Abstract
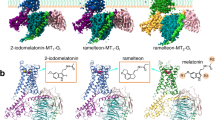
Melatonin receptors (MT1 and MT2) transduce inhibitory signaling by melatonin (N-acetyl-5-methoxytryptamine), which is associated with sleep induction and circadian rhythm modulation. Although recently reported crystal structures of ligand-bound MT1 and MT2 elucidated the basis of ligand entry and recognition, the ligand-induced MT1 rearrangement leading to Gi-coupling remains unclear. Here we report a cryo-EM structure of the human MT1–Gi signaling complex at 3.3 Å resolution, revealing melatonin-induced conformational changes propagated to the G-protein-coupling interface during activation. In contrast to other Gi-coupled receptors, MT1 exhibits a large outward movement of TM6, which is considered a specific feature of Gs-coupled receptors. Structural comparison of Gi and Gs complexes demonstrated conformational diversity of the C-terminal entry of the Gi protein, suggesting loose and variable interactions at the end of the α5 helix of Gi protein. These notions, together with our biochemical and computational analyses, highlight variable binding modes of Gαi and provide the basis for the selectivity of G-protein signaling.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Cryo-EM maps and atomic models for the MT1–Gi complex have been deposited in the Electron Microscopy Data Bank and wwPDB with the following accession numbers: EMD-30627 and PDB 7DB6, respectively. The raw images have been deposited in the Electron Microscopy Public Image Archive, under accession code EMPIAR-10714. Source data are provided with this paper.
Code availability
The Python code and data used for the contact and r.m.s.d. analysis are available on GitHub (https://github.com/raimondifranc/gpcr-gprotein-3D-comparison/).
References
Lerner, A. B., Case, J. D., Takahashi, Y., Lee, T. H. & Mori, W. Isolation of melatonin, the pineal gland factor that lightens melanocytes. J. Am. Chem. Soc. 80, 2587 (1958).
Snyder, S. H., Axelrod, J., Fischer, J. E. & Wurtman, R. J. Neural and photic regulation of 5-hydroxytryptophan decarboxylase in the rat pineal gland. Nature 203, 981–982 (1964).
Wurtman, R. J., Axelrod, J. & Phillips, L. S. Melatonin synthesis in the pineal gland: control by light. Science 142, 1071–1073 (1963).
Snyder, S. H., Zweig, M., Axelrod, J. & Fischer, J. E. Control of the circadian rhythm in serotonin content of the rat pineal. Proc. Natl Acad. Sci. USA 53, 301–305 (1965).
Ganguly, S., Coon, S. L. & Klein, D. C. Control of melatonin synthesis in the mammalian pineal gland: the critical role of serotonin acetylation. Cell Tissue Res. 309, 127–137 (2002).
Hardeland, R., Pandi-Perumal, S. R. & Cardinali, D. P. Melatonin. Int. J. Biochem. Cell Biol. 38, 313–316 (2006).
Jockers, R. et al. Update on melatonin receptors: IUPHAR Review 20. Br. J. Pharmacol. 173, 2702–2725 (2016).
Clarke, T. C., Black, L. I., Stussman, B. J., Barnes, P. M. & Nahin, R. L. Trends in the use of complementary health approaches among adults: United States, 2002–2012. Natl Health Stat. Report. 79, 1–16 (2015).
Harpsøe, N. G., Andersen, L. P. H., Gögenur, I. & Rosenberg, J. Clinical pharmacokinetics of melatonin: a systematic review. Eur. J. Clin. Pharmacol. 71, 901–909 (2015).
Kato, K. et al. Neurochemical properties of ramelteon (TAK-375), a selective MT 1/MT2 receptor agonist. Neuropharmacology 48, 301–310 (2005).
Millan, M. J. et al. The novel melatonin agonist agomelatine (S20098) is an antagonist at 5-hydroxytryptamine2C receptors, blockade of which enhances the activity of frontocortical dopaminergic and adrenergic pathways. J. Pharmacol. Exp. Ther. 306, 954–964 (2003).
Vachharajani, N. N., Yeleswaram, K. & Boulton, D. W. Preclinical pharmacokinetics and metabolism of BMS-214778, a novel melatonin receptor agonist. J. Pharm. Sci. 92, 760–772 (2003).
Patel, N. et al. Structure-based discovery of potent and selective melatonin receptor agonists. Elife 9, e53779 (2020).
Stauch, B. et al. Structural basis of ligand recognition at the human MT1 melatonin receptor. Nature 569, 284–288 (2019).
