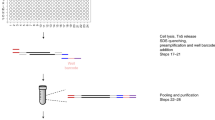
Abstract
The assay for transposase-accessible chromatin using sequencing (ATAC-seq) provides a simple and scalable way to detect the unique chromatin landscape associated with a cell type and how it may be altered by perturbation or disease. ATAC-seq requires a relatively small number of input cells and does not require a priori knowledge of the epigenetic marks or transcription factors governing the dynamics of the system. Here we describe an updated and optimized protocol for ATAC-seq, called Omni-ATAC, that is applicable across a broad range of cell and tissue types. The ATAC-seq workflow has five main steps: sample preparation, transposition, library preparation, sequencing and data analysis. This protocol details the steps to generate and sequence ATAC-seq libraries, with recommendations for sample preparation and downstream bioinformatic analysis. ATAC-seq libraries for roughly 12 samples can be generated in 10 h by someone familiar with basic molecular biology, and downstream sequencing analysis can be implemented using benchmarked pipelines by someone with basic bioinformatics skills and with access to a high-performance computing environment.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Code availability
The source code for the iterative overlap is freely available at https://github.com/corceslab/ATAC_IterativeOverlapPeakMerging88. All other ATAC-seq data analysis for the figures used in this protocol were generated using PEPATAC73 with Bulker (container version 1.0.8), available at http://pepatac.databio.org/en/latest/.
References
Ernst, J. & Kellis, M. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nat. Biotechnol. 28, 817–825 (2010).
Ernst, J. et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 473, 43–49 (2011).
Klemm, S. L., Shipony, Z. & Greenleaf, W. J. Chromatin accessibility and the regulatory epigenome. Nat. Rev. Genet. 20, 207–220 (2019).
Johnson, D. S., Mortazavi, A., Myers, R. M. & Wold, B. Genome-wide mapping of in vivo protein–DNA interactions. Science 316, 1497–1502 (2007).
Furey, T. S. ChIP–seq and beyond: new and improved methodologies to detect and characterize protein–DNA interactions. Nat. Rev. Genet. 13, 840–852 (2012).
Nakato, R. & Sakata, T. Methods for ChIP-seq analysis: a practical workflow and advanced applications. Methods 187, 44–53 (2021).
Schmid, M., Durussel, T. & Laemmli, U. K. ChIC and ChEC: genomic mapping of chromatin proteins. Mol. Cell 16, 147–157 (2004).
Skene, P. J. & Henikoff, S. An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. eLife 6, e21856 (2017).
Kaya-Okur, H. S. et al. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat. Commun. 10, 1930 (2019).
Schmidl, C., Rendeiro, A. F., Sheffield, N. C. & Bock, C. ChIPmentation: fast, robust, low-input ChIP-seq for histones and transcription factors. Nat. Methods 12, 963–965 (2015).
Wang, Q. et al. CoBATCH for high-throughput single-cell epigenomic profiling. Mol. Cell 76, 206–216.e7 (2019).
Handa, T. et al. Chromatin integration labeling for mapping DNA-binding proteins and modifications with low input. Nat. Protoc. 15, 3334–3360 (2020).
Harada, A. et al. A chromatin integration labelling method enables epigenomic profiling with lower input. Nat. Cell Biol. 21, 287–296 (2019).
Ku, W. L. et al. Single-cell chromatin immunocleavage sequencing (scChIC-Seq) to profile histone modification. Nat. Methods 16, 323–325 (2019).
Zheng, X.-Y. & Gehring, M. Low-input chromatin profiling in Arabidopsis endosperm using CUT&RUN. Plant Reprod. 32, 63–75 (2019).
Hainer, S. J., Bošković, A., McCannell, K. N., Rando, O. J. & Fazzio, T. G. Profiling of pluripotency factors in single cells and early embryos. Cell 177, 1319–1329 (2019).
Skene, P. J., Henikoff, J. G. & Henikoff, S. Targeted in situ genome-wide profiling with high efficiency for low cell numbers. Nat. Protoc. 13, 1006–1019 (2018).
Crawford, G. E. et al. Genome-wide mapping of DNase hypersensitive sites using massively parallel signature sequencing (MPSS). Genome Res. 16, 123–131 (2006).
