Abstract
Invasive fungal infections pose an important threat to public health and are an under-recognized component of antimicrobial resistance, an emerging crisis worldwide. Across a period of profound global environmental change and expanding at-risk populations, human-infecting pathogenic fungi are evolving resistance to all licensed systemic antifungal drugs. In this Review, we highlight the main mechanisms of antifungal resistance and explore the similarities and differences between bacterial and fungal resistance to antimicrobial control. We discuss the research and innovation topics that are needed for risk reduction strategies aimed at minimizing the emergence of resistance in pathogenic fungi. These topics include links between the environment and One Health, surveillance, diagnostics, routes of transmission, novel therapeutics and methods to mitigate hotspots for fungal adaptation. We emphasize the global efforts required to steward our existing antifungal armamentarium, and to direct the research and development of future therapies and interventions.
Similar content being viewed by others
Introduction
Fungi cause diverse diseases in humans, ranging from allergic syndromes to superficial, disfiguring and life-threatening invasive fungal diseases (IFDs), which together affect more than a billion people worldwide1,2. Historically, treatment has relied heavily on just four classes of systemically acting antifungal drugs: the polyenes, azoles, echinocandins and the pyrimidine analogue 5-flucytosine3. However, fungi respond nimbly to chemical attack4 and treatment failure is a common outcome. This failure is attributable to an interplay between underlying host immune defects, antifungal drug properties (pharmacokinetics, pharmacodynamics and drug–drug interactions) and fungal characteristics including diverse cell morphologies, antifungal tolerance and antifungal resistance. Resistance to antifungal drugs is an emerging concern worldwide in both space and time4, including novel resistant variants of previously susceptible pathogens (for example, the ubiquitous mould Aspergillus fumigatus5) as well as entirely new emerging species that are resistant to multiple antifungal drugs (for example, the yeast Candida auris6). The increasing public health burden is now officially recognized with the listing of both of these pathogens on the urgent antimicrobial resistance (AMR) threat list published by the US CDC in 2019 (ref.7).
Traditionally, AMR programmes excluded antifungals because fungi have been widely neglected as a threat to public health8,9. Biological differences between fungal (eukaryotic) and bacterial (prokaryotic) pathogens also complicate the integration of fungi into existing AMR programmes. Yet the emerging problem of AMR is shared across the domains of life and many parallels exist between drug-resistant microorganisms (Table 1). The widespread use of broad-spectrum antibacterial antibiotics (for example, β-lactams, cephalosporins, carbapenems, quinolones and macrolides) profoundly impacts bacterial communities by purging susceptible genotypes in favour of those harbouring polymorphisms and genes conferring resistance, the fittest examples of which can go on to become globally widespread10. Although less well studied, aspects of this evolutionary process are mirrored across the fungal kingdom, and all pathogenic fungi can acquire resistance through adaptation to drug selection pressure4.
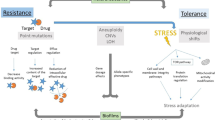
Mechanistically, antifungal resistance is usually acquired due to changes that directly or indirectly affect the drug–target interaction. Causally, resistance may arise via genetic changes to the target binding site (for example, mutation of the genes encoding lanosterol demethylase for azoles or β-glucan synthase for echinocandins)3, via overexpression of the amount of target available and/or by altering the effective drug concentration (via elevated drug efflux activity for intracellular drugs such as azoles3, or inhibition of prodrug activation for flucytosine11). Generalized fungal resistance mechanisms are shown in Fig. 1 and Box 1. In contrast to antifungal resistance, antifungal tolerance refers to the ability of drug-susceptible cells to grow at drug concentrations above the minimum inhibitory concentration (MIC) and involves a wide range of general stress response and/or epigenetic pathways (reviewed in ref.12). Tolerance is most evident with fungistatic drugs, and has been measured and characterized most extensively in Candida albicans isolates treated with fluconazole. However, its clinical importance remains an open question.
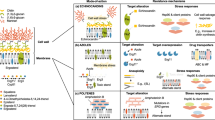
Routes to acquiring antifungal drug resistance and/or tolerance vary depending on the mode of action (MOA). a | Azole drug resistance is primarily due to increased efflux of the drug from the fungal cell (particularly in Candida spp.) and modifications to the sterol biosynthesis pathway caused by point mutations and promoter insertions in CYP51A (Aspergillus fumigatus). In other fungal species, such as Cryptococcus neoformans, overexpression of the drug target and efflux pumps caused by chromosomal aneuploidy and hypermutation is common. b | Polyenes alter cell membrane permeability by forming a complex with ergosterol, and resistance is caused by loss-of-function mutations in ergosterol biosynthesis genes (particularly in Aspergillus and Candida spp.). In Candida albicans in particular, double loss of ERG3 confers resistance. However, drug tolerance is common, via upregulation of ERG5, ERG6 and ERG25 in C. albicans. c | Cell membrane stress can also impact regulators of HSP90, conferring drug tolerance. Echinocandins inhibit 1,3-β-d-glucan synthase (FKS1), and mutations in this gene cause resistance in Candida and Fusarium spp. Echinocandin exposure can also lead to cell wall stress through inhibition of β-glucan synthase, with indirect downstream activation of Ca2+/calcineurin or HSP90/mTOR pathways, which are involved in drug tolerance. d | Pyrimidine analogues such as 5-flucytosine inhibit DNA and RNA synthesis. Resistance can arise via point mutations in the target gene FCY1, and is common in Candida spp. Hypermutation in Cryptococcus spp. is also known to cause resistance to this drug class. TR, tandem repeat.
Acquisition and emergence of antifungal drug resistance is fundamentally an evolutionary response to the selective pressure exerted by the drug. The likelihood of resistance emerging due to genetic changes is governed by the size of the population exposed to the selective pressure, the rate of cell doubling, the number of different pathways (physiological mechanisms and causal genetic changes) that confer resistance and the fitness costs associated with each of them. Importantly, antifungal drug resistance may originate in the host or in the environment. On one hand, in vivo resistance evolves de novo in individuals during antifungal therapy and causes treatment failure for a spectrum of pathogenic fungi spanning moulds13 and yeasts14. This is highly relevant for diverse Candida yeasts that are leading causes of nosocomial bloodstream infections and show widespread emergence of resistance to antifungals15,16. For instance, emergence of azole resistance in C. albicans during prolonged fluconazole therapy for oral candidiasis in individuals infected with HIV was well documented17. This phenomenon is not restricted to azole antifungals as progressive loss of echinocandin activity has also been reported during prolonged caspofungin therapy for C. albicans oesophagitis18. On the other hand, environmental resistance can emerge due to prior exposure of human pathogenic fungi to fungicides in nature5. Application of fungicides is dictated by the perennial need to defend intensively farmed animals and cultivations of solo, genetically homogeneous crops against fungal infections, as well as to preserve materials against saprotrophic decay by fungi. The environmental pressure of fungicides drives the evolution of resistance against all major classes of fungicides, including benzimidazoles, anilinopyrimidines, strobilurins, succinate dehydrogenase inhibitors and the sterol demethylation inhibitors (DMIs) including azoles4. Environmental resistance has not only necessitated the development of resistance management strategies and the breeding of more disease-resilient crops; it is also inextricably linked to the emergence of antifungal-resistant IFDs in humans as a consequence of the use of sterol 14α-DMIs both in the environment and in the clinic4,5. This emergence of drug-resistant fungi in nature and the clinic alongside expanding at-risk patient populations has prompted international funding bodies to add antifungal resistance to their research agendas. Of note, the Joint Programming Initiative on Antimicrobial Resistance (JPIAMR) consortium first included antifungal resistance in their Strategic Research and Innovation Agenda on Antimicrobial Resistance in April 2021. Their comprehensive One Health framework integrates six priority topics for addressing antifungal resistance: environment, transmission, surveillance, diagnostics, therapeutics and potential interventions19. In this Review, we focus on these priority areas with the goal of outlining current and future strategies alongside the key research that is needed to tackle the emerging public health issue of antifungal resistance.
Environment–One Health links and emerging antifungal resistance
Opportunistic pathogenic fungi are commonly found within our close living environments, and many can produce abundant airborne spores. Consequently, humans are exposed daily to diverse environmental fungal pathogens as bioaerosols. Whereas most environmental fungi cause no noticeable pathophysiological events in healthy individuals, those with compromised health or immunity are susceptible to a spectrum of disease including superficial, allergic, chronic and life-threatening IFDs. Patient populations at risk of IFDs are currently expanding and (of note) include older people20, those with immune systems compromised by HIV, cancer chemotherapy or transplant-necessitated immune suppression therapy, as well as those with severe viral infections such as influenza virus21 and COVID-19 (refs22,23). This latter group of patients has experienced surges in infection by groups of fungi, notably Aspergillus spp.24, Candida spp., including C. auris25, and in India the Mucoromycota species26, which exhibit robust intrinsic and acquired resistance to antifungal treatments.
Molecular epidemiological studies have repeatedly shown that many fungal diseases are acquired from our near environments; this is especially true for IFDs caused by Coccidioides spp.27, A. fumigatus28,29,30 and Cryptococcus spp.31. The intimate relationship between environmental populations of fungi and ensuing exposures to antifungals means that emerging environmental resistance is likely to affect the clinical management of fungal infections. In the agricultural setting, phytopathogenic fungi continually evolve resistance to the array of fungicides deployed against them. This rapid adaptation necessitates a continuous cycle of development as agribusinesses synthesize variants of existing fungicides or develop novel chemistries to thwart the accumulation of resistance4,32. However, as with licensed medical antifungals, agricultural fungicides used in agriculture have broad-spectrum activity across the fungal kingdom. As such, resistance arises not only in the crop pathogens per se but also in other environmental fungi that include potential human fungal pathogens.