Johansson, L. C. et al. XFEL structures of the human MT2 melatonin receptor reveal the basis of subtype selectivity. Nature 569, 289–292 (2019).
Stein, R. M. et al. Virtual discovery of melatonin receptor ligands to modulate circadian rhythms. Nature 579, 609–614 (2020).
Kirchhofer, A. et al. Modulation of protein properties in living cells using nanobodies. Nat. Struct. Mol. Biol. 17, 133–139 (2010).
Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife 7, e42166 (2018).
Rosenthal, P. B. & Henderson, R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 333, 721–745 (2003).
Koehl, A. et al. Structure of the μ-opioid receptor-Gi protein complex. Nature 558, 547–552 (2018).
Maeda, S., Qu, Q., Robertson, M. J., Skiniotis, G. & Kobilka, B. K. Structures of the M1 and M2 muscarinic acetylcholine receptor/G-protein complexes. Science 364, 552–557 (2019).
Kato, H. E. et al. Conformational transitions of a neurotensin receptor 1–Gi1 complex. Nature 572, 80–85 (2019).
Krishna Kumar, K. et al. Structure of a signaling cannabinoid receptor 1-G protein complex. Cell 176, 448–458.e12 (2019).
Yin, J. et al. Structure of a D2 dopamine receptor–G-protein complex in a lipid membrane. Nature 584, 125–129 (2020).
Zhuang, Y. et al. Structure of formylpeptide receptor 2-Gi complex reveals insights into ligand recognition and signaling. Nat. Commun. 11, 885 (2020).
Xing, C. et al. Cryo-EM structure of the human cannabinoid receptor CB2-Gi signaling complex. Cell 180, 645–654.e13 (2020).
Hua, T. et al. Activation and signaling mechanism revealed by cannabinoid receptor-Gi complex structures. Cell 180, 655–665.e18 (2020).
Qiao, A. et al. Structural basis of Gs and Gi recognition by the human glucagon receptor. Science 367, 1346–1352 (2020).
Morimoto, K. et al. Crystal structure of the endogenous agonist-bound prostanoid receptor EP3. Nat. Chem. Biol. 15, 8–10 (2019).
Fan, H. et al. Structural basis for ligand recognition of the human thromboxane A2 receptor. Nat. Chem. Biol. 15, 27–33 (2019).
Toyoda, Y. et al. Ligand binding to human prostaglandin E receptor EP 4 at the lipid-bilayer interface. Nat. Chem. Biol. 15, 18–26 (2019).
Kang, Y. et al. Cryo-EM structure of human rhodopsin bound to an inhibitory G protein. Nature 558, 553–558 (2018).
Inoue, A. et al. Illuminating G-protein-coupling selectivity of GPCRs. Cell 177, 1933–1947.e25 (2019).
Clement, N. et al. Importance of the second extracellular loop for melatonin MT 1 receptor function and absence of melatonin binding in GPR50. Br. J. Pharmacol. 175, 3281–3297 (2018).
Rasmussen, S. G. F. et al. Crystal structure of the β2 adrenergic receptor–Gs protein complex. Nature 477, 549–557 (2011).
Zhou, Q. et al. Common activation mechanism of class a GPCRs. Elife 8, e50279 (2019).
Dror, R. O. et al. Activation mechanism of the β2-adrenergic receptor. Proc. Natl Acad. Sci. USA 108, 18684–18689 (2011).
White, K. L. et al. Structural connection between activation microswitch and allosteric sodium site in GPCR signaling. Structure 26, 259–269.e5 (2018).
Huang, W. et al. Structural insights into μ-opioid receptor activation. Nature 524, 315–321 (2015).
Tsai, C. J. et al. Crystal structure of rhodopsin in complex with a mini-Go sheds light on the principles of G protein selectivity. Sci. Adv. 4, 7052 (2018).
Lin, X. et al. Structural basis of ligand recognition and self-activation of orphan GPR52. Nature 579, 152–157 (2020).
García-Nafría, J., Lee, Y., Bai, X., Carpenter, B. & Tate, C. G. Cryo-EM structure of the adenosine A2A receptor coupled to an engineered heterotrimeric G protein. Elife 7, e35946 (2018).
Nguyen, A. H. et al. Structure of an endosomal signaling GPCR–G protein–β-arrestin megacomplex. Nat. Struct. Mol. Biol. 26, 1123–1131 (2019).
Manglik, A. et al. Crystal structure of the μ-opioid receptor bound to a morphinan antagonist. Nature 485, 321–326 (2012).
