Abstract
Intestinal epithelial cell (IEC) damage by T cells contributes to graft-versus-host disease, inflammatory bowel disease and immune checkpoint blockade-mediated colitis. But little is known about the target cell-intrinsic features that affect disease severity. Here we identified disruption of oxidative phosphorylation and an increase in succinate levels in the IECs from several distinct in vivo models of T cell-mediated colitis. Metabolic flux studies, complemented by imaging and protein analyses, identified disruption of IEC-intrinsic succinate dehydrogenase A (SDHA), a component of mitochondrial complex II, in causing these metabolic alterations. The relevance of IEC-intrinsic SDHA in mediating disease severity was confirmed by complementary chemical and genetic experimental approaches and validated in human clinical samples. These data identify a critical role for the alteration of the IEC-specific mitochondrial complex II component SDHA in the regulation of the severity of T cell-mediated intestinal diseases.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
Transcriptomics data have been deposited in the NCBI’s GEO database under the accession code GSE158259. Source data are provided with this paper.
References
Strober, W., Fuss, I. & Mannon, P. The fundamental basis of inflammatory bowel disease. J. Clin. Invest. 117, 514–521 (2007).
Martins, F. et al. Adverse effects of immune-checkpoint inhibitors: epidemiology, management and surveillance. Nat. Rev. Clin. Oncol. 16, 563–580 (2019).
Ferrara, J. L., Levine, J. E., Reddy, P. & Holler, E. Graft-versus-host disease. Lancet 373, 1550–1561 (2009).
Neurath, M. F. Targeting immune cell circuits and trafficking in inflammatory bowel disease. Nat Immunol. 20, 970–979 (2019).
Zeiser, R. & Blazar, B. R. Acute graft-versus-host disease—biologic process, prevention, and therapy. N. Engl. J. Med. 377, 2167–2179 (2017).
O’Neill, L. A. & Pearce, E. J. Immunometabolism governs dendritic cell and macrophage function. J. Exp. Med. 213, 15–23 (2016).
Everts, B. et al. TLR-driven early glycolytic reprogramming via the kinases TBK1-IKKɛ supports the anabolic demands of dendritic cell activation. Nat. Immunol. 15, 323–332 (2014).
Buck, M. D. et al. Mitochondrial dynamics controls T cell fate through metabolic programming. Cell 166, 63–76 (2016).
Albenberg, L. et al. Correlation between intraluminal oxygen gradient and radial partitioning of intestinal microbiota. Gastroenterology 147, 1055–1063 (2014).
He, G. et al. Noninvasive measurement of anatomic structure and intraluminal oxygenation in the gastrointestinal tract of living mice with spatial and spectral EPR imaging. Proc. Natl Acad. Sci. USA 96, 4586–4591 (1999).
Fan, Y. Y. et al. A bioassay to measure energy metabolism in mouse colonic crypts, organoids, and sorted stem cells. Am. J. Physiol. Gastrointest. Liver Physiol. 309, G1–G9 (2015).
Salabei, J. K., Gibb, A. A. & Hill, B. G. Comprehensive measurement of respiratory activity in permeabilized cells using extracellular flux analysis. Nat. Protoc. 9, 421–438 (2014).
Jacquemin, G. et al. Granzyme B-induced mitochondrial ROS are required for apoptosis. Cell Death Differ. 22, 862–874 (2015).
Wang, F., Yin, Q., Chen, L. & Davis, M. M. Bifidobacterium can mitigate intestinal immunopathology in the context of CTLA-4 blockade. Proc. Natl Acad. Sci. USA 115, 157–161 (2018).
Perez-Ruiz, E. et al. Prophylactic TNF blockade uncouples efficacy and toxicity in dual CTLA-4 and PD-1 immunotherapy. Nature 569, 428–432 (2019).
Mills, E. L. et al. Succinate dehydrogenase supports metabolic repurposing of mitochondria to drive inflammatory macrophages. Cell 167, 457–470 (2016).
Bailis, W. et al. Distinct modes of mitochondrial metabolism uncouple T cell differentiation and function. Nature 571, 403–407 (2019).
Tannahill, G. M. et al. Succinate is an inflammatory signal that induces IL-1β through HIF-1α. Nature 496, 238–242 (2013).
Littlewood-Evans, A. et al. GPR91 senses extracellular succinate released from inflammatory macrophages and exacerbates rheumatoid arthritis. J. Exp. Med. 213, 1655–1662 (2016).