The One Health implications of the widespread use of broad-spectrum agricultural fungicides have been most closely studied for the DMI azoles, where these compounds (for example, difenoconazole, epoxiconazole, propiconazole and tebuconazole) are not only structurally similar to the first-line medical triazoles (isavuconazole, itraconazole, posaconazole and voriconazole) but are used in increasing quantities worldwide. Azole fungicide usage in the United States has increased by more than 400% to ~3,000 metric tons per year from 2006 to 2016 (ref.33). China uses ten times more (~30,000 metric tons per year)34 with similar patterns repeated in the European Union35. The degradation half-life of azole fungicides is long, ranging from 47 days for tebuconazole to up to 120 days for epoxiconazole. Given their annual global use, substantial azole persistence in the environment is expected and has the potential to promote resistance or tolerance in opportunistic fungi. Worldwide increases in azole-resistant human fungal pathogens have been charted, both environmentally and clinically, since azoles were widely introduced in the 1980s (ref.4) and represent a ‘smoking gun’ linking agricultural fungicide use to burgeoning resistance in the clinic.
Dual use of azoles in the environment and clinic
Potential eco-evolutionary links between environmental and clinical resistance have been widely explored for A. fumigatus, following initial reports of azole-resistant A. fumigatus occurring in the environment and in patients with no prior history of antifungal treatment36. Ecological ‘hotspots’ have been postulated, whereby biotic and abiotic conditions would converge, permitting growth of the fungus in contact with sub-MIC azole concentrations and, thereby, generating conditions that are suitable for adaptation to drug pressure5 (Fig. 2). Support for this hypothesis comes from studies of environments that support high growth rates of A. fumigatus in the presence of agricultural DMIs; these environments include both home and industrial composters37, urban environments38 and greenhouses39.
Fungi in the environment are exposed to broad-spectrum classes of antifungals that are also utilized as frontline antifungal treatments in the clinic. Ecological hotspots occur that can act as amplifiers of resistant genotypes. One example is green waste stockpiling and composting. Humans with invasive fungal diseases (IFDs) may also transmit resistant genotypes (for instance in nosocomial outbreaks); however, the extent to which humans and other animals contribute to the presence of antifungal resistance in the environment remains unknown. Multiple extrinsic factors exist that are expected to influence the incidence of antifungal resistance. These include changing patterns of fungicide use in the environment and in waste management33; changing at-risk human host groups including viral infections such as COVID-19; changing climates that may alter the geographical range of fungi and adaptive landscape for resistance50 as well as providing novel routes for infection (for example, natural disasters); changing biotic interactions that may include xenobiotic chemicals that are analogues to antifungals; and changing virulence of the fungi themselves owing to intrinsic genetic change or synergies with combinations of the above drivers47.
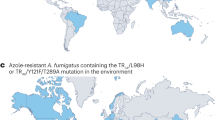
Environmental triazole resistance in A. fumigatus is characterized by hallmark genetic changes involving expression-upregulating tandem repeats (TRs) in the promoter region of CYP51A that drive increased expression of the gene, accompanied by within-gene point mutations that alter the drug target (Fig. 1). The most commonly occurring resistance alleles, TR34/L98H and TR46/Y121F/T289A, are associated with itraconazole and voriconazole resistance, respectively, both within and outside the clinic5, and are increasingly found worldwide40. Molecular epidemiological methods uncovered numerous examples of paired resistant isolates, sourced from the environment and infected individuals, with statistically significant genetic identity implying the infection source was the resistant environmental isolate30. Therefore, the topical question is no longer ‘does resistance in the environment affect patients?’. Rather, ‘where and why does resistance evolve?’ and ‘how does it disperse and what can be done to mitigate against it?’ have become crucial questions. The potential for global spread of triazole-resistant A. fumigatus through horticultural products, such as traded plant bulbs41, has been demonstrated and could be regulated. However, the dispersal of conidia on air currents is impossible to contain42. Moreover, although humans are not widely considered as an ecologically relevant source of azole-resistant A. fumigatus, the potential for certain groups of patients to acquire and to shed azole-resistant pathogens in health-care settings means that they cannot be excluded as a source of drug-resistant inoculum43 (Fig. 2).
Species-wide impacts of resistance in changing environments
The selection imposed by environmental fungicides likely has widespread effects upon the population genetic structures of human fungal pathogens and their genetically encoded phenotypic traits. The emergence of the TR34/L98H resistance-associated trait in A. fumigatus is associated with the escalating frequency of specific azole-resistant clones that carry this allele. However, scans across the genome of A. fumigatus have shown that azole selection leads to selective sweeps that operate across multiple genomic regions, and upon specific genetic backgrounds30. Accordingly, adaptation to fungicides in the environment may result in phenotypic changes beyond those encoded by the resistance mechanism. One example concerns the hypothesis that azole resistance can also drive adaptation of A. fumigatus to infection-related stress and virulence44,45. Sterol biosynthesis (the molecular target of the azoles), iron homeostasis and oxygen sensing are inextricably linked, as the production of ergosterol employs many iron-dependent enzymes and is highly oxygen-dependent46. As the host environment is both iron and oxygen limiting, any changes in the genome of A. fumigatus that increase azole resistance by enhancing iron uptake and adaptation to hypoxia have the potential to concurrently promote heightened virulence, a hypothesis that should be tested. Similarly, adaptation by Cryptococcus gattii to the broad-spectrum fungicide benomyl was linked to cross-resistance to fluconazole and increased virulence in mice, a phenotype that was attributed to MDR1 efflux pump overexpression47. In another example, the higher average temperatures expected under climate change scenarios may affect the emergence of antifungal resistance. Fungi respond to temperature by regulating cell membrane lipid composition, for example, by modulating ergosterol biosynthetic pathways48, which in turn alters antifungal resistance indirectly. The frequency of azole-resistant A. fumigatus is elevated in high-temperature environments such as composts5, greenhouses39 and tropical countries49, suggesting that synergistic interactions between temperature and antifungal resistance do occur. Further investigations, however, are needed to establish the directionality and significance of these interactions50. In parallel, synergies between temperature (thermal adaptation to warming climates) and fungicide exposure have been invoked to explain the rapid worldwide emergence of multidrug-resistant C. auris in humans, following its discovery in 2009 (ref.51).
Much remains to be learned about the genetic architecture and fitness landscapes of fungi following their adaptation to agrochemicals and how this impacts their interplay with other aspects of environmental change (Fig. 2). Thus, One Health solutions that address antifungal resistance must span site-specific local (for example, green waste composting containing chemical residues from agriculture) and global (for example, biosecurity in trade and changing climate) scales40,52. The evolution of resistance may cause wider phenotypic changes including elevated virulence, either as a direct consequence of the initial mutations or as secondary adaptation to the azole-rich environment found in patients, or in agricultural settings. Changes in fitness may ultimately affect their persistence after azole application has ceased, and future research should include assessing the ‘background’ frequency of resistance genotypes in sample sites where azoles have been discontinued, or have never been applied. These complex eco-evolutionary scenarios heighten the necessity of understanding the One Health consequences of antifungal resistance on fungal pathogens, their ecology and the outcome of our exposures to such organisms: this understanding requires heightened surveillance.
Diagnostics and surveillance
Identifying antifungal resistance
The identification of antifungal resistance (and tolerance) has relied on susceptibility testing of cultured microorganisms, identifying MICs for specific antimicrobials that, when compared with clinical break points, define susceptibility or resistance. Several methods are available for antifungal susceptibility testing: broth microdilution, disk diffusion, azole agar screening, gradient diffusion and the use of rapid automated instruments53. The Clinical Laboratory Standards Institute (CLSI) and European Committee on Antimicrobial Susceptibility Testing (EUCAST) organizations establish standards for performing susceptibility testing and determine clinical ‘break points’ for effectively treating infections. However, standardized CLSI and EUCAST broth microdilution reference methods — the gold standard for antifungal susceptibility testing — are labour-intensive, time-consuming and performed infrequently in most clinical laboratories. In addition, they require mycological culture from clinical specimens, which limits sensitivity and does not detect unculturable Pneumocystis jirovecii54,55,56. Clinical break points have only been defined for the main antifungal agents for the most common species (for example, C. albicans, Candida glabrata, Candida tropicalis, Candida parapsilosis and A. fumigatus) and there is an over-reliance on these as proxy break points for less studied species. Considerable variation between EUCAST and CLSI break points further complicates comparisons57. The application of break points relies on accurate species-level identification; this has improved for yeasts with the increasing use of MALDI-TOF mass spectrometry systems, but for moulds is still dependent on local database content58. Direct detection of antifungal resistance with the MALDI-TOF platform for yeasts59 and moulds60 is an exciting new direction; however, MALDI-TOF is too costly for many centres (thereby complicating international resistance surveillance initiatives) and reliance on culturing increases the time to diagnosis.
Resistance detection, transmission and surveillance
Molecular diagnostic approaches have the proven, but underutilized, capacity to identify genetic markers potentially associated with antifungal resistance and to also recognize fungal species that are intrinsically resistant (reviewed in ref.61). Their sensitivity allows direct application to clinical specimens, avoiding the need for culture and improving turnaround times. Species of the Aspergillus fumigati complex, such as Aspergillus lentulus and Aspergillus felis, that are difficult to differentiate using conventional methods and have potentially higher MIC values to azole antifungals can be identified by real-time PCR62. Resistant Candida spp., such as C. auris, C. glabrata and Candida krusei, can be detected and differentiated by PCR, potentially aiding infection control and patient management63. The utilization of fully automated molecular platforms (T2 Biosystems or Becton Dickinson Max) provide rapid testing systems requiring minimal specialist training comparable with the Cepheid GeneXpert platform for detecting multidrug-resistant tuberculosis. However, the range of this potential near-patient test must be expanded to include detection of mutations associated with resistance in generally susceptible fungal species.