Song, L. & Crawford, G. E. DNase-seq: a high-resolution technique for mapping active gene regulatory elements across the genome from mammalian cells. Cold Spring Harb. Protoc. 2010, pdb.prot5384 (2010).
Boyle, A. P. et al. High-resolution mapping and characterization of open chromatin across the genome. Cell 132, 311–322 (2008).
Giresi, P. G., Kim, J., McDaniell, R. M., Iyer, V. R. & Lieb, J. D. FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) isolates active regulatory elements from human chromatin. Genome Res. 17, 877–885 (2007).
Cui, K. & Zhao, K. in Chromatin Remodeling: Methods and Protocols (ed. Morse, R. H.) 413–419 (Humana, 2012).
Schones, D. E. et al. Dynamic regulation of nucleosome positioning in the human genome. Cell 132, 887–898 (2008).
Kelly, T. K. et al. Genome-wide mapping of nucleosome positioning and DNA methylation within individual DNA molecules. Genome Res. 22, 2497–2506 (2012).
Minnoye, L. et al. Chromatin accessibility profiling methods. Nat. Rev. Methods Primer 1, 1–24 (2021).
Weintraub, H. & Groudine, M. Chromosomal subunits in active genes have an altered conformation. Science 193, 848–856 (1976).
Galas, D. J. & Schmitz, A. DNAse footprinting: a simple method for the detection of protein–DNA binding specificity. Nucleic Acids Res. 5, 3157–3170 (1978).
He, H. H. et al. Refined DNase-seq protocol and data analysis reveals intrinsic bias in transcription factor footprint identification. Nat. Methods 11, 73–78 (2014).
Sung, M.-H., Baek, S. & Hager, G. L. Genome-wide footprinting: ready for prime time? Nat. Methods 13, 222–228 (2016).
Mieczkowski, J. et al. MNase titration reveals differences between nucleosome occupancy and chromatin accessibility. Nat. Commun. 7, 11485 (2016).
Chereji, R. V., Bryson, T. D. & Henikoff, S. Quantitative MNase-seq accurately maps nucleosome occupancy levels. Genome Biol. 20, 198 (2019).
Buenrostro, J. D., Giresi, P. G., Zaba, L. C., Chang, H. Y. & Greenleaf, W. J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013).
Goryshin, I. Y. & Reznikoff, W. S. Tn5 in vitro transposition. J. Biol. Chem. 273, 7367–7374 (1998).
Adey, A. et al. Rapid, low-input, low-bias construction of shotgun fragment libraries by high-density in vitro transposition. Genome Biol. 11, R119 (2010).
Gangadharan, S., Mularoni, L., Fain-Thornton, J., Wheelan, S. J. & Craig, N. L. DNA transposon Hermes inserts into DNA in nucleosome-free regions in vivo. Proc. Natl Acad. Sci. USA 107, 21966–21972 (2010).
Corces, M. R. et al. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat. Methods 14, 959–962 (2017).
Corces, M. R. et al. Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat. Genet. 48, 1193–1203 (2016).
Corces, M. R. et al. The chromatin accessibility landscape of primary human cancers. Science 362, eaav1898 (2018).
Calderon, D. et al. Landscape of stimulation-responsive chromatin across diverse human immune cells. Nat. Genet. 51, 1494–1505 (2019).
Naik, S. et al. Inflammatory memory sensitizes skin epithelial stem cells to tissue damage. Nature 550, 475–480 (2017).
Marco, A. et al. Mapping the epigenomic and transcriptomic interplay during memory formation and recall in the hippocampal engram ensemble. Nat. Neurosci. 23, 1606–1617 (2020).
Li, D. et al. Chromatin accessibility dynamics during iPSC reprogramming. Cell Stem Cell 21, 819–833 (2017).
Guo, J. et al. Chromatin and single-cell RNA-seq profiling reveal dynamic signaling and metabolic transitions during human spermatogonial stem cell development. Cell Stem Cell 21, 533–546 (2017).
Wu, J. et al. The landscape of accessible chromatin in mammalian preimplantation embryos. Nature 534, 652–657 (2016).
Daugherty, A. C. et al. Chromatin accessibility dynamics reveal novel functional enhancers in C. elegans. Genome Res. 27, 2096–2107 (2017).