Kuo, C. Y., Chiu, Y. C., Lee, A. Y. & Hwang, T. L. Mitochondrial Lon protease controls ROS-dependent apoptosis in cardiomyocyte under hypoxia. Mitochondrion 23, 7–16 (2015).
Ishizawa, J. et al. Mitochondrial ClpP-mediated proteolysis induces selective cancer cell lethality. Cancer Cell 35, 721–737 (2019).
Ghezzi, D. et al. SDHAF1, encoding a LYR complex-II specific assembly factor, is mutated in SDH-defective infantile leukoencephalopathy. Nat. Genet. 41, 654–656 (2009).
Na, U. et al. The LYR factors SDHAF1 and SDHAF3 mediate maturation of the iron–sulfur subunit of succinate dehydrogenase. Cell Metab. 20, 253–266 (2014).
Michelucci, A. et al. Immune-responsive gene 1 protein links metabolism to immunity by catalyzing itaconic acid production. Proc. Natl Acad. Sci. USA 110, 7820–7825 (2013).
Mathewson, N. D. et al. Gut microbiome-derived metabolites modulate intestinal epithelial cell damage and mitigate graft-versus-host disease. Nat. Immunol. 17, 505–513 (2016).
Suzuki, T., Yoshida, S. & Hara, H. Physiological concentrations of short-chain fatty acids immediately suppress colonic epithelial permeability. Br. J. Nutr. 100, 297–305 (2008).
Wu, S. R. & Reddy, P. Tissue tolerance: a distinct concept to control acute GVHD severity. Blood 129, 1747–1752 (2017).
Donohoe, D. R. et al. The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon. Cell Metab. 13, 517–526 (2011).
Chouchani, E. T. et al. Ischaemic accumulation of succinate controls reperfusion injury through mitochondrial ROS. Nature 515, 431–435 (2014).
Guzy, R. D., Sharma, B., Bell, E., Chandel, N. S. & Schumacker, P. T. Loss of the SdhB, but not the SdhA, subunit of complex II triggers reactive oxygen species-dependent hypoxia-inducible factor activation and tumorigenesis. Mol. Cell. Biol. 28, 718–731 (2008).
Lemarie, A., Huc, L., Pazarentzos, E., Mahul-Mellier, A. L. & Grimm, S. Specific disintegration of complex II succinate:ubiquinone oxidoreductase links pH changes to oxidative stress for apoptosis induction. Cell Death Differ. 18, 338–349 (2011).
Martinvalet, D., Zhu, P. & Lieberman, J. Granzyme A induces caspase-independent mitochondrial damage, a required first step for apoptosis. Immunity 22, 355–370 (2005).
Dotiwala, F. et al. Granzyme B disrupts central metabolism and protein synthesis in bacteria to promote an immune cell death program. Cell 171, 1125–1137 (2017).
Walch, M. et al. Cytotoxic cells kill intracellular bacteria through granulysin-mediated delivery of granzymes. Cell 161, 1229 (2015).
Zhang, Z. et al. Gasdermin E suppresses tumour growth by activating anti-tumour immunity. Nature 579, 415–420 (2020).
Lu, Y. et al. Caspase-11 signaling enhances graft-versus-host disease. Nat. Commun. 10, 4044 (2019).
Humphries, F. et al. Succination inactivates gasdermin D and blocks pyroptosis. Science 369, 1633–1637 (2020).
Luan, H. H. et al. GDF15 Is an inflammation-induced central mediator of tissue tolerance. Cell 178, 1231–1244 e1211 (2019).
Cooke, K. R. et al. Tumor necrosis factor-alpha production to lipopolysaccharide stimulation by donor cells predicts the severity of experimental acute graft-versus-host disease. J. Clin. Invest. 102, 1882–1891 (1998).
Hill, G. R. et al. Interleukin-11 promotes T cell polarization and prevents acute graft-versus-host disease after allogeneic bone marrow transplantation. J. Clin. Invest. 102, 115–123 (1998).
Mathew, A. V., Seymour, E. M., Byun, J., Pennathur, S. & Hummel, S. L. Altered metabolic profile with sodium-restricted dietary approaches to stop hypertension diet in hypertensive heart failure with preserved ejection fraction. J. Card. Fail. 21, 963–967 (2015).
Campbell, E. L. et al. Transmigrating neutrophils shape the mucosal microenvironment through localized oxygen depletion to influence resolution of inflammation. Immunity 40, 66–77 (2014).
West, A. P. et al. Mitochondrial DNA stress primes the antiviral innate immune response. Nature 520, 553–557 (2015).
