Abstract
Globe-LFMC 2.0, an updated version of Globe-LFMC, is a comprehensive dataset of over 280,000 Live Fuel Moisture Content (LFMC) measurements. These measurements were gathered through field campaigns conducted in 15 countries spanning 47 years. In contrast to its prior version, Globe-LFMC 2.0 incorporates over 120,000 additional data entries, introduces more than 800 new sampling sites, and comprises LFMC values obtained from samples collected until the calendar year 2023. Each entry within the dataset provides essential information, including date, geographical coordinates, plant species, functional type, and, where available, topographical details. Moreover, the dataset encompasses insights into the sampling and weighing procedures, as well as information about land cover type and meteorological conditions at the time and location of each sampling event. Globe-LFMC 2.0 can facilitate advanced LFMC research, supporting studies on wildfire behaviour, physiological traits, ecological dynamics, and land surface modelling, whether remote sensing-based or otherwise. This dataset represents a valuable resource for researchers exploring the diverse LFMC aspects, contributing to the broader field of environmental and ecological research.
Similar content being viewed by others
Background & Summary
Live Fuel Moisture Content (LFMC), a critical parameter in fire-related research, quantifies the vegetation water content. It is computed as:
where Wf represents the weight of fresh plant material, measured post-sample collection, Wd indicates the weight of the same sample after thorough drying, often in an oven.
Numerous studies have demonstrated LFMC’s influence on various wildfire metrics, including flammability, rate of spread, fire occurrence and cumulative burnt area1,2,3,4,5. Growing interest surrounds the exploration of LFMC dynamics in relation to ecological, meteorological and ecophysiological parameters6,7,8,9,10, especially within the context of a changing climate11.
However, conducting fieldwork, collecting measurements, and recording data can be costly, time consuming, and resource-intensive. Therefore, the convenience of having access to a readily available LFMC dataset proves beneficial for advancing research. As a result, several LFMC datasets12,13, including the 2019 version of Globe-LFMC14, have emerged online.
Globe-LFMC 2.015, presented herein and accessible at the figshare repository, represents an updated version of the 2019 release. It incorporates previously published datasets and adds more than 120,000 additional measurements hitherto unavailable to the research community.
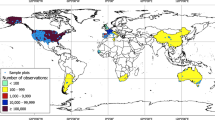
This extensive dataset comprises over 280,000 LFMC values derived from samples gathered at more than 2,000 locations across 15 countries. It includes data from more than 500 different species or combinations of species. The timeframe of the data spans from 1977 to 2023 (Tables 1, 2, Fig. 1).
Locations of sampling sites. The sampling sites are represented as coloured points on the map, with the colour intensity indicating the abundance of LFMC values collected at each location. To enhance clarity, points have been ordered on the z-axis based on the number of LFMC samples, with sites having fewer samples placed beneath those with a higher data count. Predominantly, the sampling sites and LFMC measurements are concentrated in the USA, France, and Spain. The base map for this figure is derived from NASA’s Visible Earth ‘Explorer Base Map’30.
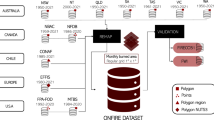
The compilation process included formatting source data, performing rigorous and recursive quality checks, merging data from co-authors, and introducing supplementary information. Notably, each data point now includes land cover type and meteorological variables, aligned with the sampling date and location. An outlier detection analysis was executed, and its findings are presented (Fig. 2).
Workflow followed to compile Globe-LFMC 2.015.
Distinguishing Globe-LFMC 2.015 from its predecessor, it presents two significant enhancements. First, it incorporates a large number of LFMC measurements from individual samples, broadening its coverage across various geographic and climatic conditions. Second, it includes additional descriptor variables per sample (Tables 3, 4) and rectifies inaccuracies and typos that may have been present in the previous version. These improvements not only increase the comprehensiveness of the dataset but also enhance its adaptability for end-users, allowing them to process the data and aggregate the samples as they see fit.
