Abstract
A major factor in the progression to heart failure in humans is the inability of the adult heart to repair itself after injury. We recently demonstrated that the early postnatal mammalian heart is capable of regeneration following injury through proliferation of preexisting cardiomyocytes1,2 and that Meis1, a three amino acid loop extension (TALE) family homeodomain transcription factor, translocates to cardiomyocyte nuclei shortly after birth and mediates postnatal cell cycle arrest3. Here we report that Hoxb13 acts as a cofactor of Meis1 in postnatal cardiomyocytes. Cardiomyocyte-specific deletion of Hoxb13 can extend the postnatal window of cardiomyocyte proliferation and reactivate the cardiomyocyte cell cycle in the adult heart. Moreover, adult Meis1–Hoxb13 double-knockout hearts display widespread cardiomyocyte mitosis, sarcomere disassembly and improved left ventricular systolic function following myocardial infarction, as demonstrated by echocardiography and magnetic resonance imaging. Chromatin immunoprecipitation with sequencing demonstrates that Meis1 and Hoxb13 act cooperatively to regulate cardiomyocyte maturation and cell cycle. Finally, we show that the calcium-activated protein phosphatase calcineurin dephosphorylates Hoxb13 at serine-204, resulting in its nuclear localization and cell cycle arrest. These results demonstrate that Meis1 and Hoxb13 act cooperatively to regulate cardiomyocyte maturation and proliferation and provide mechanistic insights into the link between hyperplastic and hypertrophic growth of cardiomyocytes.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The data that support the findings of this study are available within the paper and its Supplementary Information. Source Data for Figs. 1–4 and Extended Data Figs. 1–6, 8, 9 are provided with the online version of the paper. ChIP–seq data have been deposited in the Gene Expression Omnibus (GEO) under accession number GSE141769. Data or other materials are available from the corresponding author upon reasonable request.
References
Bui, A. L., Horwich, T. B. & Fonarow, G. C. Epidemiology and risk profile of heart failure. Nat. Rev. Cardiol. 8, 30–41 (2011).
Soonpaa, M. H. & Field, L. J. Assessment of cardiomyocyte DNA synthesis in normal and injured adult mouse hearts. Am. J. Physiol. 272, H220–H226 (1997).
Senyo, S. E. et al. Mammalian heart renewal by pre-existing cardiomyocytes. Nature 493, 433–436 (2013).
Bergmann, O. et al. Evidence for cardiomyocyte renewal in humans. Science 324, 98–102 (2009).
Eschenhagen, T. et al. Cardiomyocyte regeneration: a consensus statement. Circulation 136, 680–686 (2017).
Porrello, E. R. et al. Transient regenerative potential of the neonatal mouse heart. Science 331, 1078–1080 (2011).
Porrello, E. R. et al. Regulation of neonatal and adult mammalian heart regeneration by the miR-15 family. Proc. Natl Acad. Sci. USA 110, 187–192 (2013).
Soonpaa, M. H. et al. Cyclin D1 overexpression promotes cardiomyocyte DNA synthesis and multinucleation in transgenic mice. J. Clin. Invest. 99, 2644–2654 (1997).
Mahmoud, A. I. et al. Meis1 regulates postnatal cardiomyocyte cell cycle arrest. Nature 497, 249–253 (2013).
Eulalio, A. et al. Functional screening identifies miRNAs inducing cardiac regeneration. Nature 492, 376–381 (2012).
Puente, B. N. et al. The oxygen-rich postnatal environment induces cardiomyocyte cell-cycle arrest through DNA damage response. Cell 157, 565–579 (2014).
Heallen, T. et al. Hippo pathway inhibits Wnt signaling to restrain cardiomyocyte proliferation and heart size. Science 332, 458–461 (2011).
Chen, J. et al. mir-17-92 cluster is required for and sufficient to induce cardiomyocyte proliferation in postnatal and adult hearts. Circ. Res. 112, 1557–1566 (2013).
Shen, W. F. et al. HOXA9 forms triple complexes with PBX2 and MEIS1 in myeloid cells. Mol. Cell. Biol. 19, 3051–3061 (1999).
Shen, W. F. et al. AbdB-like Hox proteins stabilize DNA binding by the Meis1 homeodomain proteins. Mol. Cell. Biol. 17, 6448–6458 (1997).