Chacko, B. K. et al. The Bioenergetic Health Index: a new concept in mitochondrial translational research. Clin. Sci. 127, 367–373 (2014).
Acknowledgements
This work was supported by US NIH grants HL090775, CA173878, CA203542, HL149633 (to P.R.), K08HL130944 (to A.V.M.), DK081943 and DK89503 (to S.P.), JSPS Postdoctoral Fellowships for Research Abroad (to H.F.), The YASUDA Medical Foundation Grants for Research Abroad (to H.F.), JSPS KAKENHI grants JP20K22901 and JP21H02904 (to H.F.), The Kawasaki Foundation of Medical Science and Medical Welfare (to H.F.), The Ryobiteien Memorial Foundation (to H.F.), The MSD Life Science Foundation Public Interest Incorporated Foundation (to H.F.), The Okayama Medical Foundation (to H.F.), The SENSHIN Medical Research Foundation (to H.F.), The Kato Memorial Bioscience Foundation (to H.F.) and The NOVARTIS Foundation (Japan) for the Promotion of Science (to H.F.). We acknowledge use of the Microscopy & Image-analysis Laboratory (MIL) of the University of Michigan’s Biomedical Research Core Facilities for the preparation of samples and images. Support for the MIL core is provided by the University of Michigan Rogel Cancer Center (NIH grant CA46592).
Author information
Authors and Affiliations
Contributions
H.F. and P.R. conceived and designed this study. H.F., K.S. and P.R. planned and guided the research and wrote the manuscript. H.F., K.S., I.K., A.V.M., A.P., H.M., T.T., I.H., S.J.W., S.K., A.T., D.S., K.O.-W., Y.S. and J.B. performed experiments. I.K., A.V.M., H.-J.L. and J.B. analyzed LC–MS and MFA data. D.P. performed experiments, analyzed human data and edited the paper. C.L. performed experiments and histopathological analysis. M.D.B. performed transcriptome analysis. H.F., T.S. and P.R. generated the SDHA-floxed mice. P.S., Y.M., M.S.W., T.S., C.A.L., S.P., A.R. and R.P. supervised the work carried out in this study.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Immunology thanks the anonymous reviewers for their contribution to the peer review of this work. Editor Recognition Statement: N. Bernard was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Mitochondrial respiration and metabolic pathway in IECs after allo-HCT.
C57BL/6 animals received HCT (BALB/c or C57BL/6 → C57BL/6). (a) IECs of colon 7days post-HCT were isolated and incubated with medium. Cell death (AnnexinV + ) was determined before incubation and after 2 hours incubation (n = 4). (b) Mitochondrial bioenergetic profile was determined as previously described44. % OCR change of several parameters in mitochondrial and non-mitochondrial cellular respiration normalized from basal respiration in IECs of colon 21days post-HCT (n = 5). (c) Non-mitochondrial OCR of IECs 21days post HCT with or without NADH (n = 4). (d) Comprehensive measurement of respiratory activity in permeabilized IECs 21days post HCT. OCR of IECs without substrates of complexes (left). The combined OCR of syngeneic (middle) and allogeneic IECs (right) treated with each substrate (n = 3). (e) Representative BN-PAGE image of mitochondria from IECs 21days post HCT showing abundant mitochondrial protein complexes (n = 4). (f) Representative 2D-PAGE images of mitochondria from IECs on day21 post HCT showing reduced expression of SDHA protein (arrow, n = 4). (g-h) Succinate levels in kidney (g, Naive: n = 8, Syngeneic (Syn): n = 10, Allogeneic (Allo): n = 10) and lactate levels (h, Naive: n = 8, Syn: n = 9, Allo: n = 8) in IECs from syngeneic and allogeneic recipients d21 post HCT with colorimetric methods. (i) Abundance from uniformly 13C-glutamine of cis-aconitate, α-ketoglutarate, succinate, and malate after 4 h incubation in syngeneic or allogeneic isolated IECs 7 and 21days post HCT (D7 Syn: n = 4, Allo: n = 5, D21 Syn: n = 5, Allo: n = 7). All data are from biologically independent animals. Representative plots and a graph summarizing the results of at least two independent experiments are shown. One-way ANOVA analysis with Tukey post hoc test (a, c, g, h) or two-tailed unpaired t-test (b, i) were used to determine significance (mean ± s.e.m.).
Extended Data Fig. 2 Specific reduction of SDHA activity in colon and ileum post allo-HCT.
