Abstract
Enhancer and super-enhancers are master regulators of cell fate. While they act at long-distances on adjacent genes, it is unclear whether they also act on one another. The immunoglobulin heavy chain (IgH) locus is unique in carrying two super-enhancers at both ends of the constant gene cluster: the 5′Eμ super-enhancer promotes VDJ recombination during the earliest steps of B-cell ontogeny while the 3′ regulatory region (3′RR) is essential for late differentiation. Since they carry functional synergies in mature B-cells and physically interact during IgH locus DNA looping, we investigated if they were independent engines of locus remodelling or if their function was more intimately intermingled, their optimal activation then requiring physical contact with each other. Analysis of chromatin marks, enhancer RNA transcription and accessibility in Eμ- and 3′RR-deficient mice show, in mature activated B-cells, an unilateral dependence of this pair of enhancers: while the 3′RR acts in autonomy, Eμ in contrast likely falls under control of the 3′RR.
Similar content being viewed by others
Introduction
Super-enhancers (SEs) are master regulators of cell fate which differ from basic enhancers by their ten-fold higher load of chromatin marks, their binding of mediator, their long length and their impact on nuclear organisation1, 2. The immunoglobulin heavy chain (IgH) locus undergoes multiple changes along B-cell differentiation, affecting transcription and accessibility for V(D)J recombination, somatic hypermutation (SHM) and class switch recombination (CSR)3. The IgH locus is somehow unique in carrying two SEs, Eμ and the 3′ regulatory region (3′RR) at both ends of the constant gene cluster, which control locus remodelling along B-cell differentiation3. In mature B-cells, the IgH locus assumes an enigmatic loop conformation in which these two SEs are brought in close proximity despite their 200 kb distance on the chromosome (Fig. 1a)4. Two mechanistic hypothesises may explain the function of this 3D chromatin structure: a bidirectional crosstalk between these two SEs allowing reciprocal activation, or a simple chromatin arch assembly that brings enhancers, promoters and switch (S) regions into close proximity to facilitate transcription, accumulation of RNA pol II, AID targeting and S junction machinery. In a third hypothesis, the Eμ-3′RR interaction might lack any mechanistic role by passively witness transitional links, with Eμ on one side promoting Sμ-Sx synapsis and the 3′RR stimulating the Ix-Sx transcriptional unit. We investigated if these two IgH SEs were independent engines of locus remodelling simply combining their proper actions or if their functions were more strongly intermingled and required physical contact with each other. SEs are potent clusters of transcriptional enhancers and regulate the expression of key cell lineage specific genes. The 5′Eμ SE has clearly such a role in early developmental stages of B-cells through its key role on V(D)J recombination. Since 5′Eμ may be considered as a SE only in pro-B/pre-B-cells but not in mature ones, we used the qualifier of enhancer to name 5′Eμ at the mature B-cell stage during this study.
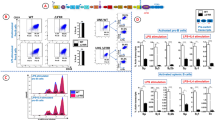
Eμ and CSR. (a) Schematic 3D conformation of the IgH locus during LPS-induced CSR. 3C experiments indicated that Eμ and the 3′RR are in close proximity in resting B cells. After appropriated stimulation the activated S acceptor region gets closer the Sμ donor region. Adapted from Wuerffel et al.4. (b) Flow cytometry analysis of IgG3 and IgG2b CSR in Eµ-deficient B-cells. Cells were stimulated 4 days with LPS. Cells were labelled with anti-B220-BV421, anti-IgG3-FITC, anti-IgG2b-PE and anti-IgM-PC7 antibodies. Cells were gated on B220+ B-cells. One representative experiment out of four is shown. (c) Quantitative analysis of IgG1, IgG3 and IgG2b CSR. Results are reported as mean ± SEM of 4 independent experiments. *p < 0.05, Mann-Whitney U-test. (d) Flow cytometry analysis of IgG1 CSR in Eμ-deficient B-cells. Cells were stimulated 4 days with LPS + IL-4. Cells were labelled with anti-B220-BV421, anti-IgG1-PE and anti-IgM-PC7 antibodies. Cells were gated on B220+ B-cells. One representative experiment out of four is shown. (e) H3K4me3 epigenetic mark in S μ and S γ1 during IgG1 CSR ChIP assays were performed with CD43− splenic B-cells from Eμ-deficient and wt mice. Cells were stimulated with LPS + IL-4 for 2 days. Background signals from mock samples with irrelevant antibody were subtracted. Values were normalized to the total input DNA. Data are the mean ± SEM of 3 independent experiments with 2 mice. (f) H3K4me3 epigenetic mark in S μ and S γ3 during IgG3 CSR. Cells were stimulated with LPS for 2 days. Same protocol as in part E.
