Abstract
Simultaneous [18 F]-fluorodeoxyglucose positron emission tomography and functional magnetic resonance imaging (FDG-PET/fMRI) provides the capability to image two sources of energetic dynamics in the brain – cerebral glucose uptake and the cerebrovascular haemodynamic response. Resting-state fMRI connectivity has been enormously useful for characterising interactions between distributed brain regions in humans. Metabolic connectivity has recently emerged as a complementary measure to investigate brain network dynamics. Functional PET (fPET) is a new approach for measuring FDG uptake with high temporal resolution and has recently shown promise for assessing the dynamics of neural metabolism. Simultaneous fMRI/fPET is a relatively new hybrid imaging modality, with only a few biomedical imaging research facilities able to acquire FDG PET and BOLD fMRI data simultaneously. We present data for n = 27 healthy young adults (18–20 yrs) who underwent a 95-min simultaneous fMRI/fPET scan while resting with their eyes open. This dataset provides significant re-use value to understand the neural dynamics of glucose metabolism and the haemodynamic response, the synchrony, and interaction between these measures, and the development of new single- and multi-modality image preparation and analysis procedures.
Measurement(s) | glucose import • haemodynamic response • functional brain measurement |
Technology Type(s) | FDG-Positron Emission Tomography • Blood Oxygen Level-Dependent Functional MRI |
Factor Type(s) | resting human brain |
Sample Characteristic - Organism | Homo sapiens |
Machine-accessible metadata file describing the reported data: https://doi.org/10.6084/m9.figshare.12994508
Similar content being viewed by others
Background & Summary
Simultaneous magnetic resonance imaging - positron emission tomography (MRI-PET) represents a significant development in human imaging neuroscience. Simultaneous BOLD-fMRI/FDG-PET (blood oxygen level-dependent functional magnetic resonance imaging/[18 F]-fluorodeoxyglucose positron emission tomography) enables the simultaneous measurement of the two of the most widely used in vivo markers of neuronal activity. The simultaneous nature of the acquisition is of particular importance, as it enables glucose uptake and haemodynamic responses to the same neuronal activity to be captured, without the confounds of intra-individual differences in attention, fatigue, motivation, nutrient intake and blood chemistry that occur in non-contemporaneous acquisitions1.
BOLD-fMRI provides a haemodynamic-based surrogate of neuronal activity with a temporal resolution of the order of seconds and spatial resolution of around a millimetre. The BOLD signal is comprised of both neuronal and non-neuronal components. The neuronal component of the BOLD signal is believed to arise from local field potentials of peri-synaptic activity2,3, whereas the non-neuronal component appears from cerebrovascular sources, including cerebral blood flow, volume, and the metabolic rate of oxygen. The BOLD signal, therefore, is confounded by intra- and inter-individual differences in heart rate variability, respiration, haemoglobin concentration and the oxygen-carrying capacity of the blood4,5,6,7,8.
BOLD-fMRI is a semi-quantitative index of neuronal function that cannot be compared across brain regions, subjects, or imaging sites3. By comparison, FDG-PET is a fully quantitative index of neuronal activity that captures cerebral glucose uptake that is primarily localised to the synapses9,10,11. FDG-PET has a spatial resolution of around 4 mm12 and, until recently, a temporal resolution that was effectively equal to the scan duration – around 10–40 minutes. However, recent developments in radiotracer delivery13,14 have resulted in substantial improvements in FDG-PET temporal resolution, reducing it down to 60 sec14,15,16,17,18 or less [12 sec19; 16 sec13,20; 30 sec21]. This method, known as ‘functional’ PET (fPET) administers the radiotracer throughout the scan, either as a constant infusion14 or hybrid bolus/infusion19. Detailed protocols for continuous infusion and hybrid bolus/infusion radiotracer administration are provided in13.
Functional connectivity measures the temporal coherence of neural signals across distributed regions of the brain. BOLD-fMRI has been incredibly useful for characterising the integrative activity of brain networks distributed across the brain22,23. Canonical resting-state networks include the default mode, dorsal attention, salience networks, and others23. This method is commonly labelled resting-state functional connectivity, but here we use the term haemodynamic connectivity, as functional connectivity can be measured by any functional neuroimaging method, including EEG24, PET25, fNIRS26, etc. Resting-state connectivity measured using static FDG-PET predates resting-state fMRI by at least a decade25. However, due to the limited temporal resolution of traditional FDG-PET, resting-state ‘connectivity’ was estimated as the covariance of FDG-PET signals across-subjects, rather than the temporal coherence of brain signals within a subject, as is the case in haemodynamic connectivity. Therefore, we use the term ‘metabolic covariance’ to refer to region-to-region, static, across-subject FDG-PET correlation from now on. With the development of fPET methodology, it is now possible to estimate the intra-subject dynamics of glucose uptake during the resting-state – that is, ‘metabolic connectivity’. Using this data, we20 have shown that resting-state fPET metabolic connectivity is similar to BOLD-fMRI haemodynamic connectivity in the frontoparietal cortex and dissimilar in other regions of the brain (subcortical, temporo-occipito regions). Furthermore, fPET metabolic connectivity was dissimilar to static FDG-PET metabolic covariance across the brain. This work forms the basis of a new field for human imaging neuroscience – the characterisation and exploration of anatomically distributed networks of coupled metabolic dynamics.