Berthelsen, J., Kilstrup-Nielsen, C., Blasi, F., Mavilio, F. & Zappavigna, V. The subcellular localization of PBX1 and EXD proteins depends on nuclear import and export signals and is modulated by association with PREP1 and HTH. Genes Dev. 13, 946–953 (1999).
Ali, S. R. et al. Existing cardiomyocytes generate cardiomyocytes at a low rate after birth in mice. Proc. Natl Acad. Sci. USA 111, 8850–8855 (2014).
Li, H. et al. Balanced interactions of calcineurin with AKAP79 regulate Ca2+–calcineurin–NFAT signaling. Nat. Struct. Mol. Biol. 19, 337–345 (2012).
Molkentin, J. D. et al. A calcineurin-dependent transcriptional pathway for cardiac hypertrophy. Cell 93, 215–228 (1998).
Kiserud, T. Physiology of the fetal circulation. Semin. Fetal Neonatal Med. 10, 493–503 (2005).
Fiedler, B. & Wollert, K. C. Targeting calcineurin and associated pathways in cardiac hypertrophy and failure. Expert Opin. Ther. Targets 9, 963–973 (2005).
Heineke, J. & Ritter, O. Cardiomyocyte calcineurin signaling in subcellular domains: from the sarcolemma to the nucleus and beyond. J. Mol. Cell. Cardiol. 52, 62–73 (2012).
Wilkins, B. J. & Molkentin, J. D. Calcium-calcineurin signaling in the regulation of cardiac hypertrophy. Biochem. Biophys. Res. Commun. 322, 1178–1191 (2004).
Colella, M. et al. Ca2+ oscillation frequency decoding in cardiac cell hypertrophy: role of calcineurin/NFAT as Ca2+ signal integrators. Proc. Natl Acad. Sci. USA 105, 2859–2864 (2008).
Parra, V. & Rothermel, B. A. Calcineurin signaling in the heart: the importance of time and place. J. Mol. Cell. Cardiol. 103, 121–136 (2017).
van Rooij, E. et al. Requirement of nuclear factor of activated T-cells in calcineurin-mediated cardiomyocyte hypertrophy. J. Biol. Chem. 277, 48617–48626 (2002).
Hogan, P. G., Chen, L., Nardone, J. & Rao, A. Transcriptional regulation by calcium, calcineurin, and NFAT. Genes Dev. 17, 2205–2232 (2003).
Schaeffer, P. J. et al. Impaired contractile function and calcium handling in hearts of cardiac-specific calcineurin b1-deficient mice. Am. J. Physiol. Heart Circ. Physiol. 297, H1263–H1273 (2009).
Rothermel, B. A. et al. Myocyte-enriched calcineurin-interacting protein, MCIP1, inhibits cardiac hypertrophy in vivo. Proc. Natl Acad. Sci. USA 98, 3328–3333 (2001).
Nakada, Y. et al. Hypoxia induces heart regeneration in adult mice. Nature 541, 222–227 (2017).
Nascimento, D. S. et al. MIQuant—semi-automation of infarct size assessment in models of cardiac ischemic injury. PLoS ONE 6, e25045 (2011).
Mahmoud, A. I., Porrello, E. R., Kimura, W., Olson, E. N. & Sadek, H. A. Surgical models for cardiac regeneration in neonatal mice. Nat. Protoc. 9, 305–311 (2014).
Kimura, W. et al. Hypoxia fate mapping identifies cycling cardiomyocytes in the adult heart. Nature 523, 226–230 (2015).
Goldman, A. et al. The calcineurin signaling network evolves via conserved kinase-phosphatase modules that transcend substrate identity. Mol. Cell 55, 422–435 (2014).
Wang, Z. et al. Mechanistic basis of neonatal heart regeneration revealed by transcriptome and histone modification profiling. Proc. Natl Acad. Sci. USA 116, 18455–18465 (2019).
Hashimoto, H. et al. Cardiac reprogramming factors synergistically activate genome-wide cardiogenic stage-specific enhancers. Cell Stem Cell 25, 69–86 (2019).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Li, Q., Brown, J. B., Huang, H. & Bickel, P. J. Measuring reproducibility of high-throughput experiments. Ann. Appl. Stat. 5, 1752–1779 (2011).
Kharchenko, P. V., Tolstorukov, M. Y. & Park, P. J. Design and analysis of ChIP–seq experiments for DNA-binding proteins. Nat. Biotechnol. 26, 1351–1359 (2008).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010).
Zhou, Y. et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 10, 1523 (2019).