Globe-LFMC 2.015 applications are manifold. Researchers can employ it to develop and validate models for LFMC estimation from remote sensing data16,17, or for other types of land surface modelling, such as those derived from climate variables3. It is equally valuable in investigating the relationship between LFMC and wildfire occurrence and behaviour, as well as its associations with other plant water status metrics, meteorological parameters and ecological drivers.
In conclusion, as we plan to keep the dataset updated and publish future versions, we invite researchers and other interested parties to contact us if they wish to contribute.
Methods
Compilation of Live Fuel Moisture Content measurements
Globe-LFMC 2.015 is the result of collaborative efforts involving international researchers and agencies, incorporating data from multiple sources, including publicly available datasets12,13,14,18,19.
The authors meticulously adapted their datasets to conform to the template spreadsheets, aligning with the structured format of Globe-LFMC 2.015 (a comprehensive breakdown of the dataset fields is available in the Data Records section). These refined spreadsheets were subsequently integrated into a unified dataset, following a rigorous visual quality check. This check was essential to verify data integrity, and rectify any typographical errors, formatting inconsistencies and obsolete information to ensure the dataset’s reliability and accuracy.
LFMC values in this dataset were derived from destructive measurements of plant materials obtained during field sampling. While sampling and weighing protocols varied among contributors, the common procedure involved weighing fresh plant material, typically leaves, either in the field or a laboratory after secure transportation in a sealed bag or container. Subsequently, the samples were oven-dried for several hours at a minimum of 60 °C and re-weighed. Sampling details, including location, date, and sometimes the time of sampling, as well as specific sampling protocols, were meticulously recorded.
Unlike the previous version of Globe-LFMC14, efforts were made to avoid data aggregation and preserve individual sample measurements wherever possible. This means that values corresponding to the same combination of species, sampling location, and date were not averaged together. In cases where data from the 2019 version of the dataset were included, averaging was replaced with the original individual measurements, when available.
Entries that remained as mean LFMC values for multiple measurements were flagged in a dedicated dataset column.
A comprehensive review of the 2019 dataset was undertaken to rectify typos and inaccuracies, encompassing species names, protocol details, and, in a limited number of instances, sampling dates (a list of the dates changed is available at the figshare repository15).
The US National Fuel Moisture Database (NFMD)19 was redownloaded from the original source, leading to differences from the previous Globe-LFMC14 version. Some data entries were added, others were removed. Dead Fuel Moisture measurements were excluded, while all LFMC values were retained, irrespective of whether they were later identified as outliers during the quality check. The decision not to delete these values was due to impracticality in contacting the original data providers for further investigation.
After compiling all data sources, extensive efforts were made to harmonize the diverse datasets, ensuring uniformity and consistency across Globe-LFMC 2.015. In cases where the same site name was associated with different coordinates, we introduced unique identifiers at the end of the name to distinguish them. Conversely, when identical coordinates were linked to multiple sites, their names were merged. This meticulous process culminated in a dataset where each site name corresponded exclusively to one set of coordinates, and vice versa, fostering data integrity and precision.
Land cover data
Land cover type information was also added to the final dataset following the IGBP classification from LP DAAC MCD12Q1.061 (MODIS/Terra + Aqua Land Cover Type Yearly L3 Global 500 m SIN Grid)20.
The process started by downloading the complete set of MCD12Q1.061 sinusoidal tiles products spanning the years 2001 to 2022. Subsequently, these tiles were mosaicked into yearly raster images at a spatial resolution of 500 m within the WGS84 reference system.
For each LFMC value, the mosaic corresponding to the respective calendar year was employed to retrieve the land cover ID by selecting the pixel value at the precise sampling location. Additionally, the descriptive land cover name (e.g., “Grasslands”) was incorporated into the dataset.
Given that the available land cover time series extended from 2001 to 2022, the land cover type of 2001 was attributed to all samples collected before 2001, as it most closely represented the respective sampling date. Similarly, for samples collected after 2022, the land cover type of the year 2022 was assigned. This method ensured consistent land cover information across all samples.
Meteorological data
Meteorological data was sourced from AgERA5 (Agrometeorological indicators from 1979 to present derived from reanalysis) AgERA5 is a high level product built upon ERA5 data, which were aggregated to obtain daily values and downscaled to 0.1° × 0.1° spatial resolution21.