Direct sequencing of genes encoding drug target proteins (for example, CYP51A in A. fumigatus or ERG11 in Candida spp.) was commonly used to identify potential resistance-associated mutations61. Subsequently, and based on the high prevalence of common mutations (for example, TR34/L98H and TR46/Y121F/T289A in A. fumigatus and dihydropteroate synthase mutations in P. jirovecii), commercial real-time PCR assays were launched64,65 and their diagnostic use is increasing owing to the high sensitivity and specificity of PCR-based approaches. With azole resistance in Candida spp. associated with a wide range of mechanisms and subsequent mutations, development of real-time PCR approaches are limited. DNA sequencing remains the best option for identifying the mutations associated with azole resistance, limiting clinical application, particularly direct sample testing66. Sequencing of ERG11 and FKS1 genes in C. auris strains with resistance to azoles and echinocandins has identified associated hotspots and specific mutations permitting the development of rapid molecular tests67. A small number of FKS1 gene mutations are associated with the majority of echinocandin resistance in Candida spp. and PCR assays have been developed68. Currently, there are no commercial PCR tests to detect mutations associated with antifungal resistance in yeasts, and to improve diagnosis it is essential that this be recognized through enhanced commercial development and regulatory body support.
Resistance detection is being facilitated by technical and computational advances. Examples here include integrating thermocycler-free DNA amplification by loop-mediated isothermal amplification onto lab-on-a-chip platforms with silicon-chip detectors and cloud connectivity to allow future point-of-care resistance detection69, or newly developed pyrosequencing techniques70. The implementation of whole-genome sequencing (WGS) holds great promise for exploring the biological basis of gene mutations more fully. Routine implementation of WGS for bacterial pathogen identification, resistance allele detection and identifying pathways of transmission is becoming commonplace. Beyond the detection of resistance alleles71, a major advantage of WGS is the ability to reconstruct the evolutionary trajectories of AMR variants across time and space10. However, in contrast to antibacterial resistance, a standardized WGS typing method is not widely used for fungi because of their larger genome sizes, frequent sexual recombination and the lack of standardized bioinformatic pipelines. Improved knowledge of antifungal resistance determinants and species genomes would support the transition to a WGS-powered understanding of fungal AMR for several human fungal pathogens72. Towards this goal, the development of rapid genomic analysis has been key to understanding the international73 and local-scale74 transmission of C. auris including the emergence of multidrug-resistant variants. Unculturable fungi present a challenge, and more targeted methods are needed. For instance, a successful consensus multilocus sequence typing scheme for P. jirovecii75 enables antifungal resistance marker analysis76. For Aspergillus spp., more knowledge of resistance mechanisms is required as many resistant isolates do not carry the few known resistance-associated alleles77. Nonetheless, WGS is increasingly being used to trace transmission of AMR in A. fumigatus for known polymorphisms30. Improvements in the ability of point-of-care WGS devices such as nanopore sequencers are accelerating our ability to detect antifungal resistance mutations and will likely transform our ability to understand pathways of nosocomial transmission in outbreak settings74 (Fig. 3).
Fungal samples can be acquired from the clinic or environment, including engaging with the public as ‘citizen scientists’42. Traditional, established microbiology methods can culture and select isolates from these samples, ready for extraction of genomic DNA. These DNA fragments are used to generate a sequencing library for whole-genome sequencing (WGS). There are many sequencing platforms available, generating both long-read and short-read sequence data. Raw sequence data need to be quality controlled prior to mapping against a reference genome, either locally or using cloud computing. Calling high-confidence single-nucleotide polymorphisms (SNPs) can help infer alleles associated with drug resistance and their evolutionary histories. Phylodynamic inference and building interactive online portals (such as Nextstrain131 or Microreact132) that are available to researchers and clinicians alike enable tracing of transmission events.
Towards global surveillance of antifungal resistance
Public health agencies have instigated systematic surveillance for bacterial AMR in many countries and have appointed reference laboratories to liaise with routine medical microbiological laboratories. Large international surveillance studies, led by the US CDC and the European Centre for Disease Prevention and Control, monitor the spread of antibiotic-resistant bacteria and broadcast early warning signals. However, fungi have, hitherto, been excluded from most AMR surveillance programmes. In 2018, the WHO (World Health Organization) launched a pilot Candida surveillance scheme to gather retrospective data on antifungal resistance for invasive Candida isolates; this was recently formally included in the Global Antimicrobial Resistance Surveillance System (GLASS) programme (Box 2). The Emerging Infections Program of the CDC currently conducts active population-based surveillance in ten state health departments in the United States, monitoring epidemiological trends in candidaemia. Globally, the SENTRY Antimicrobial Surveillance Program has at least 427 participating centres78 and antifungal resistance data are collected both indirectly (via blood culture surveillance) and directly. Unfortunately, relatively few centres contribute fungal pathogen data. Apart from these broader and more systematic surveillance programmes, nationwide surveillance data for Candida spp. are available from several countries such as Australia, Scotland, Finland, Iceland, Norway, Sweden, the United Kingdom and Denmark79. Nevertheless, surveillance of other fungal species is rare with most published data restricted to azole-resistant A. fumigatus80,81.
The rising rates of antifungal resistance and rapid global emergence of new multidrug-resistant species such as C. auris82 make it imperative to include fungal infections into existing national and international surveillance programmes. Despite the detection of azole-resistant genotypes of A. fumigatus worldwide, in most clinical settings its presence is not tested for and there are few studies exploring its association with clinical failure. Notably, as a ‘call to arms’, the WHO is currently defining a fungal pathogen priority list83 in line with its bacterial counterpart, a major step likely to trigger research and innovation in the field. A current high priority is the need to implement standardized surveillance through the collection of basic clinical and epidemiological data. This is because improved surveillance will further increase understanding of the evolution and transmission of fungal AMR alongside helping to implement modern genomic surveillance methodologies. In tandem, there is an urgent need for collaborative networks that include research, clinical and industry partners to undertake multicentre studies; these networks will also require access to shared biorepositories that collate validated samples alongside metadata, and that can distribute these rapidly and equitably when needed. Locally, accurate fungal species identification, simple resistance screening methodologies and MIC testing should be empowered at clinical laboratories in both high-resource and resource-limited countries, where there is a need for capacity building of clinical mycological expertise. When resistant isolates are identified locally, confirmation by reference laboratories in combination with the collection of essential clinical and epidemiological data will facilitate the downstream development of policy recommendations and control strategies.
Therapeutic approaches for tackling antifungal resistance
For commensal organisms, antifungal drug resistance can be acquired through drug exposure in treated individuals. For example, echinocandin resistance is more common in individuals previously treated with echinocandins84, and azole-resistant genotypes of Cryptococcus neoformans85 and A. fumigatus13 develop during long courses of treatment. For antifungal drugs to be effective, they must reach the site of infection. Each individual antifungal drug has vastly different absorption, distribution, metabolism and excretion (pharmacokinetic) properties, and even more pronounced are the differences amongst drugs in their tissue-specific penetration. Persistently low, or transiently high, drug concentrations may accelerate the evolution of resistance. However, using overly high doses of drugs carries an attendant risk of toxicity. For these reasons, regular therapeutic drug monitoring is required to optimize the dosage to maximize therapeutic potential, and to minimize the evolution of resistance whilst minimizing adverse reactions. Tissue-specific pharmacokinetics are largely unknown, although physiologically based modelling approaches have begun to shed some light on this issue86,87,88,89. Real-world studies are increasingly using therapeutic drug monitoring to explore pharmacokinetics across clinical cohorts, for example monitoring of individuals with cystic fibrosis has demonstrated a high prevalence of subtherapeutic levels of azoles alongside a high probability (>20%) of developing a resistant infection after 2 years90. For these reasons, better implementation of therapeutic drug monitoring through antifungal stewardship programmes is needed in susceptible patient cohorts. In tandem, the informed application of drug combinations may circumvent drug resistance. For example, micafungin inhibits several human and fungal efflux pumps, and thus when combined with drugs such as azoles may enhance their intracellular retention and efficacy.
Future studies will need to identify the likelihood with which resistance and tolerance mechanisms emerge. Pharmacometric approaches allow the simulation of model predictions91, and, for example, the hollow fibre model uses available pharmacokinetic data to mimic the human pharmacokinetics of antimicrobials92. Moreover, drug delivery at the site of infection remains a challenge due to extensive necrosis resulting in poor outcomes. For diseases where drug penetration at the site of infection is poor, improved pharmacodynamic models are needed to optimize dosing regimens and prevent treatment failure. Together, twinned pharmacokinetic/pharmacodynamic approaches could facilitate integrative, dynamic studies of the interplay between (unbound) drug concentrations, pathogen growth and kill kinetics in order to identify conditions that minimize the evolution of antifungal resistance in situ93.
Nurturing new therapeutic directions
An obvious solution to the allied problems of limited classes of drugs that may be compromised by dual use is to accelerate drug development. However, this is not a solution that can be achieved rapidly as it takes around 5–7 years from first initiation in human trials to approval of a novel anti-infective94,95 and can cost hundreds of millions of dollars. Timescales and costs are much higher if early development costs are accounted for. For instance, the development programme for Cresemba (isavuconazole), developed by Basilea Pharmaceutica and, subsequently, Astellas Pharma and Pfizer, took 13 years and required circa US $100 million of funding, with further downstream post-approval costs of circa US $30 million. Although isavuconazole has a broader spectrum than voriconazole, including efficacy against the Mucorales, and was similarly effective in patients with invasive aspergillosis with fewer drug-related adverse events than voriconazole96, the drug still shows cross-resistance to other azoles in both Aspergillus and Candida spp.97. The drug discovery company F2G Ltd is developing olorofim, a new mode of action (MOA) antifungal that targets dihydroorotate dehydrogenase, which has required several rounds of investment totalling more than US $213 million since their incorporation in 1998. The total time for F2G Ltd to identify the initial compound and develop their lead to phase II trials has been around 23 years. Although olorofim is not active against Candida spp., the drug shows promising activity against Aspergillus spp., including isolates with acquired azole resistance and other difficult to treat moulds such as Lomentospora prolificans98. These examples highlight the investment and risk associated with identifying and developing a novel class of antifungal drug.
