Abstract
Parkinson’s disease (PD) is an age-related neurodegenerative disease associated with the misfolding and aggregation of alpha-synuclein (aSyn). The molecular underpinnings of PD are still obscure, but nutrition may play an important role in the prevention, onset, and disease progression. Dietary (poly)phenols revert and prevent age-related cognitive decline and neurodegeneration in model systems. However, only limited attempts were made to evaluate the impact of digestion on the bioactivities of (poly)phenols and determine their mechanisms of action. This constitutes a challenge for the development of (poly)phenol-based nutritional therapies. Here, we subjected (poly)phenols from Arbutus unedo to in vitro digestion and tested the products in cell models of PD based on the cytotoxicity of aSyn. The (poly)phenol-digested metabolites from A. unedo leaves (LPDMs) effectively counteracted aSyn and H2O2 toxicity in yeast and human cells, improving viability by reducing aSyn aggregation and inducing its clearance. In addition, LPDMs modulated pathways associated with aSyn toxicity, such as oxidative stress, endoplasmic reticulum (ER) stress, mitochondrial impairment, and SIR2 expression. Overall, LPDMs reduced aSyn toxicity, enhanced the efficiency of ER-associated protein degradation by the proteasome and autophagy, and reduced oxidative stress. In total, our study opens novel avenues for the exploitation of (poly)phenols in nutrition and health.
Similar content being viewed by others
Introduction
(Poly)phenols are natural compounds found in a wide variety of foods either as dietary or nutraceutical supplements 1. The term polyphenol is used to describe a structure with at least one aromatic ring with one or more hydroxyl groups attached2. However, compounds with only one phenolic ring are also referred to as polyphenols, as hydroxycinnamates and phenolic acids. Thus, polyphenols in general, and more accurately, are referred to as (poly)phenols2. Epidemiological and clinical studies showed that (poly)phenols can reduce the incidence and prevalence of cardiovascular diseases, cancer, diabetes, inflammation and age-related disorders, as pure (poly)phenols, in extracts or through diet1,2,3,4,5,6. However, for the rational recommendation of (poly)phenols in nutrition, their bioaccessibility and the mechanism of action must be dissected. Thus, the understanding of the influence of digestion on the bioactivities of dietary (poly)phenols is imperative. Studies of digestion in humans and in animal models pose ethical, time and cost concerns. Thus, in vitro digestion models to predict phytochemical changes during digestion offer several advantages, including the suitability as a high-throughput approach amenable for mechanistic studies of dietary (poly)phenols in cellular models of disease7.
Several studies support the beneficial effects of (poly)phenols and their bioavailability and accumulation even in the brain8,9. Strikingly, (poly)phenols can reverse/prevent age-related cognitive decline and neurodegeneration10, being attractive as dietary supplements.
Parkinson’s disease (PD) is an age-related neurodegenerative disease for which we still lack effective therapies11. The misfolding and aggregation of alpha-synuclein (aSyn) is deeply associated with PD pathogenesis12, but the precise underlying molecular mechanisms remain elusive. Among these, oxidative stress and mitochondrial dysfunction are common factors, contributing to disturbances in cellular processes that result in irreversible cell damage and death13. In particular, the excessive accumulation of unfolded proteins and oxidative stress that are typical in PD, result in the failure of the endoplasmic reticulum (ER) to cope with the excess of protein load, a process denominated by ER stress14,15.
The ER plays several essential functions in the cell as it is responsible for the synthesis of one-third of the total proteome, and is involved in the folding, maturation and posttranslational modification of numerous proteins14. In response to ER stress, cells activate the unfolded protein response (UPR)14,15,16, leading to the transcription of genes related with protein homeostasis, including chaperones, and protein degradation and secretion pathways14. Sustained ER stress and UPR activation may overwhelm cellular protective mechanisms, ultimately triggering apoptosis14. The ER-associated protein degradation (ERAD) is a cellular pathway that targets misfolded proteins for ubiquitination and subsequent degradation by the ubiquitin–proteasome system (UPS) in the cytoplasm16. Macroautophagy, hereafter designated as autophagy, is also initiated in response to ER stress caused by misfolded proteins, via ER-activated autophagy (ERAA)16,17, which induces a partial UPR and a calcium-mediated signalling cascade. ERAA is a degradation pathway and serves the function of mitigating ER stress and suppressing cell death, as ERAD16.