C57BL/6 animals received 10 Gy total body irradiation and received 3×106 (BALB/c → C57BL/6) or 1 ×106 (C3H.SW → C57BL/6) T cells along with 5×106 TCD-BM cells from either syngeneic or allogeneic donors. (a) Representative images of SDH enzyme activity staining in GVHD target tissues (colon, ileum, liver and skin) and non-target tissues (heart, pancreas and kidney) from naive animals or recipients 21days post HCT (BALB/c → C57BL/6,). (b) Representative images of SDH enzyme activity staining in GVHD target tissues (colon, ileum) from recipients 21days post HCT (C3H.SW → C57BL/6). (c) Integrated intensity of SDH enzyme activity staining from colon and ileum 7days post HCT (BALB/c → C57BL/6, n = 5 biologically independent animals). Representative plots and a graph summarizing the results of at least two independent experiments are shown. Scale bar (a,b): yellow=500 µm, black=200 µm. All statistical analysis by two-tailed Mann-Whitney test (c) (mean ± s.e.m.).
Extended Data Fig. 3 Morphological mitochondrial changes but not numbers in IECs post allo-HCT.
C57BL/6 animals received HCT (BALB/c or C57BL/6 → C57BL/6). (a) Immunoblot of SDHA, complex V, III, IV, SDHB, I and TOM-20 in mitochondria from colonic IECs 21days post HCT (n = 4). (b) Immunoblot and protein density quantification of SDHA, complex I, III, IV, V, and TOM-20 in mitochondria from colonic IECs 7days post HCT (n = 4). (c) Representative images of immunofluorescence staining of kidney and liver from recipients 21days post HCT (Complex IV = green, SDHA = red, DAPI = blue, scale bar= 50 µm, n = 5). (d) Numbers of gold particles per mitochondrial from colon and ileum of naive animals or recipients 7days post HCT in transmission electron microscopy images with immune-gold staining of SDHA (total 50 mitochondria from 3 samples). (e) Representative images of transmission electron microscopy in mitochondria of colon from naive or recipients 21days post HCT (scale bar =200 nm). Arrow indicates normal cristae and arrowhead indicates abnormal cristae (n = 3). (f) Mitochondria DNA relative copy numbers of colon and ileum from syngeneic and allogeneic recipients 7 and 21days post HCT (n = 5). All data are from biologically independent animals. Representative plots and a graph summarizing the results of at least two independent experiments are shown. All statistical analysis by two-tailed Mann-Whitney test (b) or one-way ANOVA analysis with Tukey post hoc test (d) (mean ± s.e.m.).
Extended Data Fig. 4 Conditioning regimen does not affect SDHA expression in IECs.
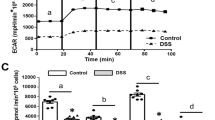
(a-b) C57BL/6 mice received 10 Gy total body irradiation followed by 1×106 T cells along with 5×106 TCD-BM cells from either syngeneic C57BL/6 or allogeneic mHA-mismatched C3H.SW donors (n = 5). (a) Representative images of immunofluorescence staining and (b) fluorescent intensity of SDHA in colon and ileum from recipients 28days post HCT are shown. (c-d) C57BL/6 mice received chemotherapy and received 1×107 T cells along with 1×107 TCD-BM cells from either syngeneic C57BL/6 or allogeneic BALB/c donors (n = 5). (c) Representative images of immunofluorescence staining and (d) fluorescent intensity of SDHA in colon and ileum from recipients 21days post HCT. (e-g) C57BL/6 mice were treated with 3% DSS or vehicle in drinking water for 7 days (n = 5). (e) Representative images of immunofluorescence staining and (f) fluorescent intensity of SDHA in colon 12days after DSS treatment. (g) Succinate levels in isolated IECs from colon and ileum of mice treated with 3% DSS at day12. (h) C57BL/6 mice receiving isotype control IgG or anti-CTLA-4 antibody were treated with 3% DSS in drinking water for 7 days. Time course of body weight changes after DSS administration. All data are from biologically independent animals. Representative plots and a graph summarizing the results of at least two independent experiments are shown. Scale bar (a, c, e) =50 µm. All statistical analysis by two-tailed Mann-Whitney test (b, d, f) or two-tailed unpaired t-test (g, h) (mean ± s.e.m.).
Extended Data Fig. 5 SDH inhibition causes ROS accumulation in IECs.