These high costs and protracted timescales have clear implications with respect to developing therapies to treat IFDs caused by antifungal-resistant species, most of which are relatively rare and are unlikely to provide a significant return on investment. Novel therapies to treat such diseases are likely to appear only as adjuncts of broad-spectrum antifungals that have been progressed primarily to treat more common fungal diseases. A key question then arises of what market size is sufficient to make an antifungal development project viable. One answer may lie with the development of the promising fungal cell wall chitin-synthase inhibitor Nikkomycin Z99, which stalled after an apparently successful phase I trial100. The developers, Valley Fever Solutions Inc., have to date been unable to secure investment to develop the compound further. This may well be related to the limited spectrum of activity of Nikkomycin Z that is most active against relatively rare endemic mycoses such as Coccidioides spp., which in turn only have a patient population of circa 25,000 (ref.1) and predicted peak sales of US $130 million per annum. Even though a large proportion of these infections occur in the United States, investors have until now considered this market size to be too small even though Nikkomycin Z had support from governmental initiatives such as orphan drug designation and fast-track designation, and promising results in combination with other antifungals99.
That the antifungal pipeline is experiencing a substantial boost suggests that the US $13 billion global market for antifungals is encouraging the development of refined pre-existing compounds alongside new MOA antifungals that have a broad spectrum of activity. Of note, the Gwt1 inhibitor fosmanogepix (newly acquired by Pfizer), the (1 → 3)-β-d-glucan synthase inhibitor ibrexafungerp (Scynexis) and olorofim (F2G Ltd) are all new MOA antifungals that will open opportunities for treating azole-resistant or echinocandin-resistant pathogens (Supplementary Fig. 1). Other new MOA antifungals under development have intracellular targets, and thus are likely to be effective against isolates that are resistant to the existing drug classes.
In addition to novel drugs that are systemically given, new strategies for delivering antifungal drugs to the site of action are currently being explored. Opelconazole (Pulmocide), a reformulated azole drug administered by nebulization, has been evaluated for treating invasive aspergillosis in a phase I trial. Owing to the far higher drug concentrations that can be achieved in the lung, local application may overcome azole resistance in A. fumigatus. The useful life of an anti-infective relative to the potential rate of resistance emergence needs to be considered with the next generations of antifungals. Therefore, estimated evolutionary risks of resistance for new antifungals should be determined at the earliest possible stage of development, as has been advocated for antibacterial pipelines101. Chronic aspergillosis and acute candidiasis models or in vitro systems that better replicate the in vivo environment are recommended for monitoring the potential for the development of resistance in vivo, both for the target organism and for commensal fungi at the site of infection and distant body sites. Combining these in vivo models with pharmacokinetic/pharmacodynamic models could facilitate dosing studies estimating the likelihood of resistance emerging and minimizing the emergence of resistance, fungal persistence and tolerance.
Use of the same drug class in agriculture and medicine is a key driver for environmental drug resistance in Aspergillus spp. Removing azoles from agriculture is not trivial nor practical, as it would have a significant effect on global food production. Yet azole resistance in plant pathogens is emerging rapidly in agricultural settings. So what is the future of antifungal development with One Health in mind? Clearly, the development of fungicides for agriculture and antifungals for pharma needs to diverge4. In agriculture, this could be achieved by developing integrated disease management in crops, including ‘evolution-smart’ disease-resistant crops with mosaics of pathogen resistance genes alongside, for instance, the development of species-specific novel antifungal treatments based on RNA interference102. Approaches that focus on targets that are crucial for pathogenicity in plants but are different to those in humans may also lead to diverging methods of controlling fungal pathogens. Towards this end, significant technological strides have been made to enable high-throughput identification of virulence determinants by combining functional genomics and next-generation sequencing103,104. Undoubtedly, accelerated development of diverse, differentiated and ring-fenced antifungal pipelines for both agribusiness and pharma are not only the key to developing new fungicidal compounds but are also key to addressing evolving antifungal resistance in the coming years.
Current and future interventions
How can we stem the tide of emerging antifungal resistance? Integrating the ‘pillars’ of the JPIAMR and WHO initiatives will protect and augment our ability to treat IFDs (Fig. 4). Currently available strategies to limit the evolution of human fungal pathogens to chemical control include boosting surveillance and antifungal stewardship programmes, both of which require improved diagnosis of IFDs and antifungal resistance; minimizing environmental–clinical dual usage of antifungals; and optimizing resilient combination therapies using existing licensed drugs. Future strategies to lessen the impact of antifungal resistance largely require treating at-risk individuals with novel antifungal compounds patented solely for clinical use. This ‘personalized medicine’ approach should include reducing the risk of acquired IFDs by addressing the weakened immunity that predisposes individuals to these diseases, by employing immunotherapies and/or vaccines against IFDs.
Synoptic integrated One Health understanding is necessary to understand not only the complex multifactorial pathways that lead to the emergence of resistance across the fungal kingdom but also potential interventions to mitigate the rate of emergence. a | Complex biotic and abiotic interactions lead to occurrence of evolutionary hotspots for antimicrobial resistance (AMR) development in environmental opportunistic fungi requiring targeted interventions in the environment. b,c | Patient exposures to environmental AMR require enhanced methods of detection with more focus on key fungal life-history factors (part b), and new and emerging drug-resistant fungal pathogens that have the potential for global nosocomial carriage and outbreaks in health-care settings require transnational surveillance (part c). A cross-cutting theme is the need for industry to separate development and use of agricultural fungicides from those antifungals that are used in the clinic to develop treatments that are resilient to the evolutionary forces at play in parts a–c. GLASS, Global Antimicrobial Resistance Surveillance System; WHO, World Health Organization.
Widespread prophylactic and empiric prescribing of antifungals to treat suspected IFDs in individuals who are chronically at risk (for example, individuals with cystic fibrosis), those who are critically ill and patients with haemato-oncology remains a concern. Effective antifungal stewardship is required to optimize antifungal use and to preserve the limited antifungal arsenal105,106. This is especially relevant for fungal infections that are highly transmissible, such as Candida spp. and skin-infecting Trichophyton spp.107.
In largely single-centre, historic cohort observational (non-randomized) studies, antifungal stewardship programmes have consistently demonstrated an improvement in measures such as timely and appropriate antifungal prescribing (guideline-driven), the use of diagnostics and drug monitoring as well as a reduction in antifungal consumption, reducing antifungal selective pressures and the development of resistance108,109,110,111. Although such studies were not designed to demonstrate improved clinical outcomes, the absence of an adverse impact of antifungal stewardship implementation on the incidence of IFDs, length of hospital stay and in-hospital mortality are important findings112. Antifungal stewardship is underpinned by access to timely and sensitive diagnostics, and although a review of various pre-emptive diagnostic versus empirical antifungal strategies confirmed the suitability of pre-emptive strategies, the optimal strategy and limits have not been defined113. Goals for future work include optimizing rapid diagnostic strategies for ‘early start–de-escalation–early stop’ antifungal strategies and better hospital infection control, as well as demonstrating the impact of antifungal stewardship on rates of antifungal resistance inpatient cohorts or the hospital environment.
Combination antimicrobial treatment is an established and effective strategy to prevent the development of secondary AMR for various bacterial and viral infections. The principle was established in the 1950s in the treatment of tuberculosis, and has been repeated, for example, for HIV treatment in the 1990s and for the treatment of hepatitis C virus more recently114. Combination therapies with amphotericin B plus flucytosine (or fluconazole plus flucytosine in settings where amphotericin B is not available) are the established standard of care in cryptococcosis115. Combining flucytosine and fluconazole can prevent the selection of fluconazole hetero-resistant fungal populations that occur in individuals with cryptococcal meningitis following initial treatment with fluconazole monotherapy115. In terms of primary, environmentally derived, antifungal resistance, combination treatment of patients may have a limited effect, but combinations could reduce treatment failure due to primary resistance and limit the development of secondary, clinical antifungal resistance. Combination treatments may be additive or synergistic in terms of antimicrobial efficacy, and further work is needed to further their potential in a wide range of life-threatening fungal infections.
For invasive aspergillosis, consistent in vitro and animal model data both suggest that combining azole and echinocandin classes increases fungal killing and improves survival116,117,118. In a randomized clinical trial, mortality in those given this combination was 19% compared with 28% for those on azole monotherapy119; although the size of this study was limited, meaning the survival benefit did not reach conventional statistical significance, the approach described is encouraging. Animal models suggest a role for combination therapy in azole-resistant invasive aspergillosis120, but more work is needed to systematically explore combinations of established and new antifungal agents in experimental models and phase II clinical studies before moving to adequately powered phase III trials. In comparison with opportunistic fungal pathogens, C. auris can persist and spread within intensive care units and other health-care settings, leading to severe and intractable nosocomial outbreaks. Echinocandin monotherapy is commonly used to treat patients with C. auris, which is generally resistant to fluconazole. As this approach may facilitate the evolution and spread of multidrug-resistant isolates16, combination therapy strategies must be evaluated systematically to mitigate risk in this now globalized fungus.
Other approaches to protect existing antifungals include exploiting host-directed approaches to manage antifungal resistance. These include immunotherapy121, fungal vaccines122 and antibodies to fungal targets123. Because IFDs are most common in immunocompromised hosts, host-directed immunotherapies, including recombinant cytokines, monoclonal antibodies and fungus-specific engineered T cells121, have been in development. The use of interferon-γ to prevent and treat invasive aspergillosis in patients with chronic granulomatous disease was the first successful host-directed antifungal immunotherapy124. Since then, patient case series describing successful use of the TLR7 agonist imiquimod in chromoblastomycosis125 and granulocyte–macrophage colony-stimulating factor (GM-CSF) therapy for central nervous system candidiasis associated with CARD9 deficiency126 have been reported. These advances highlight the potential for host-directed approaches to lessen the pressure on antifungal drugs. Moreover, cell-based therapies, including dendritic cell transfer and chimeric antigen receptor (CAR) T cell therapy, have shown promising results in vitro127 but require evaluation in clinical trials.