Here, we describe the Monash rsPET-MR dataset27: a simultaneous fMRI-fPET dataset acquired from young, healthy individuals at rest (Fig. 1). We make this data publicly available as few biomedical imaging facilities currently have the requisite technology to acquire similar data. The Monash rsPET-MR dataset has re-use value to explore the dynamics of glucose metabolic and haemodynamic signals across distributed regions of the brain. The dataset also has re-use value to support the development of new imaging processing and analysis strategies for simultaneous fMRI/fPET. Simultaneous MRI-PET, fMRI-fPET, and the fPET methodology are nascent technologies with immature processing pipelines. As new methods, these fields do not benefit from many years of work validating data preparation and signal detection optimisation, particularly in comparison to the fMRI field. Examples of important validation work yet to be performed includes test-retest reliability (which can be difficult in humans for ethical reasons due to biosafety constraints related to radioactivity exposure), and whether the FDG in a continuous infusion protocol acts as a radiotracer or is more analogous to a radioligand28. Examples of re-use of the Monash rsfPET-MR dataset may include development of data analysis techniques18, synergistic data fusion techniques29, discoveries about the relationship between glucose uptake and the haemodynamic response17, as well as revelations about the fundamental basis of energy use in the human brain21,30,31,32.
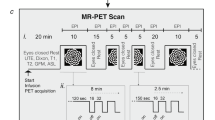
Paradigm & workflow. Panels a–d indicate the workflow for the data available in the Monash rsPET-MR dataset27 and panels e-g indicate the workflow for the results presented under Technical Validation, and in Jamadar et al.20. (a) Participants completed a demographics, safety screening and cognitive assessment an hour prior to MR-PET scanning. (b) Next, participants were prepared for scanning; a cannula was placed in the forearm vein of each arm, and then haemoglobin and blood sugar level was taken. (c) Participants then underwent a 95-minute MR-PET scan using the paradigm shown here. (d) Illustration of the data obtained for each method (left to right): Structural T1 MRI anatomical images for each subject (subj); static PET (sPET) acquired a single image per subject; functional PET (fPET) was binned into 16 sec images, resulting in a timeseries of images for each subject; fMR images were obtained with TR 2.45 sec, resulting in a timeseries for each subject. (e) Structural T1 MRI was registered to MNI space and then segmented into 82 regions of interest (ROI). This parcellation was applied to the sPET, fPET and fMRI images. (f) Illustration of example processing steps for each modality, as used in Jamadar et al. (2020). sPET was demeaned, parcellated into 82 regions (ROIs) and then correlated across subjects. Scatterplot shows an example correlation between two regions across subjects, out of a total of 82 × 82 region-wise correlations. fPET was motion corrected (moco), filtered, parcellated, and then correlated across time-series for each subject. Illustration shows 2 example timeseries of the total 82 × 82 region-by-region correlations conducted. fMRI was preprocessed, parcellated, correlated across time for each subject, then group-averaged. (g) Subject-level matrices for sPET, fPET and fMRI were then group averaged. Matrices are indicative of potential connectivity matrices, and are those that are reported in Jamadar et al.20.
Methods
All methods were reviewed by the Monash University Human Research Ethics Committee, following the Australian National Statement of Ethical Conduct in Human Research (2007). Subjects provided informed consent to participate in the study. Administration of ionising radiation was approved by the Monash Health Principal Medical Physicist, following the Australian Radiation Protection and Nuclear Safety Agency Code of Practice (2005). For participants aged over 18-yrs, the annual radiation exposure limit of 5 mSv applies, and the effective dose in this study was 4.8 mSv.
A video methods article describing the constant infusion acquisition procedure is reported in13. A comparison of static PET, fPET, and fMRI connectivity using this data is reported in20. The dataset reported here includes demographic information, anthropometry, and minimally-processed simultaneously acquired MRI and PET images acquired during the course of a more extensive study.