Results
Eμ and CSR
During CSR, a chromatin loop is found with Eμ and 3′RR in close proximity3, 4. It is generally accepted that Eµ is dispensable for CSR, although some studies suggest that its deletion impacts at minor levels CSR efficiency5,6,7. In this study, flow cytometry analysis shows normal proportion of Eμ-deficient γ3- and γ2b-expressing B-cells in response to LPS stimulation (Fig. 1b and c). In contrast, the significant decrease of Eμ-deficient γ1-expressing B-cells in response to LPS + IL4 (Fig. 1c and d) could mostly be attributed to the lack of follicular B-cells previously reported in this model7. These results are in accordance with the lowered serum IgG1 levels but normal IgG3/IgG2b levels in this model7. Specific epigenetic mark enrichment in S regions is a prerequisite for CSR8, 9. During CSR, the 3′RR SE fosters H3K4me3 histone modifications in the S acceptor but not Sμ donor region10, suggesting another cis-transcriptional enhancer for this role. This role is not devolved to Eμ since its deletion did not affect H3K4me3 histone modifications in Sμ and Sγ1 in response to LPS + IL-4 stimulation (Fig. 1e) nor in Sμ and Sγ3 in response to LPS stimulation (Fig. 1f). Altogether, these data indicate that the Eμ enhancer is mostly dispensable for CSR and did not poise Sμ for CSR. The cis-transcriptional enhancer that poised Sμ for CSR is currently unknown. One might suggest the enigmatic transcriptional enhancer located between Cγ1 and Cγ2b found to interact with both Eμ and 3′RR in pro-B cells2, 11, 12.
Eμ, 3′RR and respective epigenetic marks
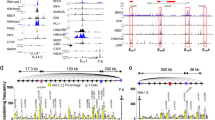
Epigenetic changes in Eμ and 3′RR are of importance during CSR13. In this study we have focussed on H3K4me3, H3K9ac and H3K27ac epigenetic marks that are associated with active regulatory regions14. Lack of the Eμ enhancer did not affect H3K4me3, H3K9ac and H3K27ac epigenetic marks in the four 3′RR transcriptional enhancers (hs3a; hs1,2; hs3b; hs4) during LPS-induced B-cell CSR (Fig. 2a). In turn, if deletion of the 3′RR SE had no effect on H3K4me3 and H3K27ac marks of Eμ (Fig. 2b), a significant decrease was found for H3K9ac. This difference between epigenetic marks may be explained by the proposed model of sequential histone modifications. Indeed, several studies suggest that tri-methylation of H3K4 is the earliest modification, and that H3K4me3 then facilitate H3 acetylation and thus establishment of chromatin openness. All these marks together may then positively regulate transcription and enhancer activation15, 16. At the mature B-cell stage, data suggest the lack of Eμ-dependence for the 3′RR SE during CSR, and the influence of the 3′RR on the Eμ enhancer, a result that fits well with their different kinetics of activation, i.e., at the immature and mature B-cell stages for Eμ and 3′RR, respectively3.
Epigenetic marks in Eμ and 3′RR during CSR. (a) H3K4me3, H3K9ac and H3K27ac epigenetic marks in the four enhancers (hs3a, hs1,2, hs3b and hs4) of the 3′RR in LPS-stimulated B-cells of Eμ-deficient and wt mice. Cells were stimulated with 5 μg/ml LPS for 2 days. Data are the means ± SEM of 3 independent experiments with 2 mice. (b) H3K4me3, H3K9ac and H3K27ac epigenetic marks in Eμ in LPS-stimulated B-cells of 3′RR-deficient and wt mice. Same experimental protocol as in part (a). Data are the means ± SEM of 3 independent experiments with 2 mice. *p < 0.05 (Mann-Whitney U-test).