Acknowledgements
We thank S. Zhang for MRI studies at the Mouse MRI Core, Advanced Imaging Research Center, University of Texas Southwestern Medical Center; J. Shelton for assistance with histology; H. I. May for conducting mouse surgeries; J. Heinrich for helping with voluntary exercise; J. Xu, X. Liu and Y. Jung Kim from the Sequencing Core Facility in Children’s Research Institute in University of Texas Southwestern Medical Center for performing the Illumina sequencing. The UTSW mouse MRI facility is supported by an NIH shared instrumentation grant 1S10OD023552-01. H.A.S. is supported by grants from the NIH (1R01HL115275 and 5R01H2131778), National Aeronautics and Space Administration (NNX-15AE06G), American Heart Association (16EIA27740034), Cancer Prevention and Research Institute of Texas (RP160520), Hamon Center for Regenerative Science and Medicine, and Fondation Leducq (Redox Regulation of Cardiomyocyte Renewal). M.S.C. and J.R. are funded by NIH R01GM119336. B.A.R. was supported by grants from the NIH (R01 HD101006, R01 HL072016) and Wellstone (U54HD087351). J.A.H. is supported by grants from NIH (R01 HL120732, R01 HL128215, R01 HL126012). N.U.N.N. is supported by AHA Postdoctoral Fellowship 19POST34450039. Z.W. was supported by a predoctoral fellowship from the American Heart Association and the Harry S. Moss Heart Trust (19PRE34380436). N.T.L. is supported by a Haberecht Wildhare-Idea Research Grant.
Author information
Authors and Affiliations
Contributions
N.U.N.N., D.C.C. and S.A.M. conducted calcineurin and Rcan1-related experiments and interpreted results. N.U.N.N., F.X., Y.N., S.L., S.A.M., J.M., D.C.C., Victor Le, K.A.Z., H.W.E.-F. and W.M.E. conducted immunofluorescent staining. F.X., D.C.C., N.U.N.N. and X.M. designed and performed phosphorylated antibody experiments and interpreted results. N.U.N.N., F.X. and M.E.H. conducted immunoprecipitation and PLA experiments. N.U.N.N., F.X. and S.L. conducted TUNEL assay. Z.W. and N.U.N.N. conducted ChIP–seq experiments. M.K., C.X., C.G.A.-N, W.L.W.T, R.S.F. and Z.W. interpreted ChIP–seq results. E.V., F.X., S.R.A. and N.U.N.N. conducted cardiac fibroblast experiments and interpreted results. N.U.N.N., I.M.-M., Victoria Le and M.S.M. conducted TAC and exercise-related experiments. N.U.N.N., M.S.A., M.S.M. and I.M.-M. conducted BrdU-labelling experiments. N.L., F.X. and N.U.N.N. conducted MADM-related experiments and interpreted results. J.R. and M.S.C. designed and performed calcineurin-binding assays. S.L., and N.U.N.N. performed mouse surgeries. S.L., D.C.C., J.J.S. and N.U.N.N. conducted echocardiography experiments and interpreted results. S.T. and N.U.N.N. managed mouse colonies. N.U.N.N. designed and conducted experiments, interpreted results and contributed to manuscript preparation. J.A.H. interpreted results. B.A.R. interpreted results and contributed to manuscript preparation. H.A.S. designed the experiments, conceived the project and contributed to manuscript preparation.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature thanks Norbert Frey and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Additional characterization of Meis1-iKO, Hoxb13-KO, and Hoxb13-iKO mice.