Participants
Participants (n = 27) were aged 18–23 years (mean 19 years), 21 female, all right-handed. Participants had between 13–18 years of education (mean 14 years), normal or corrected-to-normal vision, and no personal history of diagnosed Axis-1 mental illness, diabetes, or cardiovascular illness. Participants were screened for claustrophobia, non-MRI compatible implants, clinical, or research PET scan in the previous 12 months. Women were screened for current or suspected pregnancy.
Procedure
Figure 1 presents the acquisition and analysis workflow for the study.
Before the scan, participants were directed to consume a high protein/low sugar diet for 24hrs, fast for 6hrs, and drink 2–6 glasses of water. Participants completed a demographic assessment (10-min) and a brief cognitive battery (30-mins, data not reported here).
Participants were cannulated in a vein in each forearm with a minimum size 22-gauge cannula, and a 10 mL baseline blood sample was taken at the time of cannulation. For all participants, the left cannula was used for FDG infusion, and the right cannula was used for blood sampling. Primed extension tubing was connected to the right cannula (for blood sampling) via a three-way tap.
Participants underwent a 95-minute simultaneous MR-PET scan in a Siemens (Erlangen) 3 Tesla Biograph molecular MR (mMR) scanner (Syngo VB20 P). Quality control is performed daily on the scanner using a 68-Ge (germanium) phantom. Cross-calibration is also conducted between the scanner, dose calibrator and well counter on delivery of the radiotracer dose using an 18-F and water phantom.
Participants were positioned supine in the scanner bore with their head in a 16-channel radiofrequency (RF) head coil, and were instructed to lie as still as possible with eyes open, and think of nothing in particular. [18-F] fluorodeoxyglucose (FDG; average dose 233MBq) was infused over the course of the scan at a rate of 36 mL/hr using a BodyGuard 323 MR-compatible infusion pump (Caesarea Medical Electronics, Caesarea, Israel). One participant received a lower dose (167MBq) due to an infusion pump error. Infusion onset was locked to the start of the PET scan.
Plasma radioactivity levels were measured throughout the duration of the scan. At 10-mins post-infusion onset, a 10 mL blood sample was taken from the right forearm using a vacutainer; the time of the 5 mL mark was noted for subsequent decay correction. Subsequent blood samples were taken at 10-min intervals for a total of 10 samples for the duration of the scan. The cannula line was flushed with 10 mL of saline after every sample to minimise line clotting. Immediately following blood sampling, the sample was placed in a Heraeus Megafuge 16 centrifuge (ThermoFisher Scientific, Osterode, Germany) and spun at 2000rpm for 5 mins; 1000 μL plasma was pipetted, transferred to a counting tube and placed in a well counter for 4 mins. The count start time, the total number of counts, and the counts per minute were recorded for each sample.
Scanning protocol
PET data were acquired in list mode. Infusion of the FDG radiotracer and PET data acquisition started with the Ultrashort TE (UTE) MRI for PET attenuation correction. While the PET signal rose to detectable levels over the first 30-mins following infusion onset, non-functional MRI scans were acquired. These scans included27: T1 3D MPRAGE (TA = 7.01 mins, TR = 1640ms, TE = 2.34 ms, flip angle = 8°, FOV = 256 × 256mm2, voxel size = 1 × 1 × 1mm3, 176 slices; sagittal acquisition), gradient field map (TA = 1.02 min); and several scans not reported here: UTE (TA = 1.40 mins), T2 SPACE (TA = 5.52 min), magnetic resonance spectroscopy (MRS, TA = 2.48 min), pulsed arterial spin labelling (TA = 4.21), T2 susceptibility-weighted image (TA = 6.50 min), and left-right phase correction (TA = 0.21 min). For the remainder of the scan, six consecutive 10 min blocks of T2*-weighted echo-planar images (EPIs) were acquired (TR = 2450 ms, TE = 30 ms, FOV = 190 mm, 3 × 3 × 3mm 3 voxels, 44 slices, ascending axial acquisition).
Data Records
Table 1 defines the demographic and imaging data available for each subject on OpenNeuro. The dataset containing the demographic, fMRI, PET, T1 structural and gradient field maps is freely available in BIDS format33 from the OpenNeuro repository (http://openneuro.org) with the accession number ds00289827.
Participants.tsv is a text file reporting the demographic data for each subject ordered by subject ID.
T1 structural and fMRI data along with supporting gradient field maps and face mask were organised in sub-directories according to the Brain Imaging Data Structure (BIDS)33. Following the BIDS structure, the sub-*/anat directory contains the T1 MPRAGE data. T1 structural data were converted from DICOM to NIfTI format with JSON sidecars using the Dcm2niix converter34. Facial features were removed from the T1 structural images by applying a co-registered binary face mask35,36. The sub-*/fmap directory contains the gradient field map data: short-TE, long-TE, and phase difference images. The sub-*/func directory contains the T2* BOLD-fMRI data. fMRI data were converted from DICOM to NIfTI format with JSON sidecars using the Dcm2niix converter34.