Gury-BenAri, M. et al. The spectrum and regulatory landscape of intestinal innate lymphoid cells are shaped by the microbiome. Cell 166, 1231–1246 (2016).
Liu, Q. et al. Chromatin accessibility landscapes of skin cells in systemic sclerosis nominate dendritic cells in disease pathogenesis. Nat. Commun. 11, 5843 (2020).
Liu, Y. et al. Chromatin accessibility landscape of articular knee cartilage reveals aberrant enhancer regulation in osteoarthritis. Sci. Rep. 8, 15499 (2018).
Greenwald, W. W. et al. Pancreatic islet chromatin accessibility and conformation reveals distal enhancer networks of type 2 diabetes risk. Nat. Commun. 10, 2078 (2019).
Lee, J. et al. Activation of PDGF pathway links LMNA mutation to dilated cardiomyopathy. Nature 572, 335–340 (2019).
Schmidl, C. et al. Combined chemosensitivity and chromatin profiling prioritizes drug combinations in CLL. Nat. Chem. Biol. 15, 232–240 (2019).
Scharer, C. D. et al. Epigenetic programming underpins B cell dysfunction in human SLE. Nat. Immunol. 20, 1071–1082 (2019).
Corces, M. R. et al. Single-cell epigenomic analyses implicate candidate causal variants at inherited risk loci for Alzheimer’s and Parkinson’s diseases. Nat. Genet. 52, 1158–1168 (2020).
Banovich, N. E. et al. Impact of regulatory variation across human iPSCs and differentiated cells. Genome Res. 28, 122–131 (2018).
Forrest, M. P. et al. Open chromatin profiling in hiPSC-derived neurons prioritizes functional noncoding psychiatric risk variants and highlights neurodevelopmental loci. Cell Stem Cell 21, 305–318 (2017).
Chiou, J. et al. Interpreting type 1 diabetes risk with genetics and single-cell epigenomics. Nature 594, 398–402 (2021).
Nott, A. et al. Brain cell type-specific enhancer–promoter interactome maps and disease-risk association. Science 366, 1134–1139 (2019).
Liu, Q. et al. Genome-wide temporal profiling of transcriptome and open-chromatin of early cardiomyocyte differentiation derived from hiPSCs and hESCs. Circ. Res. 121, 376–391 (2017).
Wapinski, O. L. et al. Rapid chromatin switch in the direct reprogramming of fibroblasts to neurons. Cell Rep. 20, 3236–3247 (2017).
Denny, S. K. et al. Nfib promotes metastasis through a widespread increase in chromatin accessibility. Cell 166, 328–342 (2016).
Schep, A. N. et al. Structured nucleosome fingerprints enable high-resolution mapping of chromatin architecture within regulatory regions. Genome Res. 25, 1757–1770 (2015).
Kaya-Okur, H. S., Janssens, D. H., Henikoff, J. G., Ahmad, K. & Henikoff, S. Efficient low-cost chromatin profiling with CUT&Tag. Nat. Protoc. 15, 3264–3283 (2020).
Stark, R., Grzelak, M. & Hadfield, J. RNA sequencing: the teenage years. Nat. Rev. Genet. 20, 631–656 (2019).
Bartosovic, M., Kabbe, M. & Castelo-Branco, G. Single-cell CUT&Tag profiles histone modifications and transcription factors in complex tissues. Nat. Biotechnol. 39, 825–835 (2021).
Gao, W., Lai, B., Ni, B. & Zhao, K. Genome-wide profiling of nucleosome position and chromatin accessibility in single cells using scMNase-seq. Nat. Protoc. 15, 68–85 (2020).
Lai, B. et al. Principles of nucleosome organization revealed by single-cell micrococcal nuclease sequencing. Nature 562, 281–285 (2018).
Jin, W. et al. Genome-wide detection of DNase I hypersensitive sites in single cells and FFPE tissue samples. Nature 528, 142–146 (2015).
Takaku, M. et al. GATA3-dependent cellular reprogramming requires activation-domain dependent recruitment of a chromatin remodeler. Genome Biol. 17, 36 (2016).