The initial step involved downloading NetCDF files containing specific meteorological variables: total daily precipitation, relative humidity at 2 m above surface at four distinct times (6am, 9am, 12 pm and 3 pm), maximum daily air temperature at 2 m above surface, mean daily air temperature, mean daily vapour pressure, mean daily wind speed at 10 m above surface and mean daily dewpoint temperature at 2 m above surface.
Subsequently, the values for each meteorological variable were extracted from the downloaded files at the date and location of each entry in the dataset.
Additionally, cumulative precipitation data for the preceding 3 days, 1 week, 4 weeks, and 12 weeks before the sampling date was included in the final dataset.
Detection of possible outliers
The process of identifying potential outliers within LFMC values consisted of a two-step strategy, combining both manual inspection and the application of two distinct statistical models.
We define outliers as values that deviate notably from the norm, being either anomalously high or low. Such deviations may arise from measurement inaccuracies due to instrument or human errors. Additional context regarding the interpretation of outlier detection is available in the “Technical Validation” section.
Step 1: Manual Inspection and Data Provider-Specific Methods
In the initial phase, when possible, data providers meticulously examined each dataset comprising Globe-LFMC 2.015. Since these datasets varied significantly in structure, the authors customized outlier detection methods for each. The outcomes of this initial assessment were documented in the “Extra information/Quality Flag” column of the dataset. The methods used in this step were tailored to the specific dataset’s characteristics, involving visual inspections, percentile-based, or standard deviation-based approaches to identify outliers.
Step 2: Statistical Model-Based Outlier Detection
The second approach leveraged the Isolation Forest algorithm22, a tool that utilises binary trees to identify data points as outliers via random splits in the dataset. Fewer splits required to isolate a data point indicate a higher likelihood that it is an outlier. The implementation of this method was conducted through Python’s Scikit-Learn 1.3.0 library23 as illustrated in Fig. 3. Isolation Forest analysis was executed on separate data subsets categorised by species. The variables integrated into the models were time, latitude, longitude, and LFMC to account for both variations among local populations of the same species and fluctuations in time series data.
Due to the unsupervised nature of this model, hyperparameters were predefined with a theoretical approach as the true anomalies were unknown. Key hyperparameters included the number of trees (“n_estimators”) set at 10,000, which was considered sufficient for building a precise model without excessive computational demands. Additionally, “max_samples” was set at 75% of each subset total data points to facilitate the detection of outlier clusters. The inclusion of bootstrap, “max_features” set at 4, and a contamination ratio of 0.05 was determined based on a conservative assessment of the data following preliminary visual examination.
A potential limitation of this approach is its propensity to identify data points as anomalies when they are isolated in time or space, even if their LFMC values are within the expected range. To minimize this risk, a complementary model was simultaneously applied to the same subsets. This model specifically focused on time, latitude, and longitude, with a “max_features” setting adjusted to 3. It aimed to detect data points isolated independently of their LFMC values. The anomalies identified by this secondary model were then subtracted from those found by the LFMC-inclusive model, producing a refined list of anomalous LFMC values.
Given the stochastic nature of the Isolation Forest algorithm, five model versions were created (both including and excluding LFMC as a variable), each employing different random states. A data point was designated as a possible outlier only if all five LFMC-inclusive models identified it as isolated, and at least one of the five models without LFMC did not isolate it, as depicted in Fig. 3. Data points isolated by all models with and without LFMC were not classified as outliers, as their isolation could be attributed to time or spatial factors unrelated to their LFMC values.
The results of all ten models, along with their respective scores, are provided in the figshare repository15.
Moreover, the repository contains results from an alternative outlier detection method: Cook’s Distance24, which gauges the influence of a data point on a regression line. This analysis was conducted using the Python library statsmodels 0.14.025. It involved grouping data points by species and sampling location, calculating ordinary least squares regression, and comparing Cook’s Distance scores to the “4/n” threshold (where “n” stands for the number of observations within a group of samples), commonly used to identify influential data points26,27. An additional criterion was considered, flagging data points with Cook’s Distance values more than three times the mean Cook’s Distance of data points in the same group.