The sub-*/pet directory contains the reconstructed PET data. The 5700-second list-mode PET data for each subject were binned into 356 3D sinogram frames each at 16-second intervals. PET data were converted from DICOM to NIfTI format with JSON sidecars using the Dcm2niix converter34. To remove facial features from the PET data, a binary face mask was co-registered to static PET images created by summing the PET activity over the acquisition and applying to the dynamic PET data35,36. JSON sidecars were augmented with metadata in accordance with guidelines for the content and format of PET brain data37 (Knudsen et al.37) and the current working copy of the proposed BIDS Extension for PET v0.0.1 (BEP009).
PET motion-correction parameters estimated for each subject are included in accordance with the recommendations for derivatives in BIDS datasets. Space-delimited realignment parameters [6 (pitch, roll, yaw, x, y, z)] for each frame (225) are stored on separate rows of a text file at derivatives/mcflirt/sub-*/pet/sub-*_task_rest_pet.moco.par.
Technical Validation
The Monash rsPET-MR dataset is comprised of simultaneously acquired BOLD-fMRI and FDG-fPET data acquired during the resting-state27. Since fPET is the primary novel outcome from this dataset, this section focuses on the technical validation of the fPET data.
To validate the data, the following processing pipeline was used (Fig. 1d–f). For the T1 structural image, the brain was extracted, then registered to MNI152 space using Advanced Normalization Tools (ANTs)38. The grey matter, white matter, and brain cortex labels of T1 images were segmented using Freesurfer with the Desikan-Killiany Atlas (Diedrichsen et al. 2009).
For the fMRI data, the six blocks of EPI scans underwent brain extraction (FSL BET, Smith, 2002), N4 bias field correction (ANTs, Tustison et al., 2010), motion correction (FSL MCFLIRT, Jenkinson et al.36), slice time correction (AFNI, Cox, 1996), and high pass filtering (>0.01 Hz) to remove low frequency noise (FSL, Jenkinson et al.). Subject motion was assessed using realignment parameters from MCFLIRT.
For the PET data, the pseudoCT method39 was used to correct the attenuation for all acquired data. The Ordinary Poisson-Ordered Subset Expectation Maximization (OP-OSEM) algorithm (3 iterations, 21 subsets) with point spread function correction was used to reconstruct 3D volumes from the sinogram frames. The reconstructed DICOM slices were converted to the NIFTI format with size 344x344x127 (voxel size: 2.09 × 2.09 × 2.03 mm3) for each volume. A 5-mm FWHM Gaussian post-filter was applied to each 3D volume. All 3D volumes were temporally concatenated to form a 4D (344 × 344 x 127 × 356) NIFTI volume. A guided motion correction method using simultaneously acquired MRI images was applied to correct motion during the PET scan. We retained the 225 16-sec volumes commencing from the 30-minute time point, which matched the start of the BOLD-fMRI EPI acquisition, for further analyses. A single static PET image was derived from the sum of the 16-sec volumes. The 225 PET volumes were motion-corrected (FSL MCFLIRT); the mean PET image was brain extracted and used to mask the 4D data. The fPET data was further processed using a spatio-temporal gradient filter to estimate the short-term change in glucose uptake from the cumulative glucose uptake that was measured.
Motion parameters
For the PET data, translational motion parameters for each subject were summarised by the mean distance for each direction (x, y, z) along timepoints (225 frames) for each subject, and are shown in Fig. 2. Across subjects, average mean framewise translational motion was 0.41 mm; maximum was 1.09 mm.
Plasma radioactivity curves
Interpolated plasma radioactivity curves are shown in Fig. 3. Consistent with the known dynamics of the constant infusion approach14, plasma radioactivity increased throughout the scan, reaching a peak just prior to the end of the scan (90 mins post-infusion). After the cessation of the infusion (i.e., the last measurement point in Fig. 3), the available radioactivity declines.
Stability of the fPET connectome over the scan period
The fPET technique relies upon constant infusion radiotracer administration. The influence the administration procedure has on the obtained images is a matter of current investigation e.g.,13,19. Consequently, here we report the stability of the fPET connectome over six consecutive 10-min periods (Fig. 4), corresponding with the 6 BOLD EPI blocks (Fig. 1). As can be seen from Fig. 4, the fPET connectivity structure, characterised by primarily fronto-parietal connectivity, becomes evident around the time of the peak of plasma radioactivity (i.e., block 5). This suggests that the earlier time points contribute less to the final obtained fPET connectome structure.