Fujiwara, S., Baek, S., Varticovski, L., Kim, S. & Hager, G. L. High quality ATAC-seq data recovered from cryopreserved breast cell lines and tissue. Sci. Rep. 9, 516 (2019).
Mulqueen, R. M. et al. Improved single-cell ATAC-seq reveals chromatin dynamics of in vitro corticogenesis. Preprint at bioRxiv https://doi.org/10.1101/637256 (2019).
nf-core/atacseq. (nf-core, 2021). https://doi.org/10.5281/zenodo.2634132
ATAC-seq Data Standards and Processing Pipeline. ENCODE. https://www.encodeproject.org/atac-seq/
Smith, J. P. et al. PEPATAC: an optimized pipeline for ATAC-seq data analysis with serial alignments. NAR Genomics Bioinform. 3, lqab101 (2021).
Bajic, M., Maher, K. A. & Deal, R. B. Identification of open chromatin regions in plant genomes using ATAC-seq. Methods Mol. Biol. 1675, 183–201 (2018).
Deal, R. B. & Henikoff, S. The INTACT method for cell type-specific gene expression and chromatin profiling in Arabidopsis thaliana. Nat. Protoc. 6, 56–68 (2011).
Haines, J. E. & Eisen, M. B. Patterns of chromatin accessibility along the anterior-posterior axis in the early Drosophila embryo. PLOS Genet. 14, e1007367 (2018).
Johnson, S., Nguyen, V. & Coder, D. Assessment of cell viability. Curr. Protoc. Cytom. 64, 9.2.1–9.2.26 (2013).
Chen, X. et al. ATAC-see reveals the accessible genome by transposase-mediated imaging and sequencing. Nat. Methods 13, 1013–1020 (2016).
Swanson, E. et al. Simultaneous trimodal single-cell measurement of transcripts, epitopes, and chromatin accessibility using TEA-seq. eLife 10, e63632 (2021).
Corces, R. Isolation of nuclei from frozen tissue for ATAC-seq and other epigenomic assays. https://doi.org/10.17504/protocols.io.6t8herw (2019).
Polavarapu, V. K. et al. Profiling chromatin accessibility in formalin-fixed paraffin-embedded samples. Genome Res. https://doi.org/10.1101/gr.275269.121 (2021).
Chin, H. G. et al. Universal NicE-seq for high-resolution accessible chromatin profiling for formaldehyde-fixed and FFPE tissues. Clin. Epigenetics 12, 143 (2020).
Orchard, P., Kyono, Y., Hensley, J., Kitzman, J. O. & Parker, S. C. J. Quantification, dynamic visualization, and validation of bias in ATAC-seq data with ataqv. Cell Syst. 10, 298–306 (2020).
Ewels, P. A. et al. The nf-core framework for community-curated bioinformatics pipelines. Nat. Biotechnol. 38, 276–278 (2020).
Koohy, H., Down, T. A. & Hubbard, T. J. Chromatin accessibility data sets show bias due to sequence specificity of the DNase I enzyme. PLoS ONE 8, e69853 (2013).
Satpathy, A. T. et al. Massively parallel single-cell chromatin landscapes of human immune cell development and intratumoral T cell exhaustion. Nat. Biotechnol. 37, 925–936 (2019).
Cusanovich, D. A. et al. Multiplex single cell profiling of chromatin accessibility by combinatorial cellular indexing. Science 348, 910–914 (2015).
Granja, J. M. & Corces, M. R. ATAC_IterativeOverlapPeakMerging https://doi.org/10.5281/zenodo.5903680 (2022).
Picelli, S. et al. Tn5 transposase and tagmentation procedures for massively scaled sequencing projects. Genome Res. 24, 2033–2040 (2014).
Smith, J. P. & Sheffield, N. C. Analytical approaches for ATAC-seq data analysis. Curr. Protoc. Hum. Genet. 106, e101 (2020).
A Quality Control tool for High Throughput Sequence Data. Babraham Bioinformatics. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
Zhang, Y. et al. Model-based analysis of ChIP-seq (MACS). Genome Biol. 9, R137 (2008).
Gaspar, J. M. Genrich: Detecting Sites of Genomic Enrichment (2021).
Tarbell, E. D. & Liu, T. HMMRATAC: a Hidden Markov ModeleR for ATAC-seq. Nucleic Acids Res. 47, e91 (2019).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010).