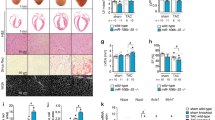
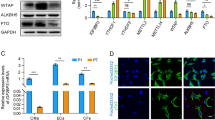
a, Echocardiographic assessment of LVEF in Meis1-iKO hearts at the indicated time points post-MI. b, Immunostaining of Meis1-interacting Hox proteins (red), cardiac troponin T (green), and nucleus (blue) at P1, P7 and 7 days after P1 MI. c, Western blot analysis of Meis1 and Hox proteins during regenerative stages. GAPDH serves as loading control. d, Coimmunoprecipitation of Meis1 from total heart extracts at P1 and P7. e, Coimmunoprecipitation from Fig. 1c, with negative controls (IgG and DKO heart). f, Coimmunoprecipitation from Fig. 1d, with negative controls (IgG and DKO heart). g, PLA of Meis1 and Hoxb13 in P7 DKO heart; serves as negative control for PLA assay in Fig. 1e. h, Schematic of targeting strategy for generation of cardiomyocyte-specific Hoxb13 mutants. Partial map of the Hoxb13 floxed allele (top), and of the knockout allele (bottom) following cardiomyocyte-specific Cre-mediated excision. Exons and loxP sites are indicated as rectangles and triangles, respectively. i, PCR analysis of heart DNA obtained by crossing Hoxb13loxP/loxP (female) mutants with MHC-cre (male) transgenic mice. Left lane shows Hoxb13loxP/loxP. Middle lane indicates cardiomyocyte-specific Cre. Right lane indicates that the deleted allele is present in cardiomyocyte DNA of this offspring. j, Top, western blot of protein extracts from hearts of constitutive (left) or inducible (right) Hoxb13 knockout and Hoxb13fl/fl littermate mice. GAPDH serves as loading control. Bottom, densitometry quantification of the above western blot. k, Control and Hoxb13-KO P14 hearts stained for Meis1 (red), cardiac troponin T (green), and nucleus (blue). Arrowheads indicated Meis1-expressing cardiomyocytes, and arrows indicate Meis1-expressing non-cardiomyocytes. l, Representative echocardiography, ejection fraction and fraction shortening in Hoxb13-KO mice P14 hearts. m, Representative images of Masson’s trichrome staining of control and Hoxb13-iKO hearts at 14 days post-deletion. n, Representative images of immunostaining for TUNEL (green), cardiac troponin T (red) and nucleus (blue) (left) and quantification (right) 14 days post-deletion. Arrow indicates TUNEL-positive cardiomyocytes. Data are mean ± s.e.m.; unpaired two-sided t-test. Data in b–f, g, i, k, m were independently repeated three times with similar results. *P < 0.05, **P < 0.01. For gel source data, see Supplementary Fig. 1. For n values, see Methods. Scale bars, 10 μm (b, k, n), 50 μm (g), 1 mm (m).
Extended Data Fig. 2 Assessment of Hoxb13-KO and Hoxb13-iKO hearts post-MI.
a, Schematic of MI model in Hoxb13-KO mice. b–d, Representative echocardiography (b), heart weight/body weight (c), and wet-to-dry lung weight ratio (d) in Hoxb13-KO mice 16 weeks after MI. e, Representative serial Masson’s trichrome staining in transversal sections (left) and quantification of fibrotic scars (right) in Hoxb13-KO MI hearts at 16 weeks after injury. f, Schematic of MI model in Hoxb13-iKO mice. g, h, Heart weight/body weight (g) and wet-to-dry lung weight ratio (h) in Hoxb13-iKO mice 16 weeks after MI. i, Serial echocardiography of Hoxb13-iKO MI and sham mice. Data are mean ± s.e.m.; unpaired two-sided t-test. **P < 0.01, ***P < 0.001; n.s., not significant. For n values, see Methods. Scale bar 1 mm (e).
Extended Data Fig. 3 Concurrent knockout of Meis1 and Hoxb13 preserves cardiomyocyte proliferative capacity.
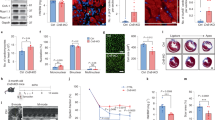
a–g, Assessment of control and DKO hearts at P28. a, Representative images of haematoxylin and eosin-stained heart sections and heart weight/body weight ratio. b, Representative images of WGA staining and cardiomyocyte cross-sectional area quantification. c, Representative images of immunostaining for PH3 (green), cardiac troponin T (red) and nucleus (blue) (left) and the percentage (right) of mitotic cardiomyocytes (arrow). Arrowhead indicates sarcomere disassembly. d, Representative images of immunostaining for aurora B kinase (green), cardiac troponin T (red) and nucleus (blue) (left) showing the percentage (right) of cardiomyocytes undergoing cytokinesis (arrow). e, f, Total number of cardiomyocytes (e) and quantification of nucleation (f). Cnx43, connexin-43. g, Representative echocardiography and LV systolic function. h–k, Assessment of control and DKO hearts at 6 months of age. h, Representative images of immunostaining for PH3 (green), cardiac troponin T (red), and nucleus (blue) (left) and the percentage (right) of mitotic cardiomyocytes (arrow). Arrowhead indicates cardiomyocytes with sarcomere disassembly. i, Representative images of immunostaining for aurora B kinase (green), cardiac troponin T (red) and nucleus (blue) (left) and the percentage (right) of cardiomyocytes undergoing cytokinesis (arrow). j, Representative images of WGA (green), cardiac troponin T (red) and nucleus (blue) staining (left) and cardiomyocyte CSA quantification (right). k, LV systolic function quantified by ejection fraction in 6-month-old DKO mice. Data are mean ± s.e.m.; unpaired two-sided t-test. *P < 0.05, **P < 0.01, ***P < 0.001. For n values, see Methods. Scale bars, 10 μm (c, d, f, h, i), 50 μm (b, j), 1 mm (a).