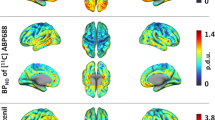
(a) Group-average fPET connectivity matrix, as reported in Jamadar et al., 2020. (b) Stability of fPET connectivity matrix over the six experimental blocks. Refer to Fig. 1 for the experimental design.
Effects of spatio-temporal filtering on fPET connectivity
The spatiotemporal filter was defined as the convolution of a 3-dimensional Gaussian filter in the spatial domain and a 1-dimensional Gaussian filter in the time domain, with the standard deviation being one voxel for the spatial Gaussian voxel and two frames for the temporal Gaussian (Fig. 5). The spatiotemporal convolution was further modified to give negative weights on prior frames (negative time) and zero weights for the current frame.
We determined the consistency of metabolic connectivity across a range of both temporal and spatial standard deviations, up to three frames and three voxels, respectively. Figure 6 shows the variability of fPET connectivity over a range of spatial and temporal widths. We found highly consistent metabolic connectivity across a range of standard deviations. We have previously reported resting-state fPET connectivity with one voxel spatial gaussian and two frames temporal gaussian20. This set of parameters provided the least amount of smoothing whilst preserving the fidelity of the observed connectivity. The main trends we observed as we explored parameter values were an increase in correlation strength with higher spatial standard deviation and an increase in mean connectivity with higher temporal standard deviation.
The spatial filter is intended to de-noise the volumes by reducing non-spatially correlated noise whilst averaging spatially correlated signals. At higher standard deviation values, the filter may begin averaging signals across ROIs. We truncated the spatio-temporal filter to a 7 × 7 × 7 × 7 voxel spatio-temporal window to avoid an artificial correlation between spatially adjacent ROIs. In the temporal domain, high values correspond to estimates of the gradient at a longer timescale. In the extreme, this approximates static connectivity as the timescale approaches that of the entire session, i.e., change in activity between the start and end of the experiment. It is possible that the global increase in connectivity, shown as a yellow-hue in the lower frames of Fig. 6, are beginning to be contaminated by these longer time-period effects. Further explorations of parameter values should be performed to assess the dependence of network correlations as a function of the spatial filter kernel, and mean connectivity as a function of temporal filtering.
Spatial variability of fMRI and fPET images
fMRI and fPET connectomes showed a low region degree (i.e., little correlation with other brain regions) in subcortical regions20. These observations contrast with the known high degree of inter-connectedness of the subcortical areas with the rest of the brain: the cortico-basal ganglia-cerebellar (Middleton and Strick40; Bostan and Strick)41 and cortico-thalamic circuits42. These results are likely attributable to the reduced sensitivity of signal detection in midbrain areas for both PET and fMRI. BOLD-fMRI echo-planar images show systematic image artefacts in midbrain areas43 and susceptibility artefacts at air-tissue interfaces44. These effects are evident in the variability maps of raw echo-planar images acquired in this study (Fig. 7). The PET images have lower signal detection in the midbrain, due to the higher attenuation of gamma rays emitted from deep brain regions. The attenuation correction corrects the signal loss but cannot fix the lower signal to noise ratio in the midbrain region of the images. We observed systematically lower signal to noise ratio and high signal variability in brain regions near the ventricles (Fig. 7). The low inter-connectedness of the FDG-fPET and BOLD-fMRI measures in the subcortical areas may be attributable to the lower neuronal signals detected in these areas.
Raw images for one individual subject showing signal intensity variation across the brain. Top panel shows mean image across a 10-minute run; middle panel shows standard deviation across the same run; and lower panel shows the coefficient of variation (standard deviation divided by the mean) across the run.
Usage Notes
Effective sharing of research products means that data, processing methods, workflows, and tools are made available, so that they can be reused and that published findings can be reproduced. Equally important is that data should be published with integration and reuse in mind, especially when using artificial intelligence and machine learning approaches. The data can be interpreted in new ways and new knowledge attained45. The FAIR principles46 have been developed to ensure that the scientific research results are findable, accessible, interoperable and reusable, for both human analysis and machine analysis. Brain imaging research is in the vanguard of neuroscience research disciplines advocating open science and compliance with the FAIR principles. Neuroimaging data can make an important contribution to building a multiscale, comprehensive, and dynamic understanding of the structure and function of the nervous system.
The primary motivation behind making the Monash rsPET-MR data publicly available is to facilitate research aiming to understand the relationship between glucose metabolism and haemodynamic signals arising from neural activity in the human brain. In particular, the data provides a multitude of opportunities to investigate the dynamic nature of metabolic and haemodynamic measures of resting-state brain activity. To our knowledge, the availability of simultaneously acquired fMR and fPET data is rare. There are only a small number of biomedical imaging research facilities that have the requisite imaging technology and expertise to obtain comparable data.