(a-d) PCECs were treated with DMSO, malonate or itaconate for 6 hours (n = 5). Cytoplasmic ROS measured by CellROX staining. (a) Representative images and (b) CellROX positive cells are shown. Mitochondria ROS measured by MitoSOX staining. (c) Representative images and (d) MitoSOX positive cells are shown. (e) PCECs were treated with DMSO or atpeninA5 for 4 hours. Cell death (left), CellROX (middle) and MitoSOX (right) levels were determined (n = 5). (f, g) Colon and ileum IECs were isolated from HCT recipients (BALB/c → C57BL/6). CellROX, MitoSOX (f) and mitochondrial membrane potential (ψm) (g) levels in (n = 5). (h, i) HCT settings are described in (f). Recipients orally received succinate every other day from day0 post HCT. Survival (h) and GVHD severity (i) are shown (n = 5). (j) Immunoblot of SUCNR1 in macrophage (MF, IC-21 cells) and IECs from BMT recipients 7days post HCT. Three independent experiments were performed. (k) Transcriptome analysis of IECs from recipients 7days post HCT. The mRNA expression of Hif1a and Cybb (CytochromeB). (n = 3). (l) Representative images of Hypoxyprobe in colon and ileum from recipients 7days after HCT (scale bar= 50 µm). Four independent experiments were performed. (m) Immunoblot image and protein density of ClpP in IECs from recipients 21days post HCT (Syn: n = 4, Allo n = 5). (n) Transcriptome analysis of IECs from recipients 7days post HCT. The mRNA expression of Clpp (n = 3). Data (f, g, k, l, m, n) are from biologically independent animals. Three independent experiments (a-e) were performed. All statistical analysis by one-way ANOVA analysis with Tukey post hoc test (b, d, e), two-tailed unpaired t-test (f, g), log-rank test (h) or two-tailed Mann-Whitney test (m) (mean ± s.e.m.). The p-values of the transcriptome analysis (k, n) obtained from DESeq-analysis.
Extended Data Fig. 6 Chemical inhibition and genetic ablation of SDHA in IECs regulate the severity of GVHD.
(a) Representative images of SDH enzyme staining of colon from naive C57BL/6 12 hours after vehicle, malonate (5 g kg-1) or itaconate (2.5 g kg-1) treatment. (Scale bar 100 µm, n = 2). (b-e) C57BL/6 or Sdhaf1-/- animals received 10 Gy total body irradiation followed by 3×106 T cells along with 5×106 TCD-BM cells from either syngeneic C57BL/6 or allogeneic BALB/c donors. (b) Survival of C57BL/6 recipients treated with vehicle or atpeninA5 (9 µg kg-1) every other day post HCT (n = 8). (c) C57BL/6 recipients treated with vehicle, malonate (5 g kg-1) or itaconate (2.5 g kg-1) every other day post HCT. Pathological GVHD scores in liver, skin and lung 7days post HCT (n = 6). (d) Succinate levels in isolated IECs from colon of naive C57BL/6 mice and Sdhaf1-/- mice (n = 4). (e) Pathological GVHD scores in liver, skin and lung 7days post HCT from C57BL/6 and Sdhaf1-/- recipients (n = 6). (f) C57BL/6 and Sdhaf1-/- animals were treated with 2.5 % DSS in drinking water for 7 days. Time course of body weight changes after DSS administration (n = 6). (g) Scheme illustrating the strategy used to generate Sdha floxed and excised alleles in IECs. (h) Expression of Sdha mRNA in CD326+ isolated IECs from six-week-old Sdha∆/IEC relative to Sdhafl/IEC mice (n = 4). Sdha expression was normalized to Gapdh expression. (i) Immunoblot of SDHA and TOM-20 in mitochondria of IECs from six-week-old Sdha∆/IEC and Sdhafl/IEC mice. (j) Representative images of immunofluorescence staining of colon, ileum, liver, skin and heart from six-week-old Sdha∆/IEC and Sdhafl/IEC mice (Complex IV = green, SDHA = red, DAPI = blue, scale bar=50 µm, n = 4). Data (a-f, h) are from biologically independent animals. All statistical analysis by log-rank test (b), Kruskal-Wallis analysis with Dunn’s post hoc test (c), two-tailed unpaired t-test (d), or two-tailed Mann-Whitney test (e, h) (mean ± s.e.m.).
Extended Data Fig. 7 Increased SDHA levels reduce T cell mediated intestinal damage.