Extended Data Fig. 4 Loss of Meis1 and Hoxb13 attenuates both pathological and physiological hypertrophic responses.
a, Schematic representing the TAC model. b, Representative images of M-mode echocardiography (top) and quantification of ejection fraction (bottom), demonstrating improved LVEF in the DKO mice compared with wild-type mice three weeks after TAC. c, Heart weights, showing significantly higher heart weight in the wild-type group. d, Tibia length was similar in both groups. e, f, The ratio of heart weight/body weight (e) and heart weight/tibial length (f) indicate that the DKO mice have a blunted response to pressure overload. g, Representative images of mouse heart sections stained with haematoxylin and eosin (H&E; top) or Masson’s trichrome (bottom) three weeks after surgery. h, Representative confocal images of cardiomyocyte WGA staining (left) and CSA quantification (right), confirming the blunted hypertrophic response in the DKO hearts in response to pressure overload. i, Schematic representation of the exercise protocol. j, Representative images of echocardiography (top) and ejection fraction quantification (bottom), demonstrating no difference in ejection fraction at baseline and after exercise. k, Running distance of control and DKO mice over four weeks, demonstrating no difference between the two groups. l, Heart weight. m, The ratio of heart weight/body weight suggests that both control and DKO hearts respond to exercise-induced cardiac hypertrophy. n, The ratio of heart weight/tibia length also indicates cardiac hypertrophy in both groups in response to exercise. o, Representative images of mouse heart sections stained with either haematoxylin and eosin (top) or Masson’s trichrome (bottom) four weeks after exercise. p, Confocal images of cardiomyocyte WGA staining and CSA quantification. Data are mean ± s.e.m.; unpaired two-sided t-test. Images in g, o are representative of six independently performed experiments with similar results. *P < 0.05, **P < 0.01, ***P < 0.001. For n values, see Methods. Scale bars, 50 μm (h, p), 1 mm (g, o).
Extended Data Fig. 5 Inducible knockout of Meis1 and Hoxb13 promotes sarcomere disassembly and cardiomyocyte division.
a, Representative images of Masson’s trichrome staining of DiKO hearts four weeks after deletion. b, c, Disassembled sarcomere cardiomyocytes in DiKO hearts one week after deletion, demonstrated by cardiac troponin T (red) (b) or sarcomeric α-actinin (red) (c). d, Representative images of immunostaining for TUNEL (green) and cardiac troponin T (red) in DiKO hearts one week and two months after deletion, as shown in Fig. 2e. e, f, Representative images of immunostaining for PH3 (green), cardiac troponin T (red) and nucleus (blue), showing additional examples of proliferating cardiomyocytes in DiKO hearts at (e) one week and (f) two months after gene deletion. g, Representative images of immunostaining for aurora B kinase (green), cardiac troponin T (red) and nucleus (blue) (left) and the percentage (right) of proliferating cardiomyocytes (arrow) one week after gene deletion. h, Additional examples of cytokinetic cardiomyocytes in DiKO hearts one week after gene deletion. Images in a–c are representative of three independently performed experiments with similar results. Scale bars, 10 μm (b, d–h), 20 μm (c), 1 mm (a).
Extended Data Fig. 6 LV function of DKO hearts after MI.
a, Schematic of MI model in DKO hearts 2 weeks post-MI. b, c, Western blot analysis (b) and quantification (c), showing decrease in expression of p21, p15/p16 in DKO hearts. d–f, Heart weight/body weight (d), heart weight (e) and body weight (f) in control and DiKO mice 16 weeks after injury. g, Representative images of the whole hearts at 16 weeks post-MI. The white arrow indicates a region with cell death and the yellow arrow indicates the dilated left atrial appendage. h, Additional representative serial Masson’s trichrome-stained transversal sections, as in Fig. 2l. i, j, LV cardiac index (i) and heart rate (j) in DiKO mice 16 weeks after MI measured by MRI. k, LVEF for sham groups. l–q, Cardiac function analysis for left-ventricular end-diastolic anterior wall thickness (l), left-ventricular end-diastolic internal diameter (m), left-ventricular end-diastolic volume (n), left-ventricular end-diastolic posterior wall thickness (o), left-ventricular end-systolic internal diameter (p), and left ventricular end-systolic volume (q) in DiKO and control mice after MI. Data are mean ± s.e.m.; unpaired two-sided t-test. *P < 0.05, **P < 0.01, ***P < 0.001. For gel source data, see Supplementary Fig. 1. For n values, see Methods. Scale bars, 1 mm (g, h).