The Monash rsPET-MR dataset has significant re-use value. One example includes quantitative exploration of network parameters and their influence on the obtained metabolic connectome. Our initial analysis of stationary fPET metabolic connectivity20 used an established method for the estimation of functional connectivity in the BOLD-fMRI literature47,48. However, dynamic estimates of brain connectivity take into account the temporal fluctuations of functional connectivity, since the temporal ordering of a time series of functional brain images is important49. Further analyses could investigate dynamic connectivity approaches, including sliding-time window approaches and models of switching between microstates48. The fPET images were reconstructed with 16-sec acquisition duration. While other groups have reported results using an acquisition duration of 12-sec19, solutions are needed for the methodological and analytical challenges in order to use FDG-fPET as a robust index of dynamic metabolic connectivity with the reduced signal-to-noise associated with shorter frames.
The Monash rsPET-MR dataset could also be re-used to develop new image processing and analysis strategies for simultaneous fMRI/fPET data. As a relatively new imaging method, tools for validating fPET data, including pre-processing and signal detection optimisation, are unavailable publicly. Several research groups have developed bespoke data analysis environments and customised analytical tools which provide a starting point for further tools development. An important technical limitation is that processing pipelines for FDG-fPET data are immature compared to the pre-processing procedures for BOLD-fMRI data. The dataset provides opportunities for further work on validating acquisition parameters, data preparation, and signal detection optimisation, in addition to the recent techniques associated with radiotracer administration e.g.,13,19, attenuation correction50, motion correction51 and data analysis18,29.
Code availability
Scripts used to insert required metadata into the published BIDS dataset are freely available at https://github.com/MonashBI/Monash_rsPET-MR_prep.
References
Chen, Z. et al. From simultaneous to synergistic MR‐PET brain imaging: A review of hybrid MR‐PET imaging methodologies. Hum. Brain Mapp. 39, 5126–5144 (2018).
Ekstrom, A. How and when the fMRI BOLD signal relates to underlying neural activity: The danger in dissociation. Brain Res. Rev. 62, 233–244 (2010).
Logothetis, N. K. What we can do and what we cannot do with fMRI. Nature 453, 869–878 (2008).
Birn, R. M. The role of physiological noise in resting-state functional connectivity. NeuroImage 62, 864–870 (2012).
Chang, C., Cunningham, J. P. & Glover, G. H. Influence of heart rate on the BOLD signal: The cardiac response function. NeuroImage 44, 857–869 (2009).
Glover, G. H., Li, T. Q. & Ress, D. Image-based method for retrospective correction of physiological motion effects in fMRI: RETROICOR. Magn. Reson. Med. 44, 162–167 (2000).
Power, J. D., Plitt, M., Laumann, T. O. & Martin, A. Sources and implications of whole-brain fMRI signals in humans. NeuroImage 146, 609–625 (2017).
Ward, P. G. D. et al. Individual differences in haemoglobin concentration influence BOLD fMRI functional connectivity and its correlation with cognition. https://doi.org/10.1101/835660 (2019).
Magistretti, P. J. & Allaman, I. A Cellular Perspective on Brain Energy Metabolism and Functional Imaging. Neuron 86, 883–901 (2015).
Sokoloff, L. The Deoxyglucose Method for The Measurement of Local Glucose Utilization and The Mapping of Local Functional Activity in The Central Nervous System. In International Review of Neurobiology vol. 22 287–333 (Elsevier, 1981).
Sokoloff, L. et al. The [14 C]Deoxyglucose Method For The Measurement Of Local Cerebral Glucose Utilization: Theory, Procedure, And Normal Values In The Conscious And Anesthetized Albino Rat. J. Neurochem. 28, 897–916 (1977).
Delso, G. et al. Performance Measurements of the Siemens mMR Integrated Whole-Body PET/MR Scanner. J. Nucl. Med. 52, 1914–1922 (2011).
Jamadar, S. D. et al. Radiotracer Administration for High Temporal Resolution Positron Emission Tomography of the Human Brain: Application to FDG-fPET. J. Vis. Exp. 60259, https://doi.org/10.3791/60259 (2019).
Villien, M. et al. Dynamic Functional Imaging of Brain Glucose Utilization using fPET-FDG. NeuroImage 100, 192–199 (2014).
Hahn, A. et al. Task-relevant brain networks identified with simultaneous PET/MR imaging of metabolism and connectivity. Brain Struct. Funct. 223, 1369–1378 (2018).