The clearance of soluble aSyn can occur both via the UPS and chaperone-mediated autophagy (CMA)18. Under pathological conditions, aSyn inhibits the proteasome and impairs CMA19,20, leading to the upregulation of autophagy21. On the other hand, the degradation of aggregated aSyn occurs by autophagy22,23. Nonetheless, the relationship between autophagy and disease is unclear.
We previously showed that (poly)phenols can protect against aSyn toxicity by promoting its clearance trough autophagy and by modulating aSyn fibrillation24. Here, we conducted a pioneering nutritional approach in order to study the bioactivities of (poly)phenol-digested metabolites (PDMs), obtained after in vitro gastro-intestinal digestion of edible fruits (FPDMs) and leaves (LPDMs)25. We tested (poly)phenols from A. unedo (strawberry tree), a native species in regions of Mediterranean climate, since we previously found remarkable protection against oxidative stress, when compared to other (poly)phenol matrices26. Additionally, it is known that A. unedo (poly)phenols hold potential for the treatment of hypertension3, cardiovascular diseases27 and cancer4,28. Nevertheless, the potential of these (poly)phenols in the context of neurodegeneration has not been explored.
The chemical alterations occurring during in vitro digestion were analyzed by LC-MS and the molecular mechanisms of PDMs protection via regulation of autophagy and other targets of aSyn toxicity were analyzed. Interestingly, we found that LPDMs displayed strong protection against aSyn and H2O2 toxicity in yeast and in human cells, respectively. In addition, these LPDMs reduced aSyn aggregation and induced its clearance through the activation of autophagic and proteasomal function. The LPDMs were able to reduce oxidative stress and aSyn-mediated mitochondrial impairment. We also found that LPDMs regulated transcription of genes involved in the unfolded protein response and Sir2 pathways. In total, our study revealed protective molecular mechanisms of the selected LPDMs in PD models, by modulating the clearance of misfolded proteins and autophagy. Altogether, our study opens novel avenues for the exploitation of nutraceuticals/therapeutics based on the use of (poly)phenols in the context of neurodegenerative diseases.
Results
PDMs from A. unedo leaves protect against aSyn-induced toxicity
We used an established model of PD based on the expression of human aSyn in yeast cells19,29 to dissect the mechanisms of protection by A. unedo PDMs. The concentration of 62 µg GAE.mL−1 was selected as non-toxic based on viability assays (Fig. S1). Yeast cells grown in raffinose medium were transferred to galactose medium, to induce aSyn expression, and treated at the same time with PDMs for 6 h. After this treatment, cellular viability was assessed by spot assays (Fig. 1A). Importantly, we observed that leaf (LPDMs) or fruit (FPDMs), had no effect on the control cells growth. Remarkably, PDMs were able to suppress toxicity induced by aSyn expression, while cells treated with LPDMs grew similarly to control cells (Fig. 1A). Protection was also observed by the reduction of PI positive cells assessed by flow-cytometry when aSyn expressing cells were treated with LPDMs (Fig. S2). Both LPDMs and FPDMs had a strong protective effect on growth (spot tests, Fig. 1A). However, the effect on membrane integrity was not as strong (PI, Fig. S2).
A. unedo PDMs reduce aSyn-induced toxicity, inclusion formation and superoxide radical levels. Yeast cells transformed with a plasmid encoding for aSyn-GFP or with the empty vector (control, ctrl), were treated with 62 μg GAE.mL−1 of LPDM or FPDMs for 6 h, in galactose liquid medium. (A) Cell viability assessed by spot assays, after treatment with PDMs in galactose liquid medium. (B) Fluorescence microscopy images of yeast cells (left panel) and respective percentage of cells with aSyn inclusions (right panel). Scale bar: 5 µm. (C) Superoxide radical levels evaluated by DHE MFI, assessed by flow cytometry. Results are represented in arbitrary units (a. u.). (D) aSyn expression levels assessed by western blot, GAPDH was used as loading control. A representative result is shown, and values represent the mean ± SD of three independent experiments. Statistically significant differences between the indicated treatments are shown, **p < 0.01, ***p < 0.001 or nd as not different for p < 0.05.