The combination of immunotherapeutics with conventional antifungal therapy also holds promise. Numerous candidate fungal vaccines have been studied in the preclinical setting122, but only the C. albicans recombinant Als3 protein vaccine has shown promising results in phase II clinical trials128. Advancing antifungal vaccines will require overcoming several hurdles, especially the ubiquitous nature of fungi in the human holobiont129, and the expected suboptimal immune response in those people most at risk for IFDs130. Also showing promise are antibodies and fungal pattern recognition receptors that potentially target antifungal agents for pathogen delivery123. Preclinical studies of dectin-2 coupled to liposomal amphotericin B have shown encouraging results in experimental pulmonary aspergillosis123 and may help reduce antifungal toxicity in the host. However, although host-directed antifungal strategies, alone or in combination with conventional antifungals, hold immense promise, furthering and financing these novel strategies from the laboratory to clinical trials will be a significant challenge in the coming decade.
Conclusions
Challenges to a clinician’s ability to manage drug-resistant IFDs today include the lack of access to sensitive and specific diagnostic tests, the lack of clinically calibrated antifungal susceptibility testing and a limited repertoire of antifungal drug classes. Furthermore, the breadth and diversity of the fungal kingdom ensures a bottomless reservoir of new pathogens, alongside endless supplies of variants of old enemies, that readily adapt and evolve when exposed to antifungal chemicals. The sheer ecological breadth of fungal species, with their unique and varied ecological trophisms, in rapidly changing environments means that human health will always be enmeshed with the complex ecology of fungal communities, whether commensal or environmental. Similarly, our simultaneous need to control fungal disease in agricultural environments and the clinic means that integrated responses take these needs into consideration. Pathogenic fungi are widely vectored both actively and passively, such that tackling antifungal resistance both in the clinic and in the field requires a coordinated global response. The current lack of transnational support for networks, infrastructures, research funding and career development must be addressed through greater coordination between policymakers, funding agencies and researchers, and include the producers and users of antifungals.
References
Bongomin, F., Gago, S., Oladele, R. O. & Denning, D. W. Global and multi-national prevalence of fungal diseases-estimate precision. J. Fungi https://doi.org/10.3390/jof3040057 (2017).
Brown, G. D. et al. Hidden killers: human fungal infections. Sci. Transl. Med. https://doi.org/10.1126/scitranslmed.3004404 (2012).
Robbins, N., Caplan, T. & Cowen, L. E. Molecular evolution of antifungal drug resistance. Annu. Rev. Microbiol. 71, 753–775 (2017).
Fisher, M. C., Hawkins, N. J., Sanglard, D. & Gurr, S. J. Worldwide emergence of resistance to antifungal drugs challenges human health and food security. Science 360, 739–742 (2018).
Verweij, P. E. et al. The one health problem of azole resistance in Aspergillus fumigatus: current insights and future research agenda. Fungal Biol. Rev. 34, 202–214 (2020).
Rhodes, J. & Fisher, M. C. Global epidemiology of emerging Candida auris. Curr. Opin. Microbiol. 52, 84–89 (2019).
CDC. Antibiotic resistance threats in the United States, 2019. Centers for Disease Control and Prevention www.cdc.gov/DrugResistance/Biggest-Threats.html (2019).
Fisher, M. C. et al. Threats posed by the fungal kingdom to humans, wildlife, and agriculture. mBio https://doi.org/10.1128/mBio.00449-20 (2020).
Rodrigues, M. L. & Nosanchuk, J. D. Fungal diseases as neglected pathogens: a wake-up call to public health officials. PLoS Negl. Trop. Dis. 14, e0007964 (2020).
Baker, S., Thomson, N., Weill, F. X. & Holt, K. E. Genomic insights into the emergence and spread of antimicrobial-resistant bacterial pathogens. Science 360, 733–738 (2018).
Edlind, T. D. & Katiyar, S. K. Mutational analysis of flucytosine resistance in Candida glabrata. Antimicrob. Agents Chemother. 54, 4733–4738 (2010).
Berman, J. & Krysan, D. J. Drug resistance and tolerance in fungi. Nat. Rev. Microbiol. 18, 319–331 (2020).
Ballard, E. et al. In-host microevolution of Aspergillus fumigatus: a phenotypic and genotypic analysis. Fungal Genet. Biol. 113, 1–13 (2018).
Shields, R. K. et al. The presence of an FKS mutation rather than MIC is an independent risk factor for failure of echinocandin therapy among patients with invasive candidiasis due to Candida glabrata. Antimicrob. Agents Chemother. 56, 4862–4869 (2012).
Steinmann, J. et al. Emergence of azole-resistant invasive aspergillosis in HSCT recipients in Germany. J. Antimicrob. Chemother. 70, 1522–1526 (2015).
Pristov, K. E. & Ghannoum, M. A. Resistance of Candida to azoles and echinocandins worldwide. Clin. Microbiol. Infect. 25, 792–798 (2019).
Johnson, E. M., Warnock, D. W., Luker, J., Porter, S. R. & Scully, C. Emergence of azole drug resistance in Candida species from HIV-infected patients receiving prolonged fluconazole therapy for oral candidosis. J. Antimicrob. Chemother. 35, 103–114 (1995).
Laverdiere, M. et al. Progressive loss of echinocandin activity following prolonged use for treatment of Candida albicans oesophagitis. J. Antimicrob. Chemother. 57, 705–708 (2006).
Joint Programming Initiative on Antimicrobial Resistance. JPIAMR Strategic Research and Innovation Agenda on Antimicrobial Resistance. JPIAMR https://www.jpiamr.eu/app/uploads/2021/06/JPIAMR_SRIA_2021.pdf (2021).
Public Health England. Laboratory Surveillance of Candidaemia in England, Wales and Northern Ireland: 2018 (Public Health England, 2019).
Wauters, J. et al. Invasive pulmonary aspergillosis is a frequent complication of critically ill H1N1 patients: a retrospective study. Intensive Care Med. 38, 1761–1768 (2012).
Armstrong-James, D. et al. Confronting and mitigating the risk of COVID-19 associated pulmonary aspergillosis. Eur. Respir. J. https://doi.org/10.1183/13993003.02554-2020 (2020).
Garg, D. et al. Coronavirus disease (COVID-19) associated mucormycosis (CAM): case report and systematic review of literature. Mycopathologia 186, 289–298 (2021).
Janssen, N. A. F. et al. Multinational observational cohort study of COVID-19-associated pulmonary aspergillosis. Emerg. Infect. Dis. 27, 2892–2898 (2021).
Arastehfar, A. et al. COVID-19-associated candidiasis (CAC): an underestimated complication in the absence of immunological predispositions? J. Fungi https://doi.org/10.3390/jof6040211 (2020).
Singh, A. K., Singh, R., Joshi, S. R. & Misra, A. Mucormycosis in COVID-19: a systematic review of cases reported worldwide and in India. Diabetes Metab. Syndr. 15, 102146 (2021).
Fisher, M. C., Rannala, B., Chaturvedi, V. & Taylor, J. W. Disease surveillance in recombining pathogens: multilocus genotypes identify sources of human Coccidioides infections. Proc. Natl Acad. Sci. USA 99, 9067–9071 (2002).
Ashu, E. E., Hagen, F., Chowdhary, A., Meis, J. F. & Xu, J. Global population genetic analysis of Aspergillus fumigatus. Msphere https://doi.org/10.1128/mSphere.00019-17 (2017).
Sewell, T. R. et al. Nonrandom distribution of azole resistance across the global population of Aspergillus fumigatus. mBio https://doi.org/10.1128/mBio.00392-19 (2019).
Rhodes, J. et al. Population genomics confirms acquisition of drug resistance Aspergillus fumigatus infection by humans from the environment Nat. Microbiol. in press.
Vanhove, M. et al. Genomic epidemiology of Cryptococcus yeasts identifies adaptation to environmental niches underpinning infection across an African HIV/AIDS cohort. Mol. Ecol. 26, 1991–2005 (2017).
Steinberg, G. et al. A lipophilic cation protects crops against fungal pathogens by multiple modes of action. Nat. Commun. 11, 1608 (2020).
Toda, M., Beer, K. D., Kuivila, K. M., Chiller, T. M. & Jackson, B. R. Trends in agricultural triazole fungicide use in the United States, 1992–2016 and possible implications for antifungal-resistant fungi in human disease. Env. Health Perspect. 129, 55001 (2021).
Chen, Y. et al. High azole resistance in Aspergillus fumigatus isolates from strawberry fields, China, 2018. Emerg. Infect. Dis. 26, 81–89 (2020).
European Centre for Disease Prevention and Control. Risk Assessment on the Impact of Environmental Usage of Triazoles on the Development and Spread of Resistance to Medical Triazoles in Aspergillus Species (ECDC, 2013).
Snelders, E. et al. Possible environmental origin of resistance of Aspergillus fumigatus to medical triazoles. Appl. Env. Microb. 75, 4053–4057 (2009).
Schoustra, S. E. et al. New Insights in the Development of Azole-resistance in Aspergillus fumigatus (RIVM: National Institute for Public Health and the Environment, 2018).
Sewell, T. R. et al. Elevated prevalence of azole-resistant aspergillus fumigatus in urban versus rural environments in the United Kingdom. Antimicrob. Agents Chemother. 63, e00548–19 (2019).