Hahn, A. et al. Quantification of Task-Specific Glucose Metabolism with Constant Infusion of 18F-FDG. J. Nucl. Med. 57, 1933–1940 (2016).
Jamadar, S. D. et al. Simultaneous task-based BOLD-fMRI and [18-F] FDG functional PET for measurement of neuronal metabolism in the human visual cortex. NeuroImage 189, 258–266 (2019).
Li, S. et al. Analysis of continuous infusion functional PET (fPET) in the human brain. https://doi.org/10.1101/778357. (2019).
Rischka, L. et al. Reduced task durations in functional PET imaging with [18F]FDG approaching that of functional MRI. NeuroImage 181, 323–330 (2018).
Jamadar, S. D. et al. Metabolic and haemodynamic resting-state connectivity of the human brain: a high-temporal resolution simultaneous BOLD-fMRI and FDG-fPET multimodality study. https://doi.org/10.1101/2020.05.01.071662 (2020).
Hahn, A. et al. Reconfiguration of functional brain networks and metabolic cost converge during task performance. eLife 9, e52443 (2020).
Biswal, B., Yetkin, F. Z., Haughton, V. M. & Hyde, J. S. Functional connectivity in the motor cortex of resting human brain using echo-planar mri. Magn. Reson. Med. 34, 537–541 (1995).
Yeo, B. T. et al. The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J. Neurophysiol. 106, 1125–1165 (2011).
Khanna, A., Pascual-Leone, A., Michel, C. M. & Farzan, F. Microstates in resting-state EEG: Current status and future directions. Neurosci. Biobehav. Rev. 49, 105–113 (2015).
Horwitz, B., Duara, R. & Rapoport, S. I. Intercorrelations of Glucose Metabolic Rates between Brain Regions: Application to Healthy Males in a State of Reduced Sensory Input. J. Cereb. Blood Flow Metab. 4, 484–499 (1984).
Lu, C.-M. et al. Use of fNIRS to assess resting state functional connectivity. J. Neurosci. Methods 186, 242–249 (2010).
Jamadar, S. et al. Monash rsPET-MR. OpenNeuro, https://doi.org/10.18112/OPENNEURO.DS002898.V1.1.0 (2020).
Matthews, P. M., Rabiner, E. A., Passchier, J. & Gunn, R. N. Positron emission tomography molecular imaging for drug development: PET for drug development. Br. J. Clin. Pharmacol. 73, 175–186 (2012).
Sudarshan, V. P., Egan, G. F., Chen, Z. & Awate, S. P. Joint PET-MRI image reconstruction using a patch-based joint-dictionary prior. Med. Image Anal. 62, 101669 (2020).
Shiyam Sundar, L. K. et al. Fully Integrated PET/MR Imaging for the Assessment of the Relationship Between Functional Connectivity and Glucose Metabolic Rate. Front. Neurosci. 14, 252 (2020).
Shokri-Kojori, E. et al. Correspondence between cerebral glucose metabolism and BOLD reveals relative power and cost in human brain. Nat. Commun. 10, 690 (2019).
Tomasi, D. G. et al. Dynamic brain glucose metabolism identifies anti-correlated cortical-cerebellar networks at rest. J. Cereb. Blood Flow Metab. 37, 3659–3670 (2017).
Gorgolewski, K. J. et al. The brain imaging data structure, a format for organizing and describing outputs of neuroimaging experiments. Sci. Data 3, 160044 (2016).
Li, X., Morgan, P. S., Ashburner, J., Smith, J. & Rorden, C. The first step for neuroimaging data analysis: DICOM to NIfTI conversion. J. Neurosci. Methods 264, 47–56 (2016).
Gulban, O. F. et al. poldracklab/pydeface: v2.0.0. Zenodo https://doi.org/10.5281/ZENODO.3524401 (2019).
Jenkinson, M., Bannister, P., Brady, M. & Smith, S. Improved Optimization for the Robust and Accurate Linear Registration and Motion Correction of Brain Images. NeuroImage 17, 825–841 (2002).
Knudsen, G. M. et al. Guidelines for the content and format of PET brain data in publications and archives: A consensus paper. J. Cereb. Blood Flow Metab. 0271678X2090543, https://doi.org/10.1177/0271678X20905433 (2020).
Avants, B. B. et al. A reproducible evaluation of ANTs similarity metric performance in brain image registration. NeuroImage 54, 2033–2044 (2011).
Burgos, N. et al. Attenuation Correction Synthesis for Hybrid PET-MR Scanners: Application to Brain Studies. IEEE Trans. Med. Imaging 33, 2332–2341 (2014).
Middleton, F. & Strick, P. Basal ganglia and cerebellar loops: motor and cognitive circuits. Brain Res. Rev. 31, 236–250 (2000).