Extended Data Fig. 7 Genome-wide identification of Meis1 and Hoxb13 binding sites in heart using ChIP–seq.
a, Venn diagram showing Meis1 and Hoxb13 binding-site identification. Three replicate sequencing runs were performed for each factor, and the enriched regions were selected only if they were detected in three biological replicates. b, An example of a Meis1 binding site in the cell cycle arrest gene Cdkn1b. c, Gene ontology terms enriched in Meis1-, Hoxb13- and Meis1–Hoxb13-bound peak regions identified from a.
Extended Data Fig. 8 Interaction and dephosphorylation status of Meis1 and Hoxb13 with calcineurin.
a–c, Coimmunoprecipitation of Fig. 3b–d with negative controls (IgG and DKO heart). d, PLA of CnA and Hoxb13 at P7 of DKO heart; serves as negative control for PLA assay in Fig. 3e. e, PLA assay using only one antibody; serves as negative control for the assay. f, PLA of CnA and Meis1 at P7 heart showed Meis1–CnA association (red dot). g, h, Schematic representation of Meis1 (g, top) and Hoxb13 (h, top) proteins showing the phosphorylatable serine and threonine residues, amino acid sequence of Meis1 (g, bottom) and Hoxb13 (h, bottom) with potentially phosphorylatable serine and threonine residues in red. Box indicates the predicted nuclear localization signal. Numbers indicate amino acid positions. i, j, Densitometry quantification of western blot analysis in Fig. 3j for Hoxb13 (i) and phosphorylated Hoxb13 S204 (j). k, Western blot analysis of phosphorylated Meis1 protein at P1 and P7. GAPDH serves as loading control. l, Western blot analysis from stable DKO cardiac fibroblasts confirms the loss of Hoxb13 in Cre-treated cells. Data are mean ± s.e.m.; unpaired two-sided t-test. Experiments in a–f, k–m were repeated independently three times with similar results. *P < 0.05, **P < 0.01. For gel source data, see Supplementary Fig. 1. For n values, see Methods. Scale bars, 10 μm (f), 50 μm (d, e).
Extended Data Fig. 9 Increased calcineurin activity suppresses postnatal cardiomyocyte proliferation.
a, Heart weight/body weight in wild-type, CnA-Tg, Rcan1-Tg, and double-Tg mice at P2 (a) and P21 (b). c, d, Representative images of WGA staining for CSA quantification in Fig. 4a (c) and Fig. 4b (d). e, f, Representative images of immunostaining for PH3 (green) and cardiac troponin T (red) for mitotic cardiomyocytes quantification in Fig. 4c (e) and Fig. 4d (f). g, Schematic of apical resection model in P1 mice. Scale bars, 10 µm (f), 50 µm (c, d). Data are mean ± s.e.m.; unpaired two-sided t-test. ***P < 0.001. For n values, see Methods.
Supplementary information
Supplementary Figure
This file contains uncropped western blot images with molecular weight markers and indication of how the gels were cropped.
Source data
Rights and permissions
About this article
Cite this article
Nguyen, N.U.N., Canseco, D.C., Xiao, F. et al. A calcineurin–Hoxb13 axis regulates growth mode of mammalian cardiomyocytes. Nature 582, 271–276 (2020). https://doi.org/10.1038/s41586-020-2228-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-020-2228-6
This article is cited by
-
Toward drug-induced heart regeneration
Nature Cardiovascular Research (2024)
-
ATF3 coordinates the survival and proliferation of cardiac macrophages and protects against ischemia–reperfusion injury
Nature Cardiovascular Research (2024)
-
N-Acetyltransferase 10 represses Uqcr11 and Uqcrb independently of ac4C modification to promote heart regeneration
Nature Communications (2024)
-
Identification of FDA-approved drugs that induce heart regeneration in mammals
Nature Cardiovascular Research (2024)
-
Regeneration of the heart: from molecular mechanisms to clinical therapeutics
Military Medical Research (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.