Mutual activation of transcription of Eμ and 3′RR SEs
The 3′RR SE controls CSR by acting on germline transcription and histone modifications8, 17, that are hallmarks of CSR accessibility. We investigated the effect of the lack of the Eμ enhancer or of the 3′RR SE on IgH constant gene transcription units (CH) in response to LPS-induced stimulation in vitro. RNAseq experiments showed that, except for Cμ, CH sense and antisense transcripts were dramatically reduced in 3′RR-deficient mice (Fig. 3a and Supplementary Figs 1–3). In contrast, Eμ deletion had no effect on CH transcription at the IgH locus. RNASeq data are presented in a quantitative way with statistics in the Supplementary Fig. 4. Non coding RNAs (ncRNAs) play an important role in the targeting of the CSR machinery and contribute to chromosomal looping10, 18. Among these ncRNAs, enhancer RNA (eRNA) are transcribed from DNA sequences of enhancer including the 3′RR and contribute to their enhancer function19, 20, and chromosomal looping4. RNAseq experiments did not highlight any effect of Eμ deletion on both sense and antisense 3′RR eRNA levels (Fig. 2a). In contrast both sense and antisense transcription around the Eμ enhancer were consistently lowered in 3′RR-deficient activated B-cells. This decrease is reminiscent of the notable effect of the 3′RR SE deletion on μ transcription evidenced with quantitative PCR in resting mature B-cells21. Physical Eμ-3′RR interactions were also documented in resting B-cells4. RNAseq data comparing splenic resting B-cells indicate that enhancer activity was, at this stage, already dominated by the 3′RR: while Eμ deletion showed no obvious effect on CH transcription and 3′RR eRNAs, the 3′RR deletion does not modifies Eμ eRNA but still decreases μ transcripts and abrogates basal transcription of all downstream CH (Fig. 2b). This confirmed a passive role for Eμ at the resting stage while H chain production originated from pVH mostly rely on 3′RR, and more precisely on hs422.
Influence of Eµ and 3′RR on IgH transcription during CSR. (a) IgH transcription on LPS-stimulated B-cells of wt, Eμ-deficient and 3′RR-deficient mice. CD43-depleted splenocytes were cultured for 2 days with 5 μg LPS. RNAseq experiments were done after depletion of rRNA. Data are the mean of two independent experiments with 3 mice per genotype. (b) IgH transcription on resting B-cells of wt, Eμ-deficient and 3′RR-deficient mice. RNAseq experiments were done with CD43-depleted splenocytes after depletion of rRNA. Data are the mean of two independent experiments with 3 mice per genotype. Precise locations of Iμ, Sμ, Cμ, Cδ, Iγ3, Sγ3, Iγ2b and Sγ2b are reported in Supplementary Figs 1–3.
Discussion
Studies highlighted different roles and kinetics of activation for IgH SEs during B-cell development. The Eμ SE regulates V(D)J recombination in pro-B cells6, 7, but is not crucial for SHM and CSR in mature B-cells23. The 3′RR SE regulates SHM24, and conventional CSR in mature B-cells10, 25, 26, but is dispensable for V(D)J recombination27. GFP transgenic mice reported that the Eμ SE is active at pro-B/pre-B cell stages28, while the 3′RR SE is active at immature/mature B-cell stages29. Despite different roles and kinetics, the IgH locus assumes in resting B-cells and CSR an enigmatic loop conformation with the Eμ enhancer and the 3′RR SE in close proximity4, suggesting a potential transcriptional cross-talk between these two enhancer entities. Present results strongly suggest that cross-talk is unidirectional. The 3′RR SE stands as a fully autonomous module in mature B-cells. The Eμ enhancer by contrast appears mostly dispensable at this stage, its deletion neither abrogating CSR-required sense/antisense germline transcription or 3′RR SE eRNA expression and activation-associated epigenetic mark enrichment. If the Eμ enhancer has no role on 3′RR activation during CSR, it itself shows at least partial 3′RR-dependence. Deletion of the 3′RR SE impacts Eμ H3K9ac activation-associated epigenetic marks and sense/anti-sense eRNA transcription.