Yu, G., Wang, L.-G. & He, Q.-Y. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 31, 2382–2383 (2015).
Zhu, L. J. et al. ChIPpeakAnno: a Bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinformatics 11, 237 (2010).
Mumbach, M. R. et al. Enhancer connectome in primary human cells identifies target genes of disease-associated DNA elements. Nat. Genet. 49, 1602–1612 (2017).
Gontarz, P. et al. Comparison of differential accessibility analysis strategies for ATAC-seq data. Sci. Rep. 10, 10150 (2020).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Yan, F., Powell, D. R., Curtis, D. J. & Wong, N. C. From reads to insight: a hitchhiker’s guide to ATAC-seq data analysis. Genome Biol. 21, 22 (2020).
Risso, D., Ngai, J., Speed, T. P. & Dudoit, S. Normalization of RNA-seq data using factor analysis of control genes or samples. Nat. Biotechnol. 32, 896–902 (2014).
UCSC Genome Browser Home. https://genome.ucsc.edu/index.html
Robinson, J. T. et al. Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 (2011).
WashU Epigenome Browser. http://epigenomegateway.wustl.edu/browser/
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
genomecov. bedtools 2.30.0 documentation https://bedtools.readthedocs.io/en/latest/content/tools/genomecov.html
bamCoverage. deepTools 3.5.0 documentation https://deeptools.readthedocs.io/en/develop/content/tools/bamCoverage.html
Khan, A. et al. JASPAR 2018: update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res. 46, D260–D266 (2018).
Weirauch, M. T. et al. Determination and inference of eukaryotic transcription factor sequence specificity. Cell 158, 1431–1443 (2014).
Kheradpour, P. & Kellis, M. Systematic discovery and characterization of regulatory motifs in ENCODE TF binding experiments. Nucleic Acids Res. 42, 2976–2987 (2014).
Tan, G. & Lenhard, B. TFBSTools: an R/bioconductor package for transcription factor binding site analysis. Bioinformatics 32, 1555–1556 (2016).
Fast Motif Matching in R. motifmatchr. https://greenleaflab.github.io/motifmatchr/index.html
Bailey, T. L. et al. MEME Suite: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208 (2009).
Schep, A. N., Wu, B., Buenrostro, J. D. & Greenleaf, W. J. chromVAR: inferring transcription-factor-associated accessibility from single-cell epigenomic data. Nat. Methods 14, 975–978 (2017).
Berest, I. et al. Quantification of differential transcription factor activity and multiomics-based classification into activators and repressors: diffTF. Cell Rep. 29, 3147–3159 (2019).
Hesselberth, J. R. et al. Global mapping of protein–DNA interactions in vivo by digital genomic footprinting. Nat. Methods 6, 283–289 (2009).
Buenrostro, J. D. et al. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 523, 486–490 (2015).
Martins, A. L., Walavalkar, N. M., Anderson, W. D., Zang, C. & Guertin, M. J. Universal correction of enzymatic sequence bias reveals molecular signatures of protein/DNA interactions. Nucleic Acids Res. 46, e9 (2018).
Baek, S., Goldstein, I. & Hager, G. L. Bivariate genomic footprinting detects changes in transcription factor activity. Cell Rep. 19, 1710–1722 (2017).
Bentsen, M. et al. ATAC-seq footprinting unravels kinetics of transcription factor binding during zygotic genome activation. Nat. Commun. 11, 4267 (2020).
Singh, A. K. & Mueller-Planitz, F. Nucleosome positioning and spacing: from mechanism to function. J. Mol. Biol. 433, 166847 (2021).
Lai, W. K. M. & Pugh, B. F. Understanding nucleosome dynamics and their links to gene expression and DNA replication. Nat. Rev. Mol. Cell Biol. 18, 548–562 (2017).
Belton, J.-M. et al. Hi-C: a comprehensive technique to capture the conformation of genomes. Methods 58, 268–276 (2012).
Mumbach, M. R. et al. HiChIP: efficient and sensitive analysis of protein-directed genome architecture. Nat. Methods 13, 919–922 (2016).