Bostan, A. C. & Strick, P. L. The Cerebellum and Basal Ganglia are Interconnected. Neuropsychol. Rev. 20, 261–270 (2010).
Parent, A. & Hazrati, L.-N. Functional anatomy of the basal ganglia. I. The cortico-basal ganglia-thalamo-cortical loop. Brain Res. Rev. 20, 91–127 (1995).
Limbrick-Oldfield, E. H. et al. Identification and characterisation of midbrain nuclei using optimised functional magnetic resonance imaging. NeuroImage 59, 1230–1238 (2012).
Bernstein, M. A., Huston, J. & Ward, H. A. Imaging artifacts at 3.0T. J. Magn. Reson. Imaging 24, 735–746 (2006).
Ferguson, A. R., Nielson, J. L., Cragin, M. H., Bandrowski, A. E. & Martone, M. E. Big data from small data: data-sharing in the ‘long tail’ of neuroscience. Nat. Neurosci. 17, 1442–1447 (2014).
Wilkinson, M. D. et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data 3, 160018 (2016).
Liégeois, R., Laumann, T. O., Snyder, A. Z., Zhou, J. & Yeo, B. T. T. Interpreting temporal fluctuations in resting-state functional connectivity MRI. NeuroImage 163, 437–455 (2017).
Preti, M. G., Bolton, T. A. & Van De Ville, D. The dynamic functional connectome: State-of-the-art and perspectives. NeuroImage 160, 41–54 (2017).
Calhoun, V. D., Miller, R., Pearlson, G. & Adalı, T. The Chronnectome: Time-Varying Connectivity Networks as the Next Frontier in fMRI Data Discovery. Neuron 84, 262–274 (2014).
Baran, J. et al. Accurate hybrid template–based and MR-based attenuation correction using UTE images for simultaneous PET/MR brain imaging applications. BMC Med. Imaging 18, 41 (2018).
Chen, Z. et al. MR-PET head motion correction based on co-registration of multicontrast MR images. Hum. Brain Mapp. https://doi.org/10.1002/hbm.24497 (2019).
Acknowledgements
This work was supported by an Australian Research Council (ARC) Linkage Project (LP170100494) that includes financial support from Siemens Healthineers. SDJ, PGDW & GFE are supported by the ARC Centre of Excellence for Integrative Brain Function (CE140100007). SDJ is supported by an ARC Discovery Early Career Researcher Award (DE150100406) and National Health and Medical Research Council Fellowship (APP1174164). The authors acknowledge Alexandra Carey & Richard McIntyre from Monash Biomedical Imaging for assistance with data collection and assistance with BIDS preparation. We also acknowledge Disha Sasan, Winnie Orchard and Irene Graafsma for their assistance in acquiring data.
Author information
Authors and Affiliations
Contributions
S.D.J., G.F.E., P.G.D.W., Z.C., N.J.S., M.P. and A.F. designed the research question. Data was analysed by P.G.D.W. Data was prepared for release by T.G.C., P.G.D.W. & S.D.J. Funding was obtained by G.F.E., S.D.J., Z.C., A.F., M.P., K.O’B., D.S. & N.J.S. Manuscript was written by S.D.J., G.F.E., P.G.D.W. & T.C. All authors have contributed to manuscript preparation.
Corresponding author
Ethics declarations
Competing interests
Siemens Healthineers contributed financial support to the ARC Linkage project held by G.F.E., S.D.J., Z.C., A.F., M.P. & N.J.S. K.O’B. and D.S. are employees and shareholders of Siemens Healthcare Pty Ltd. P.G.D.W. & T.G.C. have no competing or conflicting interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
The Creative Commons Public Domain Dedication waiver http://creativecommons.org/publicdomain/zero/1.0/ applies to the metadata files associated with this article.
About this article
Cite this article
Jamadar, S.D., Ward, P.G.D., Close, T.G. et al. Simultaneous BOLD-fMRI and constant infusion FDG-PET data of the resting human brain. Sci Data 7, 363 (2020). https://doi.org/10.1038/s41597-020-00699-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-020-00699-5
This article is cited by
-
Decoupling of regional neural activity and inter-regional functional connectivity in Alzheimer’s disease: a simultaneous PET/MR study
European Journal of Nuclear Medicine and Molecular Imaging (2022)
-
Deep Learning for Image Enhancement and Correction in Magnetic Resonance Imaging—State-of-the-Art and Challenges
Journal of Digital Imaging (2022)
-
Task-evoked simultaneous FDG-PET and fMRI data for measurement of neural metabolism in the human visual cortex
Scientific Data (2021)