(a, b) Immunoblot and protein density quantification of SDHA (a) and succinate levels (b) in colonic IECs from naive C57BL/6 and Irg1-/-mice (n = 4). (c-f) C57BL/6 animals Irg1-/- animals received HCT (BALB/c → C57BL/6 or Irg1-/-) (c) Images (left) and fluorescent intensity (right) of immunofluorescence staining of colon on 21 days after post allo-HCT (Complex IV = green, SDHA = red, DAPI = blue, n = 3). (d) Succinate levels in colon IECs at day21 post allo-HCT (n = 3). Clinical GVHD severity (e) and pathological GVHD score (f) of ileum and colon after post HCT (Clinical GVHD severity, Syn C57BL/6: n = 8, Allo C57BL/6 and Irg1-/-: n = 15, pathological GVHD score, n = 3). (g) Colonic IECs from C57BL/6, Sdha∆/IEC and Irg1-/- were used as target cells for CTL assay (2 hr co-culture with effector T cells). Cell death was analyzed (n = 3). (h, i) PCECs mediated by CRISPR-Cas9 targeting IRG1 were treated with LPS (100 ng/ml)/ IFNγ(10 ng/ml) for 6 hr. (h) Irg1 gene expression levels (normalized to Gapdh expression, n = 4 cells examined over 3 independent experiments). (i) Control and IRG1 KO PCECs were used as target cells for CTL assay (7 hr co-culture with effector T cells). Data are from 1 of 3 similar experiments) (j, k) C57BL/6 animals received HCT (BALB/c → C57BL/6) and treated with vehicle or butyrate. (j) Representative images (left) of immunofluorescence staining (Complex IV = green, SDHA = red, DAPI = blue) (left) and fluorescent intensity (right) of SDHA in colon on day21 post allo-HCT (n = 5). (k) Succinate levels in IECs from colon and ileum on day7 (top) and day21 (bottom) post allo-HCT (n = 5). (l) ChIP assay of 1 mM butyrate-treated isolated colonic IECs from naive C57BL/6 binding control IgG, pol II or acetylated histone H4 in the promoter regions of Sdha and Sdhb (n = 5). Data (a-d, f, g, j-l) are from biologically independent animals. Scale bar (c, j) = 50 µm. All statistical analysis by two-tailed Mann-Whitney test (a, h, j), two-tailed unpaired t-test (b, d, f, i, k, l), Kruskal-Wallis analysis with Dunn’s post hoc test (e) or ANOVA analysis with Tukey post hoc test (g) (mean ± s.e.m.).
Extended Data Fig. 8 Transcriptome analysis of IECs after allo-HCT.
(a-c) C57BL/6 animals received 10 Gy total body irradiation followed by injection of 3×106 T cells along with 5×106 TCD-BM cells from either syngeneic C57BL/6 or allogeneic BALB/c donors. Transcriptome analysis of colonic IECs from syngeneic and allogeneic recipients 7days post HCT. Heatmap of OXPHOS pathway gene (a) and TCA cycle related gene (b). (c) The mRNA expression of Sdha (left), Sdhb (middle) and Sdhc (right) in IECs from syngeneic and allogeneic recipients (n = 3 biologically independent animals). The p-values of the transcriptome analysis (c) obtained from DESeq-analysis (mean ± s.e.m.).
Extended Data Fig. 9 SDH expression in IECs from GVHD and non-GVHD patients.
Other representative images of immunofluorescence staining of colonic biopsy samples from patients suspected as having clinical GVHD after HCT not shown in Fig. 7c (Complex IV = green, SDHA = red, DAPI = blue, scale bar= 50 µm). Two independent experiments are performed.
Supplementary information
Supplementary Information
Supplementary Figs. 1 and 2, Tables 1 and 2 and Methods
Source data
Source Data Extended Data Fig. 1
Unprocessed immunoblots and gels.
Source Data Extended Data Fig. 3
Unprocessed immunoblots.
Source Data Extended Data Fig. 5
Unprocessed immunoblots.
Source Data Extended Data Fig. 6
Unprocessed immunoblots.
Rights and permissions
About this article
Cite this article
Fujiwara, H., Seike, K., Brooks, M.D. et al. Mitochondrial complex II in intestinal epithelial cells regulates T cell-mediated immunopathology. Nat Immunol 22, 1440–1451 (2021). https://doi.org/10.1038/s41590-021-01048-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-021-01048-3
This article is cited by
-
Acute graft-versus-host disease
Nature Reviews Disease Primers (2023)
-
Challenges and opportunities targeting mechanisms of epithelial injury and recovery in acute intestinal graft-versus-host disease
Mucosal Immunology (2022)
-
SDHA: a key player in T cell-mediated intestinal disease severity
Cellular & Molecular Immunology (2022)