Cherezov, V. et al. High-resolution crystal structure of an engineered human β2-adrenergic G protein–coupled receptor. Science 318, 1258–1265 (2007).
Draper-Joyce, C. J. et al. Structure of the adenosine-bound human adenosine A1 receptor-Gi complex. Nature 558, 559–565 (2018).
Yuan, D. et al. Activation of the α2B adrenoceptor by the sedative sympatholytic dexmedetomidine. Nat. Chem. Biol. 16, 507–512 (2020).
García-Nafría, J., Nehmé, R., Edwards, P. C. & Tate, C. G. Cryo-EM structure of the serotonin 5-HT1B receptor coupled to heterotrimeric Go. Nature 558, 620–623 (2018).
Dror, R. O. et al. Structural basis for nucleotide exchange in heterotrimeric G proteins. Science 348, 1361–1365 (2015).
Whitford, M., Stewart, S., Kuzio, J. & Faulkner, P. Identification and sequence analysis of a gene encoding gp67, an abundant envelope glycoprotein of the baculovirus Autographa californica nuclear polyhedrosis virus. J. Virol. 63, 1393–1399 (1989).
Maeda, S. et al. Development of an antibody fragment that stabilizes GPCR/G-protein complexes. Nat. Commun. 9, 3712 (2018).
Mastronarde, D. N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005).
Rohou, A. & Grigorieff, N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015).
Zivanov, J., Nakane, T. & Scheres, S. H. W. A Bayesian approach to beam-induced motion correction in cryo-EM single-particle analysis. IUCrJ 6, 5–17 (2019).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of COOT. Acta Crystallogr. Sect. D Biol. Crystallogr. 66, 486–501 (2010).
Wang, R. Y. R. et al. Automated structure refinement of macromolecular assemblies from cryo-EM maps using Rosetta. Elife 5, e17219 (2016).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D Biol. Crystallogr. 66, 213–221 (2010).
Afonine, P. V. et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. Sect. D Struct. Biol. 74, 531–544 (2018).
Goddard, T. D. et al. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018).
Flock, T. et al. Universal allosteric mechanism for Gα activation by GPCRs. Nature 524, 173–179 (2015).
Šali, A. & Blundell, T. L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234, 779–815 (1993).
Phillips, J. C. et al. Scalable molecular dynamics with NAMD. J. Computational Chem. 26, 1781–1802 (2005).
Humphrey, W., Dalke, A. & Schulten, K. VMD: visual molecular dynamics. J. Mol. Graph. 14, 33–38 (1996).
Best, R. B. et al. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ1 and χ2 dihedral angles. J. Chem. Theory Comput. 8, 3257–3273 (2012).
Klauda, J. B. et al. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B 114, 7830–7843 (2010).
Feller, S. E., Zhang, Y., Pastor, R. W. & Brooks, B. R. Constant pressure molecular dynamics simulation: the Langevin piston method. J. Chem. Phys. 103, 4613–4621 (1995).
Darden, T., York, D. & Pedersen, L. Particle mesh Ewald: an N·log(N) method for Ewald sums in large systems. J. Chem. Phys. 98, 10089–10092 (1993).
Ehlert, F. J., Griffin, M. T., Sawyer, G. W. & Bailon, R. A simple method for estimation of agonist activity at receptor subtypes: comparison of native and cloned M3 muscarinic receptors in guinea pig ileum and transfected cells. J. Pharmacol. Exp. Ther. 289, 981–982 (1999).
Altschul, S. F. et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997).
Isberg, V. et al. GPCRdb: an information system for G protein-coupled receptors. Nucl. Acids Res. 44, D356–D364 (2016); erratum 45, 2936 (2017).
Cock, P. J. A. et al. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 25, 1422–1423 (2009).
Sievers, F. & Higgins, D. G. in Multiple Sequence Alignment Methods. Methods in Molecular Biology (Methods and Protocols) Vol. 1079 (ed. Russell, D.) 105–116 (Humana Press, 2014).
Mistry, J. et al. Pfam: the protein families database in 2021. Nucl. Acids Res. 49, D412–D419 (2021).