Kumasaka, N., Knights, A. J. & Gaffney, D. J. Fine-mapping cellular QTLs with RASQUAL and ATAC-seq. Nat. Genet. 48, 206–213 (2016).
Gate, R. E. et al. Genetic determinants of co-accessible chromatin regions in activated T cells across humans. Nat. Genet. 50, 1140–1150 (2018).
Örd, T. et al. Single-cell epigenomics and functional fine-mapping of atherosclerosis GWAS loci. Circ. Res. 129, 240–258 (2021).
Kia, A. et al. Improved genome sequencing using an engineered transposase. BMC Biotechnol. 17, 6 (2017).
Green, B., Bouchier, C., Fairhead, C., Craig, N. L. & Cormack, B. P. Insertion site preference of Mu, Tn5, and Tn7 transposons. Mob. DNA 3, 3 (2012).
Ason, B. & Reznikoff, W. S. DNA sequence bias during Tn5 transposition. J. Mol. Biol. 335, 1213–1225 (2004).
Lazarovici, A. et al. Probing DNA shape and methylation state on a genomic scale with DNase I. Proc. Natl Acad. Sci. USA 110, 6376–6381 (2013).
Dingwall, C., Lomonossoff, G. P. & Laskey, R. A. High sequence specificity of micrococcal nuclease. Nucleic Acids Res. 9, 2659–2673 (1981).
Hörz, W. & Altenburger, W. Sequence specific cleavage of DNA by micrococcal nuclease. Nucleic Acids Res. 9, 2643–2658 (1981).
Wu, S. J. et al. Single-cell CUT&Tag analysis of chromatin modifications in differentiation and tumor progression. Nat. Biotechnol. 39, 819–824 (2021).
Patty, B. J. & Hainer, S. J. Transcription factor chromatin profiling genome-wide using uliCUT&RUN in single cells and individual blastocysts. Nat. Protoc. 16, 2633–2666 (2021).
Carter, B. et al. Mapping histone modifications in low cell number and single cells using antibody-guided chromatin tagmentation (ACT-seq). Nat. Commun. 10, 3747 (2019).
Chang, P., Gohain, M., Yen, M.-R. & Chen, P.-Y. Computational methods for assessing chromatin hierarchy. Comput. Struct. Biotechnol. J. 16, 43–53 (2018).
Mo, A. et al. Epigenomic signatures of neuronal diversity in the mammalian brain. Neuron 86, 1369–1384 (2015).
Yang, A. C. et al. Dysregulation of brain and choroid plexus cell types in severe COVID-19. Nature 595, 565–571 (2021).
Drokhlyansky, E. et al. The human and mouse enteric nervous system at single-cell resolution. Cell. 182, 1606–1622 (2020).
Deal, R. B. & Henikoff, S. A simple method for gene expression and chromatin profiling of individual cell types within a tissue. Dev. Cell 18, 1030–1040 (2010).
Bhattacharyya, S., Sathe, A. A., Bhakta, M., Xing, C. & Munshi, N. V. PAN-INTACT enables direct isolation of lineage-specific nuclei from fibrous tissues. PLoS ONE 14, e0214677 (2019).
Nuclei Isolation for Single Cell ATAC Sequencing. https://support.10xgenomics.com/single-cell-atac/sample-prep/doc/demonstrated-protocol-nuclei-isolation-for-single-cell-atac-sequencing
Lawler, A. J. et al. Cell type-specific oxidative stress genomic signatures in the globus pallidus of dopamine-depleted mice. J. Neurosci. 40, 9772–9783 (2020).
Kiseleva, E. et al. A protocol for isolation and visualization of yeast nuclei by scanning electron microscopy (SEM). Nat. Protoc. 2, 1943–1953 (2007).
Niepel, M., Farr, J. C., Rout, M. P. & Strambio-De-Castillia, C. Rapid isolation of functionally intact nuclei from the yeast Saccharomyces. Preprint at bioRxiv https://doi.org/10.1101/162388 (2017).
Nott, A., Schlachetzki, J. C. M., Fixsen, B. R. & Glass, C. K. Nuclei isolation of multiple brain cell types for omics interrogation. Nat. Protoc. 16, 1629–1646 (2021).
Fullard, J. F. et al. An atlas of chromatin accessibility in the adult human brain. Genome Res. 28, 1243–1252 (2018).