During CSR the IgH locus assumes a loop conformation. While this conformation has obviously a major interest to simultaneously bring enhancer, SE, S regions and promoters into close proximity, our data suggest another unknown functional role, which is to place Eμ under the authority of the 3′RR SE. The Eμ/Sμ/Sx/3′RR hub might have several successive purposes to facilitate CSR. This hub brings the Sx acceptor region in close proximity to the 3′RR SE, allowing for its efficient activation by acting notably on transcription and AID targeting10. It support physical/functional interactions between 3′RR elements hereby building their synergy3, and finally facilitates Sμ-Sx synapsis, increasing the probability of a legitimate junction between donor and acceptor S regions and, in the meantime, reducing the risk of potentially oncogenic translocation30,31,32. Previously reported phenotypes of Eμ- and 3′RR-deficient mice, now completed by the present study, show that both IgH enhancers can behave as completely autonomous elements with regards to chromatin marks, eRNA transcription and accessibility, with Eμ solely controlling early rearrangements of immature B-cells, and the 3′RR being both necessary and sufficient for late B-cell remodelling events. However in mature B-cells, this ends with Eμ falling under the control of the 3′RR SE and then undergoing 3′RR-dependent transcription and chromatin remodelling as almost all basic promoters of the locus. Finally, three SEs have been reported in pro-B cells: Eμ, 3′RR and an enigmatic region between Cγ1 and Cγ2 2, 11, 12. Despite that 3′RR has little role on V(D)J recombination except for silencing early transcription in pro-B cells33, investigation of the cross-talk between these three SEs in pro-B cells would be of interest to reinforce our knowledge of their role at the immature B-cell maturation stage.
Material and Methods
Mice
129 wt mice (from Charles Rivers Laboratories, France), EμMAR-deficient mice7, and 3′RR-deficient mice25 were used. EμMAR-deficient mice and 3′RR-deficient mice were in a 129 background. Our research has been approved by our local ethics committee review board (Comité Régional d’Ethique sur l′Expérimentation Animale du Limousin, Limoges, France) and carried according the European guidelines for animal experimentation.
Spleen cell cultures for B-cell activation
Single-cell suspensions of CD43− spleen cells of wt, EμMAR-deficient mice and 3′RR-deficient mice (8–12 week old, male and female) were cultured at 1 × 106 cells per ml in RPMI 1640 with 10% fetal calf serum with 5 μg per ml LPS with or without 20 ng/ml IL-4. Two days LPS-stimulated cells were used for RNAseq and Sμ/Sγ3 ChIP experiments. Two days LPS + IL4 stimulated cells were then used for Sμ/Sγ1 ChIP experiments. Four days LPS- and LPS + IL4 stimulated cells were used for γ3, γ2b and γ1 CSR flow cytometry analysis, respectively.
Flow cytometry analysis
Cultured splenic B-cells were labelled with anti-B220-BV421, anti-IgG1-PE, anti-IgG3-FITC, anti-IgG2b-PE and anti-IgM-PC7 antibodies for 30 min at 4 °C. Labelled cells were analyzed on a Fortessa LSR2 (Beckman Coulter) with Kaluza.