In cases where Cook’s Distance resulted in NaN (not a number) or infinite values, “NA” (not available) was assigned to all data points within the same group.
Data Records
The Globe-LFMC 2.0 dataset15 is available in an MS Excel file containing three sheets: “Contact” (Table 5), “LFMC Data” (Tables 3, 4) and “Protocol” (Table 6). The primary dataset is located within the “LFMC Data” sheet, which contains the core LFMC values along with associated information. The “Contact” sheet offers supplementary details regarding the contact person responsible for each sub dataset, facilitating direct communication and inquiries related to the data. In the “Protocol” sheet, a comprehensive description of the sampling and weighing procedures employed to obtain the LFMC measurements is presented, providing essential context for data interpretation.
Accompanying the dataset, additional files are provided for reference and extra data. In these files it is possible to retrieve all the outcomes generated from the outlier detection procedures, offering transparency and insight into data quality control, as well as the references to the original sources and datasets incorporated into Globe-LFMC 2.015. The files are equipped with column descriptions where needed, enhancing the accessibility and usability of the dataset.
Technical Validation
A rigorous quality check of the LFMC data was conducted individually by each contributing author, as outlined in the Methods section. Furthermore, to ensure data integrity, two outlier detection methods, the Isolation Forest and the Cook’s Distance, were applied across the entire dataset (see Usage Notes for details).
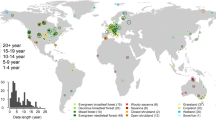
Upon removal of data points flagged as potential anomalies by the Isolation Forest method, the LFMC values generally fell within expected ranges, as demonstrated in Fig. 4 and detailed in Table 7, which provides example LFMC distributions and descriptive statistics for some of the most common species in the dataset.
Box plots and violin plots illustrating the seasonal variability and statistical distribution of LFMC for eight common species found in Globe-LFMC 2.015. The species include Quercus gambelii, a deciduous oak (sampled in USA); Quercus coccifera, an evergreen oak (sampled in France, Spain, Türkiye); Pinus edulis, a medium sized pine (sampled in USA); Pinus taeda, a tall pine (sampled from the USA); Cistus monspeliensis, an evergreen shrub with narrow leaves (sampled in France, Italy, Spain, Tunisia); Arctostaphylos patula, an evergreen shrub with round leaves (sampled in USA); Rosmarinus officinalis, an evergreen shrub with narrow leaves (sampled in France, Italy, Spain, Tunisia); and unidentified grass encompassing various unidentified grass species collected in grasslands (sampled in Argentina, Australia, China, Portugal, Senegal, Spain). The seasons were defined based on time ranges between astronomical equinoxes and solstices. (Figure created using seaborn31).
Notably low LFMC values may be attributed to samples that contain a combination of live and dead plant material or, in some cases, exclusively dead material from living vegetation. Similarly, very high LFMC values not identified as potential outliers could originate from juvenile leaves, fleshy plant species, or samples influenced by waterlogged soil conditions. Whenever available, this contextual information was included in the dataset.
It is important to acknowledge that certain data points may not have been identified as anomalies by the method depicted in Fig. 3, potentially due to isolation in time or space, irrespective of their LFMC value.
Moreover, although efforts were made to detect outliers, it is possible that a small number of very high values remain unidentified due to the stochastic nature of the method applied (Isolation Forest).
The correctness of the land cover and meteorological values added to the dataset was verified visually by comparing the output of the Python scripts with the source raster images in a Geographic Information System (GIS) software. This validation process was conducted on a small randomly selected subset consisting of 15 data points (one per country).
Usage Notes
The “LFMC data” sheet contains various attributes that can be utilized for data filtering and categorization as per research requirements. Additionally, it offers valuable meteorological and land cover data that can support the study of LFMC dynamics. Tables 3, 4 provide detailed explanations for each column, but further guidance on how to effectively use some of the more intricate attributes is provided below.