Hauberg, M. E. et al. Common schizophrenia risk variants are enriched in open chromatin regions of human glutamatergic neurons. Nat. Commun. 11, 5581 (2020).
Haines, J. ATAC-seq on nuclei from frozen, sliced, Drosophila melanogaster embryo halves. https://doi.org/10.17504/protocols.io.kj5cuq6 (2017).
Steiner, F. A., Talbert, P. B., Kasinathan, S., Deal, R. B. & Henikoff, S. Cell-type-specific nuclei purification from whole animals for genome-wide expression and chromatin profiling. Genome Res. 22, 766–777 (2012).
Han, M., Wei, G., McManus, C. E., Hillier, L. W. & Reinke, V. Isolated C. elegans germ nuclei exhibit distinct genomic profiles of histone modification and gene expression. BMC Genomics 20, 500 (2019).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, 10–12 (2011).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Jiang, H., Lei, R., Ding, S.-W. & Zhu, S. Skewer: a fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinformatics 15, 182 (2014).
Krueger, F. Trim Galore. (2021).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
broadinstitute/picard (Broad Institute, 2021) https://github.com/broadinstitute/picard
Faust, G. G. & Hall, I. M. SAMBLASTER: fast duplicate marking and structural variant read extraction. Bioinformatics 30, 2503–2505 (2014).
Boyle, A. P., Guinney, J., Crawford, G. E. & Furey, T. S. F-Seq: a feature density estimator for high-throughput sequence tags. Bioinformatics 24, 2537–2538 (2008).
Granja, J. M. et al. ArchR is a scalable software package for integrative single-cell chromatin accessibility analysis. Nat. Genet. 53, 403–411 (2021).
Buenrostro, J. D., Wu, B., Chang, H. Y. & Greenleaf, W. J. ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol. 109, 21.29.1–21.29.9 (2015).
Acknowledgements
This work was supported by NIH R00-AG059918, U01-AG072573, P01-AG073082, UM1-HG012076 and a gift from the Ray and Dagmar Dolby Family Fund (to the Gladstone Institutes). F.C.G. is an Alan Kaganov Scholar. M.R.C. is additionally supported by the Farmer Family Foundation Parkinson’s Research Initiative and an American Society of Hematology Scholar Award.
Author information
Authors and Affiliations
Contributions
All authors contributed to developing this protocol. All experiments were performed by H.M., F.C.G. and L.K. with supervision from M.R.C. The manuscript was written by F.C.G., L.K. and H.M. with input from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Protocols thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Corces, M. R. et al. Nat. Methods 14, 959–962 (2017): https://doi.org/10.1038/nmeth.4396
Corces, M. R. et al. Science 362, 6413 (2018): https://doi.org/10.1126/science.aav1898
Corces, M. R. et al. Nat. Genet. 52, 1158–1168 (2020): https://doi.org/10.1038/s41588-020-00721-x
Supplementary information
Supplementary Information
Supplementary Figs. 1–6, Supplementary Notes 1–4, Supplementary Protocol 1 (Nuclei Isolation), Supplementary Methods and Supplementary References.
Supplementary Tables 1 and 2
Alignment statistics for different ATAC-seq library read lengths. ATAC-seq dual-indexing adapters and barcode sequences.
Rights and permissions
About this article
Cite this article
Grandi, F.C., Modi, H., Kampman, L. et al. Chromatin accessibility profiling by ATAC-seq. Nat Protoc 17, 1518–1552 (2022). https://doi.org/10.1038/s41596-022-00692-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-022-00692-9
This article is cited by
-
Cryopreservation of cerebrospinal fluid cells preserves the transcriptional landscape for single-cell analysis
Journal of Neuroinflammation (2024)
-
Inference of genomic landscapes using ordered Hidden Markov Models with emission densities (oHMMed)
BMC Bioinformatics (2024)
-
MCT1-governed pyruvate metabolism is essential for antibody class-switch recombination through H3K27 acetylation
Nature Communications (2024)
-
Nano-CUT&Tag for multimodal chromatin profiling at single-cell resolution
Nature Protocols (2024)
-
Genetic variation across and within individuals
Nature Reviews Genetics (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.