Zhou, D. et al. Extensive genetic diversity and widespread azole resistance in greenhouse populations of Aspergillus fumigatus in Yunnan, China. Msphere https://doi.org/10.1128/mSphere.00066-21 (2021).
Burks, C., Darby, A., Gomez Londono, L., Momany, M. & Brewer, M. T. Azole-resistant Aspergillus fumigatus in the environment: identifying key reservoirs and hotspots of antifungal resistance. PLoS Pathog. 17, e1009711 (2021).
Dunne, K., Hagen, F., Pomeroy, N., Meis, J. F. & Rogers, T. R. Intercountry transfer of triazole-resistant Aspergillus fumigatus on plant bulbs. Clin. Infect. Dis. 65, 147–149 (2017).
Shelton, J. M. G., Fisher, M. C. & Singer, A. S. Campaign-based citizen science for environmental mycology: the science solstice and summer soil-stice projects to assess drug resistance in air- and soil-borne Aspergillus fumigatus. Citiz. Sci. Theory Pract. 5, 1–13 (2020).
Rocchi, S. et al. Molecular epidemiology of azole-resistant Aspergillus fumigatus in France shows patient and healthcare links to environmentally occurring genotypes. Front. Cell Infect. Microbiol. 11, 729476 (2021).
Hagiwara, D. et al. A novel Zn2-Cys6 transcription factor AtrR plays a key role in an azole resistance mechanism of Aspergillus fumigatus by co-regulating cyp51A and cdr1B expressions. PLoS Pathog. 13, e1006096 (2017).
Paul, S. et al. AtrR is an essential determinant of azole resistance in Aspergillus fumigatus. mBio https://doi.org/10.1128/mBio.02563-18 (2019).
Yasmin, S. et al. Mevalonate governs interdependency of ergosterol and siderophore biosyntheses in the fungal pathogen Aspergillus fumigatus. Proc. Natl Acad. Sci. USA 109, E497–E504 (2012).
Carneiro, H. C. S. et al. Hypervirulence and cross-resistance to a clinical antifungal are induced by an environmental fungicide in Cryptococcus gattii. Sci. Total Environ. https://doi.org/10.1016/j.scitotenv.2020.140135 (2020).
Kamthan, A., Kamthan, M. & Datta, A. Expression of C-5 sterol desaturase from an edible mushroom in fisson yeast enhances its ethanol and thermotolerance. PLoS ONE 12, e0173381 (2017).
Duong, T.-M. N., Le, T.-V., Tran, K.-L. H. & Beardsley, J. Azole-resistant Aspergillus fumigatus is highly prevalent in the environment of Vietnam, with marked variability by land use type. Environ. Microbiol. https://doi.org/10.1111/1462-2920.15660 (2021).
Van Rhijn, N. & Bromley, M. The consequences of our changing environment on life threatening and debilitating fungal diseases in humans. J. Fungi https://doi.org/10.3390/jof7050367 (2021).
Casadevall, A., Kontoyiannis, D. P. & Robert, V. On the emergence of Candida auris: climate change, azoles, swamps, and birds. mBio https://doi.org/10.1128/mBio.01397-19 (2019).
Fisher, M. C., Gow, N. A. R. & Gurr, S. J. Tackling emerging fungal threats to animal health, food security and ecosystem resilience. Philos. Trans. R. Soc. B Lond. B Biol. Sci. https://doi.org/10.1098/rstb.2016.0332 (2016).
Berkow, E. L., Lockhart, S. R. & Ostrosky-Zeichner, L. Antifungal susceptibility testing: current approaches. Clin. Microbiol. Rev. https://doi.org/10.1128/CMR.00069-19 (2020).
Clancy, C. J. & Nguyen, M. H. Finding the “missing 50%” of invasive candidiasis: how nonculture diagnostics will improve understanding of disease spectrum and transform patient care. Clin. Infect. Dis. 56, 1284–1292 (2013).
Levy, H., Horak, D. A., Tegtmeier, B. R., Yokota, S. B. & Forman, S. J. The value of bronchoalveolar lavage and bronchial washings in the diagnosis of invasive pulmonary aspergillosis. Respir. Med. 86, 243–248 (1992).
White, P. L., Price, J. S. & Backx, M. Pneumocystis jirovecii pneumonia: epidemiology, clinical manifestation and diagnosis. Curr. Fungal Infect. Rep. 13, 260–273 (2019).
Johnson, E. M. in Antifungal Susceptibility Testing and Resistance Ch. 47 (eds Kibbler, C. C. et. al.) (Oxford Univ. Press, 2017).
Bader, O. Fungal species identification by MALDI-ToF mass spectrometry. Methods Mol. Biol. 1508, 323–337 (2017).
Vatanshenassan, M. et al. Proof of concept for MBT ASTRA, a rapid matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS)-based method to detect caspofungin resistance in Candida albicans and Candida glabrata. J. Clin. Microbiol. https://doi.org/10.1128/JCM.00420-18 (2018).
Zvezdanova, M. E. et al. Detection of azole resistance in Aspergillus fumigatus complex isolates using MALDI-TOF mass spectrometry. Clin. Microbiol. Infect. https://doi.org/10.1016/j.cmi.2021.06.005 (2021).
Garcia-Effron, G. Molecular markers of antifungal resistance: potential uses in routine practice and future perspectives. J. Fungi https://doi.org/10.3390/jof7030197 (2021).
Chong, G. M. et al. Interspecies discrimination of A. fumigatus and siblings A. lentulus and A. felis of the Aspergillus section Fumigati using the AsperGenius® assay. Diagn. Microbiol. Infect. Dis. 87, 247–252 (2017).
Leach, L., Russell, A., Zhu, Y., Chaturvedi, S. & Chaturvedi, V. A rapid and automated sample-to-result Candida auris real-time PCR assay for high-throughput testing of surveillance samples with the BD max open system. J. Clin. Microbiol. https://doi.org/10.1128/JCM.00630-19 (2019).
Chong, G. M. et al. PCR-based detection of Aspergillus fumigatus Cyp51A mutations on bronchoalveolar lavage: a multicentre validation of the AsperGenius assay® in 201 patients with haematological disease suspected for invasive aspergillosis. J. Antimicrob. Chemother. 71, 3528–3535 (2016).
Montesinos, I. et al. Evaluation of a new commercial real-time PCR assay for diagnosis of Pneumocystis jirovecii pneumonia and identification of dihydropteroate synthase (DHPS) mutations. Diagn. Microbiol. Infect. Dis. 87, 32–36 (2017).
Perlin, D. S. & Wiederhold, N. P. Culture-independent molecular methods for detection of antifungal resistance mechanisms and fungal identification. J. Infect. Dis. 216, S458–S465 (2017).
Hou, X. et al. Rapid detection of ERG11-associated azole resistance and FKS-associated echinocandin resistance in Candida auris. Antimicrob. Agents Chemother. https://doi.org/10.1128/AAC.01811-18 (2019).
Pham, C. D., Bolden, C. B., Kuykendall, R. J. & Lockhart, S. R. Development of a Luminex-based multiplex assay for detection of mutations conferring resistance to echinocandins in Candida glabrata. J. Clin. Microbiol. 52, 790–795 (2014).
Yu, L. S. et al. Rapid detection of azole-resistant Aspergillus fumigatus in clinical and environmental isolates by use of a lab-on-a-chip diagnostic system. J. Clin. Microbiol. https://doi.org/10.1128/JCM.00843-20 (2020).
Novak-Frazer, L. et al. Deciphering Aspergillus fumigatus cyp51A-mediated triazole resistance by pyrosequencing of respiratory specimens. J. Antimicrob. Chemother. 75, 3501–3509 (2020).
Walker, T. M. et al. Tuberculosis is changing. Lancet Infect. Dis. 17, 359–361 (2017).
Brackin, A. P., Hemmings, S. J., Fisher, M. C. & Rhodes, J. Fungal genomics in respiratory medicine: what, how and when? Mycopathologia 186, 589–608 (2021).
Chow, N. A. et al. Tracing the evolutionary history and global expansion of Candida auris using population genomic analyses. mBio https://doi.org/10.1128/mBio.03364-19 (2020).
Rhodes, J. et al. Genomic epidemiology of the UK outbreak of the emerging human fungal pathogen Candida auris. Emerg. Microbes Infect. 7, 43 (2018).
Pasic, L. et al. Consensus multilocus sequence typing scheme for Pneumocystis jirovecii. J. Fungi https://doi.org/10.3390/jof6040259 (2020).
Ponce, C. A. et al. High prevalence of Pneumocystis jirovecii dihydropteroate synthase gene mutations in patients with a first episode of pneumocystis pneumonia in Santiago, Chile, and clinical response to trimethoprim–sulfamethoxazole therapy. Antimicrob. Agents Chemother. https://doi.org/10.1128/AAC.01290-16 (2017).
Bueid, A. et al. Azole antifungal resistance in Aspergillus fumigatus: 2008 and 2009. J. Antimicrob. Chemother. 65, 2116–2118 (2010).
IDSA. SENTRY program participating sites (1997–2016). Open Forum Infect. Dis. 6, S95–S102 (2019).
Astvad, K. M. T. et al. Update from a 12-year nationwide fungemia surveillance: increasing intrinsic and acquired resistance causes concern. J. Clin. Microbiol. https://doi.org/10.1128/JCM.01564-17 (2018).
Escribano, P. et al. Azole resistance survey on clinical Aspergillus fumigatus isolates in Spain. Clin. Microbiol. Infect. https://doi.org/10.1016/j.cmi.2020.09.042 (2020).
Rivero-Menendez, O., Alastruey-Izquierdo, A., Mellado, E. & Cuenca-Estrella, M. Triazole resistance in Aspergillus spp.: a worldwide problem? J. Fungi https://doi.org/10.3390/jof2030021 (2016).
Chowdhary, A., Sharma, C. & Meis, J. F. Candida auris: a rapidly emerging cause of hospital-acquired multidrug-resistant fungal infections globally. PLoS Pathog. 13, e1006290 (2017).