Acknowledgements
We thank T. Nakane for assistance with the single-particle analysis; Y. Nomura and Y. Sato for assistance with the scFv16 expression and purification; K. Sato, S. Nakano and A. Inoue at Tohoku University for assistance with the cell culture and the cell-based GPCR assays; and the staff scientists at The University of Tokyo’s cryo-EM facility, especially K. Kobayashi, H. Yanagisawa, A. Tsutsumi, M. Kikkawa and R. Danev, for help with cryo-EM data collection. This work was supported by the MEXT Grant-in-Aid for Specially Promoted Research (grant no. 16H06294) to O.N.; by the JST CREST program (no. 20344981) ‘Atomic resolution dynamics of supramolecular complexes essential for cellular functions’ to O.N.; by PRIME (JP19gm5910013) and LEAP (JP20gm0010004) grants from the Japan Agency for Medical Research and Development to A.I.; by the JSPS KAKENHI grant nos. 17K08264 (to A.I.) and 17H05000 and 20H03216 (to T.N.) and by the Platform Project for Supporting Drug Discovery and Life Science Research (Basis for Supporting Innovative Drug Discovery and Life Science Research) from AMED under grant numbers JP19am0101115 (support number 1111 to O.N.), 19am0101079 (support number 1819; to O.N.) and JP20am0101095 (to A.I.). F.R. thanks the Italian Ministry of University and Research for funding the Department of Excellence Faculty of Sciences of Scuola Normale Superiore. A.I. thanks the Takeda Science Foundation for funding along with The Uehara Memorial Foundation and Ono Medical Research Foundation. R.S. and T.K. acknowledge the Takeda Science Foundation (19H03428), and T.K. thanks the AMED grant (no. JP18gm0910007).
Author information
Authors and Affiliations
Contributions
H.H.O. prepared the cryo-EM sample of MT1–Gi–scFv16, and W.S. helped with the construction of the Gi-heterotrimer expression. N.N. and S.I. helped with the preparation of scFv16. H.H.O., T.K. and N.T. collected the cryo-EM data. H.H.O. and T.N. processed the cryo-EM data and built the structure of MT1–Gi–scFv16. K.Y. assisted with the structure building and refinement of MT1–Gi–scFv16. H.M. performed MD simulations and analyzed the results. A.I. performed the NanoBiT and flow cytometry assays of the MT1-mutants. H.T., R.S. and T.K. performed radioligand experiments. F.R. performed the contact analysis and clustering of the reported GPCR–G-protein complexes. H.H.O., T.N. and O.N. mainly wrote the manuscript. A.I., T.N. and O.N. supervised the research.
Corresponding authors
Ethics declarations
Competing interests
O.N. is a cofounder and scientific advisor for Curreio. All other authors declare no competing interests.
Additional information
Peer review information Nature Structural & Molecular Biology thanks Alisa Glukhova, Ralf Jockers and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Cryo-EM analysis procedure.
a, Flow-chart depicting the single particle analysis of the MT1-Gi complex. Fourier Shell Correlation (FSC) curve shows 3.3 Å global resolution with the gold standard criteria (FSC=0.143). Local resolution is indicated on the cryo-EM map. b, A representative micrograph of the MT1-ramelteon-Gαi-Gβ1-Gγ2-scFv16 complex. Similar images (3537 in total) were deposited to the EMPIAR (EMPIAR-10714). c, Representative 2D-averaged images of the MT1-ramelteon-Gαi-Gβ1-Gγ2-scFv16 complex. d, The map versus model FSC curves.
Extended Data Fig. 2 Cryo-EM density-map and comparison of “lid” structures.
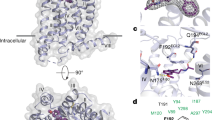
a, Cryo-EM density-map of the MT1-ramelteon-Gαi-Gβ1-Gγ2-scFv16 complex (light blue: MT1, yellow: Gαi, green: Gβ1, purple: Gγ2, red: scFv16). The Fab fragment scFv16 binds and stabilizes the interface of Gαi and Gβ1. b, Top view of MT1-Gi and reported lipid GPCRs, viewed from the extracellular side. Receptors (light blue: MT1-Gi, red: TP, blue: EP3, green: EP4) are shown as cylinder models, and ligands are shown as CPK models. MT1-Gi shows the “lid” structure, which is similar to those in the reported crystal structures of lipid GPCRs.
Extended Data Fig. 3 Functional characterization of MT1-mutants.