ChIP experiments
ChIP experiments were done on LPS- and LPS + IL4 stimulated CD43− spleen cells as previously described10. In brief, 20 × 106 B-cells were cross-linked at room temperature for 15 min in 15 ml PBS with 1% formaldehyde. The reaction was quenched with 0.125 M glycine. After lysis, chromatin was sonicated to 0.5–1 kb using a Vibracell 75043 (Thermo Fisher Scientific). After dilution in ChIP buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl, pH 8.1, and 167 mM NaCl), chromatin was precleared by rotating for 2 h at 4 °C with 50 ml of 50% protein A/G slurry (0.2 mg per ml sheared salmon sperm DNA, 0.5 mg per ml BSA, and 50% protein A/G; Sigma). 1 × 106 cell equivalents were saved as input, and 10 × 106 cell equivalents were incubated overnight with anti-H3K4me3, anti-H3K9ac, anti-H3K27ac or control antibodies. Immune complexes were precipitated by the addition of protein A/G. Cross-linking was reversed by overnight incubation (70 °C) in TE buffer with 0.02% SDS and chromatin was phenol/chloroform extracted. Anti-H3K4me3 and antu-H3K9ac were obtained from Millipore (ref: 07473 and 06942) and anti-H3K27ac was obtained from Abcam (clone ab5131). PCR probes were the following: Eμ-forward: GGGAGTGAGGCTCTCTCATA; Eμ-reverse: ACCACAGCTACAAGTTTACCTA; hs3a-forward: GGGTAGGGCAGGGATGCTCACAT; hs3a-reverse: GCTCTGGTTTGGGGC ACCTGTGC; hs1,2-forward: AGCATAGGCCACTGGGACTGG; hs1,2-reverse: CTCTCA CTTCCCTGGGGTGTT; hs3b-forward: TGGTTTGGGGCACCTGTGCTGAG; hs3b-reverse: GGGTAGGGCAGGGATGTTCACAT; hs4-forward: CCATGGGACTGAAAC TCAGGGAACCAGAAC; hs4-reverse: CTCTGTGACTCGTCCTTAGG. PCR probes for Sμ Cμ, Sγ1, Cγ1, Sγ3 and Cγ3 have been reported in a previous study10.
RNAseq experiments
CD43− splenocytes were obtained from 4 wt, 4 3′RR-deficient mice and 4 EμMAR-deficient mice before and after 48 h of in vitro stimulation (1 × 106 cells per ml in RPMI 1640 with 10% fetal calf serum) with 5 μg per ml LPS. RNA was extracted using miRNeasy kit from QIAGEN, according to the manufacturer instructions. Two pooled RNA (with two samples) were obtained for each genotype. RNA libraries were obtained using TruSeq Stranded Total RNA with Ribo-Zero Gold (Illumina), according to the manufacturer instruction. Libraries were sequenced on a NextSeq500 sequencer, using NextSeq 500/550 High Output Kit (Illumina). Illumina NextSeq500 paired-end 2 × 150 nt reads were mapped with STAR release v2.4.0a versus mm10 with gene model from Ensembl release 77 with default parameters. The long length of the reads allowed for their precise mapping on switch regions, previously reported as error prone with shorter reads due to the highly repetitive structure of these sequences34. Quantification of genes was then performed using feature Counts release subread-1.4.6-p1-Linux-x86_64 with “–primary -g gene_name -p -s 1 -M” options based on Ensembl GTF release 77 annotations.
Accession number
Data were deposited in Gene Expression Omnibus under the accession number GSE90760.
References
Qian, J. et al. B cell super-enhancers and regulatory clusters recruit AID tumorigenic activity. Cell 159, 1524–1537 (2014).
Whyte, W. A. et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153, 307–319 (2013).
Pinaud, E. et al. The IgH locus 3′ regulatory region: pulling the strings from behind. Adv Immunol. 110, 27–70 (2011).
Wuerffel, R. et al. S-S synapsis during class switch recombination is promoted by distantly located transcriptional elements and activation-induced deaminase. Immunity. 27, 711–722 (2007).
Sakai, E., Bottaro, A. & Alt, W. The Ig heavy chain intronic enhancer core region is necessary and sufficient to promote efficient class switch recombination. Int Immunol. 11, 1709–1713 (1999).
Perlot, T., Alt, F. W., Bassing, C. H., Suh, H. & Pinaud, E. Elucidation of IgH intronic enhancer functions via germ-line deletion. Proc Natl Acad Sci USA 42, 14362–14367 (2005).
Marquet, M. et al. The Eμ enhancer region influences H chain expression and B cell fate without impacting IgVH repertoire and immune response in vivo. J Immunol. 193, 1171–1183 (2014).