The “Species functional type” column provides a generic classification of the sampled species. It is particularly valuable for understanding the vertical structure of the collected species within the plot, especially when different species are sampled from the same location. The functional types were assigned by data providers based on their expertise. Hence, intermediate-size plants were occasionally categorised using different terms depending upon each author’s judgement (e.g., “small tree” and “large shrub” could refer to plants having analogous size).
Functional type information is especially useful for optical remote sensing studies, particularly in closed forests, where the canopy may obstruct visibility of lower vegetation layers. In such cases, it is advisable to select only measurements from trees.
For remote sensing applications, it is recommended to average the LFMC measurements taken on the same date and located within the same pixel of the product employed in the study. The choice of which functional type to include in the average can be guided by the land cover type of that pixel. For example, in open canopy forests, both trees and shrubs (or grass) could be included.
However, caution is advised when utilising land cover information, given the 500 m spatial resolution and inherent uncertainties in the satellite-based product, which may compromise the accuracy of land cover classification.
The “protocol” column and accompanying protocol sheet can be used to filter the data based on specific research requirements. For instance, selecting only LFMC values retrieved following a specific sampling and weighing criteria or excluding samples that might have included flowers or buds.
Land cover type and meteorological data are provided to aid preliminary studies and hypothesis testing regarding LFMC dynamics or investigation of reasons behind anomalous LFMC values, or retrieval of information about the plant type sampled.
The “Extra information/Quality Flag” column contains additional miscellaneous information provided by data providers to enhance the understanding of the data. It may include markers for suspected anomalies, explanations for unusual LFMC values, or information about the plant type sampled.
“Isolated data point” reports the output of the Isolation Forest models (Fig. 3). Users can employ this column to filter the dataset by removing “isolated” data points, which could be potential outliers (by only selecting the “FALSE” values; i.e., not isolated).
Instead of removing potential outliers from the dataset, adding flags enables each user to employ the data in the way that best suits their research needs.
It is important to note that if a data point is identified as isolated (value “TRUE”), it may not necessarily be a true outlier, as the algorithm compares it only to other data points in the same subset without prior knowledge of LFMC variability of a given species.
Moreover, it is possible that anomalous LFMC values were not flagged as outliers because those data points were selected as isolated in time or space by all the models without LFMC (see Methods for details), and they were subtracted from the potential outliers.
Lastly, due to the random nature of this method, both false positives and false negatives are possible.
Further outlier detection criteria are provided in the figshare repository15, including columns reporting the results from Cook’s Distance method. The columns “Above 4/n Cook Distance” and “Above 3xMean Cook Distance” are two ready-to-use quality flags that can help identify influential data points. Cook’s Distance methods tended to detect a much higher number of outliers; hence they appear to be more conservative than the Isolation Forest. However, it can also sometimes fail to identify possible outliers with suspiciously high (or low) LFMC values, if there are other values in the same subset that are even higher (or lower).
Moreover, additional output data from both outlier detection methods are also shared, providing dataset users with the flexibility to create customized filters to suit their specific requirements. For instance, users can employ algorithms’ scores to establish custom thresholds. Alternatively, in the context of the Isolation Forest method, they can flag a data entry as a potential outlier even if does not meet the consensus of five models.
Furthermore, it is possible to employ a combination of different methods. For example, the Cook’s Distance metrics can be used to cross-verify LFMC values of data points that were not classified as anomalies by the Isolation Forest method only because they were detected as isolated in time and space.
Finally, it is strongly recommended to use the most recent version of the dataset, as it incorporates corrections for occasional inaccuracies and typos. Continued use of the 2019 version is discouraged.
Code availability
The code for the detection of potential outliers and the extraction of land cover and meteorological data was developed using Python 3.9.7. The corresponding Jupyter Notebooks are available at the figshare repository15.
The outlier detection code uses the Globe-LFMC-2.015 file as input.
When executing the land cover and meteorological data extraction code, it is essential to have downloaded the required input data first.
References
Dennison, P. E. & Moritz, M. A. Critical live fuel moisture in chaparral ecosystems: a threshold for fire activity and its relationship to antecedent precipitation. International Journal of Wildland Fire 18, 1021 (2009).