WHO. First meeting of the WHO Antifungal Expert Group on Identifying Priority Fungal Pathogens: Meeting Report (World Health Organization, 2020).
Alexander, B. D. et al. Increasing echinocandin resistance in Candida glabrata: clinical failure correlates with presence of FKS mutations and elevated minimum inhibitory concentrations. Clin. Infect. Dis. 56, 1724–1732 (2013).
Rhodes, J. et al. A population genomics approach to assessing the genetic basis of within-host microevolution underlying recurrent cryptococcal meningitis infection. G3 7, 1165–1176 (2017).
Hens, B. et al. In silico modeling approach for the evaluation of gastrointestinal dissolution, supersaturation, and precipitation of posaconazole. Mol. Pharm. 14, 4321–4333 (2017).
Li, X. et al. A physiologically based pharmacokinetic model of voriconazole integrating time-dependent inhibition of CYP3A4, genetic polymorphisms of CYP2C19 and predictions of drug-drug interactions. Clin. Pharmacokinet. 59, 781–808 (2020).
Gerhart, J. G. et al. Physiologically-based pharmacokinetic modeling of fluconazole using plasma and cerebrospinal fluid samples from preterm and term infants. CPT Pharmacomet. Syst. Pharmacol. 8, 500–510 (2019).
Campoli, P. et al. Pharmacokinetics of posaconazole within epithelial cells and fungi: insights into potential mechanisms of action during treatment and prophylaxis. J. Infect. Dis. 208, 1717–1728 (2013).
Di Paolo, M. et al. A retrospective ‘real-world’ cohort study of azole therapeutic drug monitoring and evolution of antifungal resistance in cystic fibrosis. JAC Antimicrob. Resist. 3, dlab026 (2021).
Hope, W., Drusano, G. L. & Rex, J. H. Pharmacodynamics for antifungal drug development: an approach for acceleration, risk minimization and demonstration of causality. J. Antimicrob. Chemother. 71, 3008–3019 (2016).
Tangden, T. et al. The role of infection models and PK/PD modelling for optimising care of critically ill patients with severe infections. Intensive Care Med. 43, 1021–1032 (2017).
Chen, G. et al. Targeting the adaptability of heterogeneous aneuploids. Cell 160, 771–784 (2015).
Ward, D. J., Hammond, E., Linden-Phillips, L. & Stevens, A. J. Trends in clinical development timeframes for antiviral drugs launched in the UK, 1981–2014: a retrospective observational study. BMJ Open 5, e009333 (2015).
Jorda, A. & Zeitlinger, M. Preclinical pharmacokinetic/pharmacodynamic studies and clinical trials in the drug development process of EMA-approved antibacterial agents: a review. Clin. Pharmacokinet. 59, 1071–1084 (2020).
Maertens, J. A. et al. Isavuconazole versus voriconazole for primary treatment of invasive mould disease caused by Aspergillus and other filamentous fungi (SECURE): a phase 3, randomised-controlled, non-inferiority trial. Lancet 387, 760–769 (2016).
Jorgensen, K. M., Astvad, K. M. T., Hare, R. K. & Arendrup, M. C. EUCAST susceptibility testing of isavuconazole: MIC data for contemporary clinical mold and yeast isolates. Antimicrob. Agents Chemother. https://doi.org/10.1128/AAC.00073-19 (2019).
Buil, J. B. et al. In vitro activity of the novel antifungal compound F901318 against difficult-to-treat Aspergillus isolates. J. Antimicrob. Chemother. 72, 2548–2552 (2017).
Larwood, D. J. Nikkomycin Z-ready to meet the promise? J. Fungi https://doi.org/10.3390/jof6040261 (2020).
Nix, D. E., Swezey, R. R., Hector, R. & Galgiani, J. N. Pharmacokinetics of Nikkomycin Z after single rising oral doses. Antimicrob. Agents Chemother. 53, 2517–2521 (2009).
Brockhurst, M. A. et al. Assessing evolutionary risks of resistance for new antimicrobial therapies. Nat. Ecol. Evol. 3, 515–517 (2019).
Wang, M. et al. Bidirectional cross-kingdom RNAi and fungal uptake of external RNAs confer plant protection. Nat. Plants 2, 16151 (2016).
Macdonald, D. et al. Inducible cell fusion permits use of competitive fitness profiling in the human pathogenic fungus Aspergillus fumigatus. Antimicrob. Agents Chemother. 63, e01615–e01618 (2019).
Lee, K. T. et al. Systematic functional analysis of kinases in the fungal pathogen Cryptococcus neoformans. Nat. Commun. https://doi.org/10.1038/ncomms12766 (2016).
Logan, C., Martin-Loeches, I. & Bicanic, T. Invasive candidiasis in critical care: challenges and future directions. Intensive Care Med. 46, 2001–2014 (2020).
Michallet, M. et al. Antifungal stewardship in hematology: reflection of a multidisciplinary group of experts. Clin. Lymphoma Myeloma Leuk. 21, 35–45 (2021).
Kano, R. et al. Trichophyton indotineae sp. nov.: a new highly terbinafine-resistant anthropophilic dermatophyte species. Mycopathologia 185, 947–958 (2020).
Bienvenu, A. L. et al. A systematic review of interventions and performance measures for antifungal stewardship programmes. J. Antimicrob. Chemother. 73, 297–305 (2018).
Hart, E., Nguyen, M., Allen, M., Clark, C. M. & Jacobs, D. M. A systematic review of the impact of antifungal stewardship interventions in the United States. Ann. Clin. Microbiol. Antimicrob. 18, 24 (2019).
Rautemaa-Richardson, R. et al. Impact of a diagnostics-driven antifungal stewardship programme in a UK tertiary referral teaching hospital. J. Antimicrob. Chemother. 73, 3488–3495 (2018).
Talento, A. F., Qualie, M., Cottom, L., Backx, M. & White, P. L. Lessons from an educational invasive fungal disease conference on hospital antifungal stewardship practices across the UK and Ireland. J. Fungi https://doi.org/10.3390/jof7100801 (2021).
Whitney, L. et al. Effectiveness of an antifungal stewardship programme at a London teaching hospital 2010–16. J. Antimicrob. Chemother. 74, 234–241 (2019).
Fung, M., Kim, J., Marty, F. M., Schwarzinger, M. & Koo, S. Meta-analysis and cost comparison of empirical versus pre-emptive antifungal strategies in hematologic malignancy patients with high-risk febrile neutropenia. PLoS ONE 10, e0140930 (2015).
Naggie, S. & Muir, A. J. Oral combination therapies for hepatitis C virus infection: successes, challenges, and unmet needs. Annu. Rev. Med. 68, 345–358 (2017).
Molloy, S. F. et al. Antifungal combinations for treatment of cryptococcal meningitis in Africa. N. Engl. J. Med. 378, 1004–1017 (2018).
Kirkpatrick, W. R., Perea, S., Coco, B. J. & Patterson, T. F. Efficacy of caspofungin alone and in combination with voriconazole in a guinea pig model of invasive aspergillosis. Antimicrob. Agents Chemother. 46, 2564–2568 (2002).
Petraitis, V. et al. Combination therapy in treatment of experimental pulmonary aspergillosis: synergistic interaction between an antifungal triazole and an echinocandin. J. Infect. Dis. 187, 1834–1843 (2003).
Petraitis, V. et al. Combination therapy with isavuconazole and micafungin for treatment of experimental invasive pulmonary aspergillosis. Antimicrob. Agents Chemother. https://doi.org/10.1128/AAC.00305-17 (2017).
Marr, K. A. et al. Combination antifungal therapy for invasive aspergillosis: a randomized trial. Ann. Intern. Med. 162, 81–89 (2015).
Seyedmousavi, S. et al. Efficacy and pharmacodynamics of voriconazole combined with anidulafungin in azole-resistant invasive aspergillosis. J. Antimicrob. Chemother. 68, 385–393 (2013).
Armstrong-James, D. et al. Immunotherapeutic approaches to treatment of fungal diseases. Lancet Infect. Dis. 17, e393–e402 (2017).
Oliveira, L. V. N., Wang, R., Specht, C. A. & Levitz, S. M. Vaccines for human fungal diseases: close but still a long way to go. NPJ Vaccines 6, 33 (2021).
Ambati, S. et al. Antifungal liposomes directed by dectin-2 offer a promising therapeutic option for pulmonary aspergillosis. mBio https://doi.org/10.1128/mBio.00030-21 (2021).
International Chronic Granulomatous Disease Cooperative Study Group. A controlled trial of interferon gamma to prevent infection in chronic granulomatous disease. N. Engl. J. Med. 324, 509–516 (1991).
de Sousa Mda, G. et al. Topical application of imiquimod as a treatment for chromoblastomycosis. Clin. Infect. Dis. 58, 1734–1737 (2014).
Gavino, C. et al. CARD9 deficiency and spontaneous central nervous system candidiasis: complete clinical remission with GM-CSF therapy. Clin. Infect. Dis. 59, 81–84 (2014).
Kumaresan, P. R. et al. Bioengineering T cells to target carbohydrate to treat opportunistic fungal infection. Proc. Natl Acad. Sci. USA 111, 10660–10665 (2014).
Edwards, J. E. Jr. et al. A fungal immunotherapeutic vaccine (NDV-3A) for treatment of recurrent vulvovaginal candidiasis — a phase 2 randomized, double-blind, placebo-controlled trial. Clin. Infect. Dis. 66, 1928–1936 (2018).
Seed, P. C. The human mycobiome. Cold Spring Harb. Perspect. Med. 5, a019810 (2014).
Eades, C. P. & Armstrong-James, D. P. H. Invasive fungal infections in the immunocompromised host: mechanistic insights in an era of changing immunotherapeutics. Med. Mycol. 57, S307–S317 (2019).