Li, G. et al. Combinatorial H3K9acS10ph histone modification in IgH locus S regions targets 14-3-3 adaptors and AID to specify antibody class-switch DNA recombination. Cell Rep. 5, 702–714 (2013).
Vaidyanathan, B. & Chaudhuri, J. Epigenetic codes programing class switch recombination. Front Immunol. 6, 405 (2015).
Saintamand, A., Rouaud, P., Saad, F., Rios, G., Cogné, M. & Denizot, Y. Elucidation of IgH 3′ region regulatory role during class switch recombination via germline deletion. Nature Commun. 6, 7084 (2015).
Medvedovic, J. et al. Flexible long-range loops in the VH gene region of the IgH Locus facilitate the generation of a diverse antibody repertoire. Immunity. 39, 229–244 (2013).
Predeus, A. V. et al. Targeted chromatin profiling reveals novel enhancers in IgH and IgL chain loci. J Immunol. 192, 1064–1070 (2014).
Zan, H. & Casali, P. Epigenetics of peripheral B-cell differentiation and the antibody response. Front Immunol. 6, 631 (2015).
Ernst, J. et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 473, 43–49 (2011).
Karmodiya, K., Krebs, A. R., Oulad-Abdelghani, M., Kimura, H. & Tora, L. H3K9 and H3K14 acetylation co-occur at many gene regulatory elements, while H3K14ac marks a subset of inactive inducible promoters in mouse embryonic stem cells. BMC Genomics. 13, 424 (2012).
Roh, T. Y., Cuddapah, S., Cui, K. & Zhao, K. The genomic landscape of histone modifications in human T cells. Proc. Natl. Acad. Sci. USA 103, 15782–15787 (2006).
Saintamand, A. et al. Deciphering the importance of the palindromic architecture of the immunoglobulin heavy chain 3′ regulatory region. Nature Commun. 7, 10730 (2016).
Pefanis, E. et al. RNA exosome-regulated long non-coding RNA transcription controls super-enhancer activity. Cell 161, 774–789 (2015).
Lam, M. T. Y., Li, W., Rosenfeld, M. G. & Glass, C. K. Enhancer RNAs and regulated transcriptional programs. Trends Biochem Sci. 39, 170–182 (2014).
Péron, S. et al. AID-driven deletion causes immunoglobulin heavy chain “locus suicide recombination” in B cells. Science 336, 931–934 (2012).
Saintamand, A. et al. The IgH 3′ regulatory region governs μ chain transcription in mature B lymphocytes and the B cell fate. Oncotarget 6, 4845–4852 (2015).
Vincent-Fabert, C., Truffinet, V., Fiancette, R., Cogné, N., Cogné, M. & Denizot, Y. Ig synthesis and class switching do not require the presence of the hs4 enhancer in the 3′ IgH regulatory region. J Immunol. 182, 6926–6932 (2009).
Li, F., Yan, Y., Pieretti, J., Feldman, D. A. & Eckhardt, L. A. Comparison of identical and functional IgH alleles reveals a nonessential role for Eμ in somatic hypermutation and class switch recombination. J Immunol. 185, 6049–6057 (2010).
Rouaud, P. et al. The IgH 3′ regulatory region controls AID-induced somatic hypermutation in germinal centre B-cells in mice. J Exp Med. 210, 1501–1507 (2013).
Vincent-Fabert, C. et al. Genomic deletion of the whole IgH 3′ regulatory region (hs3a, hs1,2, hs3b, hs4) dramatically affects class switch recombination and Ig secretion to all isotypes. Blood 116, 1895–1898 (2010).
Rouaud, P. et al. Elucidation of the enigmatic IgD class switch recombination via germ-line deletion of the IgH 3′ regulatory region. J Exp Med. 211, 975–985 (2014).
Rouaud, P., Vincent-Fabert, C., Fiancette, R., Cogné, M., Pinaud, E. & Denizot, Y. Enhancers located in heavy chain regulatory region (hs3a, hs1,2, hs3b and hs4) are dispensable for diversity of VDJ recombination. J Biol Chem. 287, 8356–8360 (2012).
