, M. Multiple origins of prokaryotic and eukaryotic single-stranded DNA viruses from bacterial and archaeal plasmids. Nat. Commun. 10, 3425 (2019).
Redrejo-Rodríguez, M. et al. Primer-independent DNA synthesis by a family B DNA polymerase from self-replicating mobile genetic elements. Cell Rep. 21, 1574–1587 (2017).
Kim, J. G. et al. Spindle-shaped viruses infect marine ammonia-oxidizing thaumarchaea. Proc. Natl Acad. Sci. USA 116, 15645–15650 (2019).
Iyer, L. M., Koonin, E. V., Leipe, D. D. & Aravind, L. Origin and evolution of the archaeo-eukaryotic primase superfamily and related palm-domain proteins: structural insights and new members. Nucleic Acids Res. 33, 3875–3896 (2005).
Kazlauskas, D. et al. Novel families of archaeo-eukaryotic primases associated with mobile genetic elements of bacteria and archaea. J. Mol. Biol. 430, 737–750 (2018).
Weigel, C. & Seitz, H. Bacteriophage replication modules. FEMS Microbiol. Rev. 30, 321–381 (2006).
Brezellec, P. et al. Domestication of lambda phage genes into a putative third type of replicative helicase matchmaker. Genome Biol. Evol. 9, 1561–1566 (2017).
Pfister, P., Wasserfallen, A., Stettler, R. & Leisinger, T. Molecular analysis of methanobacterium phage psiM2. Mol. Microbiol. 30, 233–244 (1998).
Iro, M. et al. The lysogenic region of virus phiCh1: identification of a repressor-operator system and determination of its activity in halophilic Archaea. Extremophiles 11, 383–396 (2007).
Dellas, N., Snyder, J. C., Bolduc, B. & Young, M. J. Archaeal viruses: diversity, replication, and structure. Annu. Rev. Virol. 1, 399–426 (2014).
Zhaxybayeva, O. & Gogarten, J. P. Cladogenesis, coalescence and the evolution of the three domains of life. Trends Genet. 20, 182–187 (2004).
Mendler, K. et al. AnnoTree: visualization and exploration of a functionally annotated microbial tree of life. Nucleic Acids Res. 47, 4442–4448 (2019).
Roux, S., Hallam, S. J., Woyke, T. & Sullivan, M. B. Viral dark matter and virus-host interactions resolved from publicly available microbial genomes. eLife 4, e08490 (2015).
Krupovic, M. & Bamford, D. H. Putative prophages related to lytic tailless marine dsDNA phage PM2 are widespread in the genomes of aquatic bacteria. BMC Genomics 8, 236 (2007).
Pawlowski, A., Rissanen, I., Bamford, J. K., Krupovic, M. & Jalasvuori, M. Gammasphaerolipovirus, a newly proposed bacteriophage genus, unifies viruses of halophilic archaea and thermophilic bacteria within the novel family Sphaerolipoviridae. Arch. Virol. 159, 1541–1554 (2014).
Wang, F. et al. Structure of a filamentous virus uncovers familial ties within the archaeal virosphere. Virus Evol. 6, veaa023 (2020).
Weidenbach, K. et al. Methanosarcina spherical virus, a novel archaeal lytic virus targeting methanosarcina strains. J. Virol. 91, e00955-17 (2017).
Liu, Y. et al. Identification and characterization of SNJ2, the first temperate pleolipovirus integrating into the genome of the SNJ1-lysogenic archaeal strain. Mol. Microbiol. 98, 1002–1020 (2015).
Wang, J. et al. A novel family of tyrosine integrases encoded by the temperate pleolipovirus SNJ2. Nucleic Acids Res. 46, 2521–2536 (2018).
Altschul, S. F. et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997).
Acknowledgements
E.V.K. is supported by the funds of the Intramural Research Program of the National Institutes of Health of the USA. M.K. was supported by l’Agence Nationale de la Recherche grant ANR-17-CE15-0005-01.
Author information
Authors and Affiliations
Contributions
M.K. and E.V.K. researched data for article. M.K., V.V.D. and E.V.K. substantially contributed to discussion of content. M.K. and E.V.K. wrote the manuscript. M.K., V.V.D. and E.V.K. reviewed and edited the manuscript before submission.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information
Nature Reviews Microbiology thanks J. P. Gogarten, P. López-García and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Krupovic, M., Dolja, V.V. & Koonin, E.V. The LUCA and its complex virome. Nat Rev Microbiol 18, 661–670 (2020). https://doi.org/10.1038/s41579-020-0408-x
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41579-020-0408-x
This article is cited by
-
The virome of the last eukaryotic common ancestor and eukaryogenesis
Nature Microbiology (2023)
-
Mirusviruses link herpesviruses to giant viruses
Nature (2023)
-
MArVD2: a machine learning enhanced tool to discriminate between archaeal and bacterial viruses in viral datasets
ISME Communications (2023)
-
A compendium of viruses from methanogenic archaea reveals their diversity and adaptations to the gut environment
Nature Microbiology (2023)
-
Cyanophages as an important factor in the early evolution of oxygenic photosynthesis
Scientific Reports (2022)