Hadfield, J. et al. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics 34, 4121–4123 (2018).
Argimon, S. et al. Microreact: visualizing and sharing data for genomic epidemiology and phylogeography. Microb. Genom. 2, e000093 (2016).
Stone, N. R. et al. Dynamic ploidy changes drive fluconazole resistance in human cryptococcal meningitis. J. Clin. Invest. 129, 999–1014 (2019).
Balaban, N. Q. et al. Definitions and guidelines for research on antibiotic persistence. Nat. Rev. Microbiol. 17, 441–448 (2019).
Selmecki, A., Forche, A. & Berman, J. Aneuploidy and isochromosome formation in drug-resistant Candida albicans. Science 313, 367–370 (2006).
Suwunnakorn, S., Wakabayashi, H. & Rustchenko, E. Chromosome 5 of human pathogen Candida albicans carries multiple genes for negative control of caspofungin and anidulafungin susceptibility. Antimicrob. Agents Chemother. 60, 7457–7467 (2016).
Kwon-Chung, K. J. & Chang, Y. C. Aneuploidy and drug resistance in pathogenic fungi. PLoS Pathog. 8, e1003022 (2012).
Ksiezopolska, E. et al. Narrow mutational signatures drive acquisition of multidrug resistance in the fungal pathogen Candida glabrata. Curr. Biol. 31, 5314–5326.e10 (2021).
Forche, A. et al. Stress alters rates and types of loss of heterozygosity in Candida albicans. mBio https://doi.org/10.1128/mBio.00129-11 (2011).
Healey, K. R. et al. Prevalent mutator genotype identified in fungal pathogen Candida glabrata promotes multi-drug resistance. Nat. Commun. 7, 11128 (2016).
Billmyre, R. B., Clancey, S. A. & Heitman, J. Natural mismatch repair mutations mediate phenotypic diversity and drug resistance in Cryptococcus deuterogattii. eLife https://doi.org/10.7554/eLife.28802 (2017).
Singh, A. et al. Absence of azole or echinocandin resistance in Candida glabrata isolates in india despite background prevalence of strains with defects in the DNA mismatch repair pathway. Antimicrob. Agents Chemother. https://doi.org/10.1128/AAC.00195-18 (2018).
Boyce, K. J. et al. Mismatch repair of DNA replication errors contributes to microevolution in the pathogenic fungus Cryptococcus neoformans. mBio https://doi.org/10.1128/mBio.00595-17 (2017).
Gerstein, A. C. & Berman, J. Candida albicans genetic background influences mean and heterogeneity of drug responses and genome stability during evolution in fluconazole. mSphere https://doi.org/10.1128/mSphere.00480-20 (2020).
Liu, J., Gefen, O., Ronin, I., Bar-Meir, M. & Balaban, N. Q. Effect of tolerance on the evolution of antibiotic resistance under drug combinations. Science 367, 200–204 (2020).
Windels, E. M., Van den Bergh, B. & Michiels, J. Bacteria under antibiotic attack: different strategies for evolutionary adaptation. PLoS Pathog. 16, e1008431 (2020).
Moosa, M. Y., Alangaden, G. J., Manavathu, E. & Chandrasekar, P. H. Resistance to amphotericin B does not emerge during treatment for invasive aspergillosis. J. Antimicrob. Chemother. 49, 209–213 (2002).
Zarnowski, R. et al. Candida albicans biofilm-induced vesicles confer drug resistance through matrix biogenesis. PLoS Biol. 16, e2006872 (2018).
Smith, W. L. & Edlind, T. D. Histone deacetylase inhibitors enhance Candida albicans sensitivity to azoles and related antifungals: correlation with reduction in CDR and ERG upregulation. Antimicrob. Agents Chemother. 46, 3532–3539 (2002).
Li, X. et al. The Rpd3/Hda1 family of histone deacetylases regulates azole resistance in Candida albicans. J. Antimicrob. Chemother. 70, 1993–2003 (2015).
Acknowledgements
M.C.F., D.C.S. and S.J.G. are fellows in the Canadian Institute for Advanced Research (CIFAR) ‘Fungal Kingdom’ programme. M.C.F. acknowledges funding from the Natural Environment Research Council (NERC) and the Medical Research Council (MRC) Centre for Global Infectious Disease Analysis (reference MR/R015600/1), jointly funded by the UK MRC and the UK Foreign, Commonwealth & Development Office (FCDO), under the MRC/FCDO Concordat agreement, and is also part of the EDCTP2 programme supported by the European Union. J.B. is supported by the Israel Science Foundation (#997/18) and European Research Council (ERC) Synergy Fungal Tolerance (#951475). A.W. and E.M.B. are supported by the MRC Centre for Medical Mycology (grant MR/N006364/2). S.J.G. is supported by the Biotechnology and Biological Sciences Research Council (BBSRC) (grant no. BB/PO18335) and the Bill and Melinda Gates Foundation. The contribution of B.Z. and P.E.V. is supported by the project ‘One health consequences of circularity. What lessons to learn from the saprophytic and human pathogenic fungus Aspergillus fumigatus?’ (project number GROEN.2019.002), which is financed by the Dutch Research Council (NWO). The authors thank L. Schouls, Centre for Infectious Diseases Research, National Institute for Public Health and the Environment (RIVM), for comments. This Review was conceived as a result of the Joint Programming Initiative on Antimicrobial Resistance (JPIAMR) Strategic Research and Innovation Agenda (SRIA) update consultation.
Author information
Authors and Affiliations
Contributions
The authors contributed equally to all aspects of the article.
Corresponding authors
Ethics declarations
Competing interests
M.C.F. and P.E.V. receive speaker fees from Gilead Scientific. O.A.C. reports grants or contracts from Amplyx, Basilea, BMBF, Cidara, DZIF, EU-DG RTD (101037867), F2G Ltd, Gilead, Matinas, MedPace, MSD, Mundipharma, Octapharma, Pfizer and Scynexis; consulting fees from Amplyx, Biocon, Biosys, Cidara, Da Volterra, Gilead, Matinas, MedPace, Menarini, Molecular Partners, MSG-ERC, Noxxon, Octapharma, PSI, Scynexis and Seres; honoraria for lectures from Abbott, Al-Jazeera Pharmaceuticals, Astellas, Grupo Biotoscana/United Medical/Knight, Hikma, MedScape, MedUpdate, Merck/MSD, Mylan and Pfizer; payment for expert testimony from Cidara; participation on a Data Safety Monitoring Board or Advisory Board from Actelion, Allecra, Cidara, Entasis, IQVIA, Jannsen, MedPace, Paratek, PSI and Shionogi; a patent at the German Patent and Trade Mark Office (DE 10 2021 113 007.7); and other interests from DGHO, DGI, ECMM), ISHAM, MSG-ERC and Wiley. The other authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Microbiology thanks Yong-Sun Bahn, David Perlin and Ilan Schwartz for their contribution to the peer review of this work.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Glossary
- Antifungal tolerance
-
A characteristic of drug-susceptible genotypes to grow slowly at or above inhibitory drug concentrations. Characteristically, only a proportion of cells manifest tolerance.
- Antifungal resistance
-
Defined as the ability to grow at antifungal drug concentrations above a defined antifungal susceptibility break point, normally (but not exclusively) owing to a defined causal molecular change following adaptation to drug exposure. It is expressed as a minimum inhibitory concentration (MIC).
- Minimum inhibitory concentration
-
(MIC). The lowest concentration of an antifungal drug that inhibits fungal growth and, in the context of defined susceptibility break points, defines resistance.
- Fungicides
-
Antifungal compounds used in the environment to inhibit fungal growth; widely used in agriculture, horticulture and timber industries as well as components of antifouling agents and paints.
- Saprotrophic decay
-
Heterotrophic nutrition provided by extracellular digestion of organic matter in the environment.
- Intrinsic resistance
-
Species of fungi that have not obviously evolved resistance in response to drug pressure.
- Acquired resistance
-
Species of fungi that have evolved resistance in response to drug pressure.
- Aneuploidies
-
Increase in the numbers of copies of chromosomes, often resulting in phenotypic changes to drug resistance and/or tolerance profiles.
- Hypermutator
-
Genotypes that manifest accelerated mutation rates because of mutations to genes involved in nucleic acid repair mechanisms.
- Fungistatic
-
Exposure to a chemical that halts the growth of, but does not kill, the fungus.
- Antifungal susceptibility testing
-
An in vitro measure of susceptibility and resistance to the drug concentrations required to inhibit fungal growth, measured by the minimum inhibitory concentration (MIC).
- Loop-mediated isothermal amplification
-
Enzymatic nucleic acid amplification at a single temperature.
- Therapeutic drug monitoring
-
The pharmacological practice of measuring drug concentrations at specific intervals in order to optimize individual dosage regimens.
Rights and permissions
About this article
Cite this article
Fisher, M.C., Alastruey-Izquierdo, A., Berman, J. et al. Tackling the emerging threat of antifungal resistance to human health. Nat Rev Microbiol 20, 557–571 (2022). https://doi.org/10.1038/s41579-022-00720-1
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41579-022-00720-1
This article is cited by
-
Fungal coexistence in the skin mycobiome: a study involving Malassezia, Candida, and Rhodotorula
AMB Express (2024)
-
Deciphering antifungal and antibiofilm mechanisms of isobavachalcone against Cryptococcus neoformans through RNA-seq and functional analyses
Microbial Cell Factories (2024)
-
Transcriptomic meta-analysis to identify potential antifungal targets in Candida albicans
BMC Microbiology (2024)
-
Synergistic potential of α-Phellandrene combined with conventional antifungal agents and its mechanism against antibiotic resistant Candida albicans
CABI Agriculture and Bioscience (2024)
-
Chemical disinfection as a simple and reliable method to control the amphibian chytrid fungus at breeding points of endangered amphibians
Scientific Reports (2024)