Guglielmi, L. et al. The 5′HS4 insulator element is an efficient tool to analyse the transient expression of an Eμ-GFP vector in a transgenic mouse model. Trans Res. 14, 361–364 (2005).
Guglielmi, L., Le Bert, M., Truffinet, V., Cogné, M. & Denizot, Y. Insulators to improve expression of a 3′IgH LCR-driven reporter gene in transgenic mouse models. Biochem Biophys Res Commun. 307, 466–471 (2003).
Klein, I. A. et al. Translocation-capture sequencing reveals the extent and nature of chromosomal rearrangements in B lymphocytes. Cell 147, 95–106 (2011).
Zhang, Y. et al. Spatial organization of the mouse genome and its role in recurrent chromosomal translocations. Cell 148, 908–921 (2012).
Ghazzaui, N., Saintamand, A., Issaoui, H., Vincent-Fabert, C. & Denizot, Y. The IgH 3′ regulatory region and c-myc induced B-cell lymphomagenesis. Oncotarget, 2016 Oct 8, doi:10.18632/oncotarget.12535 [Epub ahead of print].
Braikia, F. Z. et al. A developmental switch in the transcriptional activity of a long-range regulatory element. Mol. Cell. Biol 35, 3370–3380 (2015).
Pavri, R. et al. Activation-induced cytidine deaminase targets DNA at sites of RNA polymerase II stalling by interaction with Spt5. Cell 143, 122–133 (2010).
Acknowledgements
This work was supported by grants from Comité d’Orientation de la Recherche en Cancérologie du Limousin (CORC:FJA/NP-2015-109 and CORC:FJA/NP-2014-126), Association pour la Recherche sur le Cancer (PJA 20141201649), Ligue Contre le Cancer (Comité de la Corrèze et Haute Vienne), Agence Nationale de la Recherche (ANR: projet EpiSwitch-3′RR 2016). Epigenetic experiments were supported by a specific grant from INCa-Cancéropôle GSO (2014). A. Saintamand was supported by a grant from fondation ARC (DOC20150602943). N. Ghazzaui was supported by a grant from Association de Spécialisation et d’Orientation Scientifique (Lebanon) and the municipality of Khiam (Lebanon). M. Cogné is supported by Institut Universitaire de France and Fondation pour la Recherche Médicale. We acknowledge the technological expertise of the Nice Sophia-Antipolis Functional Genomics Platform, supported by MICROENVIMET, FP7-HEALTHF2-2008-201279, the ARC, and the INCa. We thank Dr. P. Barbry for helpful discussions and support during this work. We thank S. Desforges and B. Remerand for help with animal care.
Author information
Authors and Affiliations
Contributions
A.S., C.V.F., M.M., N.G., E.P., M.C. and Y.D. actively participated to the experimental design of the study. V.M. performed RNAseq experiments. E.P., M.C. and Y.D. participated to the scientific discussion for manuscript writing, obtained financial grants and agreement of the ethic committee of our institution to perform the study. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Electronic supplementary material
Accession codes: GEO GSE90760.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Saintamand, A., Vincent-Fabert, C., Marquet, M. et al. Eμ and 3′RR IgH enhancers show hierarchic unilateral dependence in mature B-cells. Sci Rep 7, 442 (2017). https://doi.org/10.1038/s41598-017-00575-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-00575-0
This article is cited by
-
The overlap of suitable tea plant habitat with Asian elephant (Elephus maximus) distribution in southwestern China and its potential impact on species conservation and local economy
Environmental Science and Pollution Research (2022)
-
Class switch recombination junctions are not affected by the absence of the immunoglobulin heavy chain Eμ enhancer
Cellular & Molecular Immunology (2019)
-
RETRACTED ARTICLE: 3′RR and 5′Eμ immunoglobulin heavy chain enhancers are independent engines of locus remodeling
Cellular & Molecular Immunology (2019)
-
Deletion of the immunoglobulin heavy chain 3′ regulatory region super-enhancer affects somatic hypermutation in B1 B cells
Cellular & Molecular Immunology (2019)
-
The IgH 3′ regulatory region super-enhancer does not control IgA class switch recombination in the B1 lineage
Cellular & Molecular Immunology (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.