Dimitrakopoulos, A. & Papaioannou, K. Flammability Assessment of Mediterranean Forest Fuels. Fire Technology; Norwell 37, 143 (2001).
Park, I., Fauss, K. & Moritz, M. A. Forecasting Live Fuel Moisture of Adenostema fasciculatum and Its Relationship to Regional Wildfire Dynamics across Southern California Shrublands. Fire 5, 110 (2022).
Pimont, F., Ruffault, J., Martin-StPaul, N. K. & Dupuy, J.-L. A Cautionary Note Regarding the Use of Cumulative Burnt Areas for the Determination of Fire Danger Index Breakpoints. Int. J. Wildland Fire 28, 254 (2019).
Rossa, C. G., Veloso, R. & Fernandes, P. M. A laboratory-based quantification of the effect of live fuel moisture content on fire spread rate. Int. J. Wildland Fire 25, 569 (2016).
Bar-Massada, A. & Lebrija-Trejos, E. Spatial and temporal dynamics of live fuel moisture content in eastern Mediterranean woodlands are driven by an interaction between climate and community structure. Int. J. Wildland Fire 30, 190 (2021).
Boving, I. et al. Live fuel moisture and water potential exhibit differing relationships with leaf-level flammability thresholds. Functional Ecology, https://doi.org/10.1111/1365-2435.14423 (2023).
Griebel, A. et al. Specific leaf area and vapour pressure deficit control live fuel moisture content. Functional Ecology 37, 719–731 (2023).
Nolan, R. H. et al. Drought-related leaf functional traits control spatial and temporal dynamics of live fuel moisture content. Agricultural and Forest Meteorology 319, 108941 (2022).
Pivovaroff, A. L. et al. The Effect of Ecophysiological Traits on Live Fuel Moisture Content. Fire 2, 12 (2019).
Ma, W. et al. Assessing climate change impacts on live fuel moisture and wildfire risk using a hydrodynamic vegetation model. Biogeosciences 18, 4005–4020 (2021).
Gabriel, E. et al. Live fuel moisture content time series in Catalonia since 1998. Annals of Forest Science 78, 44 (2021).
Martin-StPaul, N. et al. Live fuel moisture content (LFMC) time series for multiple sites and species in the French Mediterranean area since 1996. Annals of Forest Science 75, 57 (2018).
Yebra, M. et al. Globe-LFMC, a global plant water status database for vegetation ecophysiology and wildfire applications. Sci Data 6, 155 (2019).
Yebra, M. et al. Globe-LFMC 2.0, An enhanced and updated dataset for Live Fuel Moisture Content research, figshare, https://doi.org/10.6084/m9.figshare.c.6980418 (2024).
Cunill Camprubí, À., González-Moreno, P. & Resco De Dios, V. Live Fuel Moisture Content Mapping in the Mediterranean Basin Using Random Forests and Combining MODIS Spectral and Thermal Data. Remote Sensing 14, 3162 (2022).
Miller, L. et al. Projecting live fuel moisture content via deep learning. Int. J. Wildland Fire 32, 709–727 (2023).
République Française - Conservatoire de la Forêt Méditerranéenne, Office National des Forêts. Réseau Hydrique http://www.reseauhydrique.dpfm.fr.
United States Government. National Fuel Moisture Database https://www.wfas.net/nfmd/public/about.php.
Friedl, M. & Sulla-Menashe, D. MCD12Q1.061 MODIS/Terra+Aqua Land Cover Type Yearly L3 Global 500m SIN Grid V061 [Data set]. NASA EOSDIS Land Processes Distributed Active Archive Center. https://doi.org/10.5067/MODIS/MCD12Q1.061 (2022).
Boogaard, H. et al. Agrometeorological indicators from 1979 to present derived from reanalysis. Copernicus Climate Change Service (C3S) Climate Data Store (CDS). https://doi.org/10.24381/cds.6c68c9bb (2020).
Liu, F. T., Ting, K. M. & Zhou, Z.-H. Isolation-Based Anomaly Detection. ACM Transactions on Knowledge Discovery from Data 6, 1–39 (2012).