a–c, NanoBiT-G-protein dissociation assays for the constructs used in the cryo-EM analysis and the MT1 mutants. Concentration–response curves for melatonin-dependent G-protein dissociation signals for MT1. To match the expression of MT1-WT to that of mutants with lower expression, 1:5 volume [WT (1:5)] or 1:10 volume [WT (1:10)] MT1-WT plasmid was used. Asterisks denote an increased volume (2.5-fold) of plasmid transfection. Symbols and error bars represent mean and s.e.m. of the indicated independent numbers of experiments (n = 3–5; see Extended Data Table S1). Note that, in most data points, error bars are smaller than the size of the symbols and thus are not visualized in the panels. The results for the construct used in the cryo-EM analysis (a), the F1965.47A residue that is important in MT1-activation (b), and the mutants to test Gi-selectivity (c) are shown. d, Cell surface expression levels of the MT1-constructs. Cells transiently expressing the FLAG epitope-tagged MT1-constructs were labeled with an anti-FLAG tag antibody, and then with an Alexa 488-conjugated secondary antibody. The fluorescent signals from individual cells were measured by a flow cytometer (See Methods). Data from each experiment are shown as dots with the mean value presented as a bar (n = 3–9). For the multiple comparison analysis, two-way ANOVA followed by the Dunnett’s test (WT (1:1) vs. mutant) was performed: ns, not significantly different, *, P < 0.05, *** P < 0.001. Source data for (a–d) are available online.
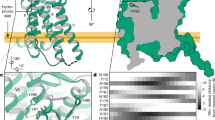
Extended Data Fig. 4 Lateral pore and molecular dynamics (MD) simulations.
a,b, Structural comparison in the sagittal cross section (left panels), the lateral pore (top panels) and the transverse cross section (right panels) between MT1-Gi (light blue) and MT1 alone (crystal structure)14 (green, PDB 6ME2). The cross section and focused view on the lateral pore in MT1-Gi are shown. c, Molecular dynamics (MD) simulations in the presence (left, top) and absence (right, top) of ramelteon. The temporal changes in the distance of the Cα atom between Ala1584.56 and His1955.46 in the presence (left, top) and the absence (right, top) of ramelteon are shown. The transverse cross sections at the ligand-binding site at 200 ns and 400 ns for each simulation in the presence (left, below) and absence (right, below) of ramelteon.
Extended Data Fig. 5 Structural comparison on the G-protein interface.
Structural comparison of the G-protein interfaces among the GPCR-Gi and GPCR-Gs complexes. Structures are shown as ribbon models. Residues are shown as stick models. Hydrogen bonds and ionic interactions are indicated by black dashed lines. Focused views at the C-terminus of Gα (top panels) and the hydrophobic pocket of Gα (lower panels) are shown for CB1-Gi23 (a; cyan, PDB 6N4B), CB2-Gi27 (b; yellow, PDB 6KPF), NTS1-Gi C-state22 (c; green, PDB 6OS9), NTS1-Gi NC-state22 (d; brown, PDB 6OSA), D2R-Gi24 (e; yellow, PDB 6VMS), GPR52-miniGs41 (f; green, PDB 6LI3), β2AR-Gs43 (g; red, PDB 6NI3), and A2AR-miniGs42 (h; blue, PDB 6GDG).
Extended Data Fig. 6 Functional characterization of the MT1-signaling.
The NanoBiT-G-protein dissociation assays for concentration-response curves of G-protein dissociation signals are shown for MT1 (red) and mock (grey), using melatonin (top panels) and ramelteon (bottom panels). Symbols and error bars represent mean and s.e.m. of three independent experiments. Note that, in most data points, error bars are smaller than the size of the symbols and thus are not visualized in the panels. Source data are available online.
Extended Data Fig. 7 Cryo-EM density map and the structural model of the MT1-Gi complex.
Cryo-EM density maps and structural models are shown for selected regions: Ligand-binding pocket, Lateral gate, Toggle switch and PIF core, Na+ site and NPxxY motif, DRY motif, TM6 (lower part) and Gi-interface.
Supplementary information
Supplementary Information
Supplementary Figs. 1–3 and Tables 1 and 2.
Supplementary Dataset 1
Source data for Supplementary Fig. 3.
Source data
Source Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Rights and permissions
About this article
Cite this article
Okamoto, H.H., Miyauchi, H., Inoue, A. et al. Cryo-EM structure of the human MT1–Gi signaling complex. Nat Struct Mol Biol 28, 694–701 (2021). https://doi.org/10.1038/s41594-021-00634-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-021-00634-1
This article is cited by
-
Structural basis for lysophosphatidylserine recognition by GPR34
Nature Communications (2024)
-
Structural and dynamic insights into supra-physiological activation and allosteric modulation of a muscarinic acetylcholine receptor
Nature Communications (2023)
-
GPCRome-wide analysis of G-protein-coupling diversity using a computational biology approach
Nature Communications (2023)
-
An inverse agonist of orphan receptor GPR61 acts by a G protein-competitive allosteric mechanism
Nature Communications (2023)
-
Cryo-EM structures of human bradykinin receptor-Gq proteins complexes
Nature Communications (2022)