Pedregosa, F. et al. Scikit-learn: Machine Learning in Python. Journal of Machine Learning Research 12, 2825–2830 (2011).
Cook, R. D. Detection of Influential Observation in Linear Regression. Technometrics 19, 15–18 (1977).
Seabold, S. & Perktold, J. statsmodels: Econometric and statistical modeling with python. in 92–96, https://doi.org/10.25080/Majora-92bf1922-011 (Austin, Texas, 2010).
Altman, N. & Krzywinski, M. Analyzing outliers: influential or nuisance? Nature Methods 13, 281–282 (2016).
Van der Meer, T., Te Grotenhuis, M. & Pelzer, B. Influential Cases in Multilevel Modeling: A Methodological Comment. American Sociological Review 75, 173–178 (2010).
Briottet, X. et al. BIODIVERSITY – A new space mission to monitor Earth ecosystems at fine scale. RFPT 224, 33–58 (2022).
Adeline, K. et al. Multi-scale datasets for monitoring Mediterranean oak forests from optical remote sensing during the SENTHYMED/MEDOAK experiment in the north of Montpellier (France). Data in Brief 53, 110185 (2024).
Stevens, J. Explorer Base Map, NASA Earth Observatory map by Joshua Stevens using data from NASA’s MODIS Land Cover, the Shuttle Radar Topography Mission (SRTM), the General Bathymetric Chart of the Oceans (GEBCO), and Natural Earth boundaries. (2020).
Waskom, M. L. seaborn: statistical data visualization. Journal of Open Source Software 6, 3021 (2021).
Acknowledgements
The early stages of dataset compilation were made possible through the Australian Government Research Training Program Domestic Scholarship. Subsequent efforts received financial support from the Bushfire Research Centre of Excellence, supported by the Australian National University and Optus. The data provided by the Hawkesbury Institute for the Environment was funded by the NSW Department of Planning and Environment via the NSW Bushfire and Risk Management Research Hub. CNES, focused on BIODIVERSITY space mission under the APR project named SentHyMED28 supported the work with A. Karine, J.B. Féret and J.M. Limousin as contacts and the raw dataset29. Portuguese FCT – Fundação para a Ciência e Teconologia in the framework of the researcher contract DL57/2016/CP1442/CP0005 supported the work of A. Monteiro. The data provided by the James Hutton Institute was funded by the Scottish Government via NatureScot. We acknowledge the incorporation of sections of the datasets at zenodo.org/records/162978 (CC BY 4.0)13 and zenodo.org/records/4694854 (CC BY 4.0)12 into Globe-LFMC 2.015. The meteorological data was generated using Copernicus Climate Change Service information 2023 (AgERA5) (https://doi.org/10.24381/cds.6c68c9bb, Accessed in May, June and July 2023). The land cover type data was retrieved from the online Data Pool, courtesy of the NASA EOSDIS Land Processes Distributed Active Archive Center (LP DAAC), USGS Earth Resources Observation and Science (EROS) Center, Sioux Falls, South Dakota, https://lpdaa.usgs.gov/tools/data-pool/ (Accessed in Aug and Sep 2023).
Author information
Authors and Affiliations
Contributions
M.Y. conceived the idea, supervised the data collection and the dataset compilation, provided data included in the dataset and feedback on the dataset format, wrote the first version of the manuscript, and contributed to the final version the article. G.S. coordinated the data collection, harmonised the dataset, provided feedback on the dataset format, wrote the first version of the manuscript, produced the figures and tables, and contributed to the final version the article. K.A., A.B.M., M.E.B., E.C., R.D.D., P.D., C.D.B., F.G., E.Gr., A.G., I.K., M.P.M., A.T.M., R.H.N., G.P., N.Y.C. provided data included in the dataset and contributed to the final versions of the dataset and the article. All remaining authors provided data included in the dataset and contributed to the final version of the dataset.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yebra, M., Scortechini, G., Adeline, K. et al. Globe-LFMC 2.0, an enhanced and updated dataset for live fuel moisture content research. Sci Data 11, 332 (2024). https://doi.org/10.1038/s41597-024-03159-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-024-03159-6