aSyn toxicity in yeast cells is associated with the formation of cytosolic inclusions19. Thus, we then assessed if the enhancement in cell viability was accompanied by alterations in the inclusion formation of aSyn, by fluorescence microscopy. Interestingly, treatment with LPDMs significantly reduced the percentage of cells displaying aSyn inclusions (Fig. 1B). However, the treatment with FPDMs had no effect on inclusions formation (Fig. 1B).
Oxidative stress is another central event in PD being a known trigger of aSyn misfolding and aggregation30. Conversely, aSyn aggregation induces mitochondrial dysfunction and ROS production31. Accordingly, we then analyzed the effect of PDMs treatment on ROS levels in yeast cells expressing aSyn. Superoxide radical levels were evaluated by flow cytometry (FCM), using dihydroethidium (DHE), which is an indicator of mitochondrial metabolic function32, a known target of aSyn-mediated toxicity33. As observed before, aSyn expression led to an increase in superoxide production (Fig. 1C)24. Treatment with LPDMs substantially reduced the levels of superoxide in cells expressing aSyn (Fig. 1C).
We next assessed whether the differences in toxicity and inclusion formation were due to different expression levels of aSyn induced by the PDMs treatments. Using western blot analyses, we found that the PDMs did not affect aSyn protein expression levels (Fig. 1D).
Overall, these results suggest that LPDMs is cytoprotective in a yeast model of PD by reducing inclusion formation and improving redox homeostasis. For this reason, we continued our study focusing on LPDMs and the chemical profile of its (poly)phenols was evaluated.
Chemical characterization of leaf fractions
LC-MS analysis of the A. unedo leaf extracts before (original) and after (LPDMs) digestion (Table 1) confirmed that they were mainly composed of (poly)phenols previously described as having bioactivities2,3,8,24. The chemical composition of the LPDMs used has been partially previously described26. The major class represented was the flavonols, with quercetin, myricetin, and kaempferol glycosides present (Table 1). As described flavonols are relatively stable to digestion25. The galloylated flavonols and the flavanols, epigallocatechin and epicatechin, were less stable and their levels were reduced in the LPDMs. Therefore, the main effect of digestion was the concentration of certain components, and removal of other less stable components from the digested samples. No significant breakdown products were noted, although it is possible that the apparent increase in certain flavonol glycosides was influenced by the drop in galloylated components.
The component with m/z 423 was increased by digestion and was subjected to further analysis by QTOF-NMR for identification. The mass and formula are consistent with a gallic acid ester of arbutin, which naturally occurs in three different structural combinations. The 1H NMR showed signals consistent with aromatic groups corresponding of hydroquinone, gallic acid and also the sugar moiety (Table 1, Fig. S3). Additionally, three other compounds were increased after digestion (Table 1) and initial MS data suggests that these may be roseoside, a megastigmane glucoside and rehmaionoside C a monoterpenoid with potential bioactivity34,35 and hydroxybenzoyl arbutin previously reported in A. unedo36. However further analysis is required to confirm these identifications.
LPDMs modulate the oligomerization of aSyn
In order to determine the effect of LPDMs on the biochemical nature of the aSyn species formed in yeast cells, we performed centrifugation in sucrose gradients, as previously described37. Briefly, protein fractions corresponding to different molecular weights (numbered from 1 to 9 in the Fig. 2A) were collected from the sucrose gradient and analyzed by SDS-PAGE and immunoblots. The amount of aSyn in each fraction was then evaluated by densitometry (Fig. 2A). aSyn oligomeric species were found dispersed throughout the various sucrose gradient fractions (Fig. 2A). Interestingly, upon treatment with LPDMs, we found that fraction 3 was enriched in aSyn species, indicating the presence of smaller molecular weight species, while the levels of aSyn in fraction 9 (oligomers with molecular weights larger than 200 kDa) were reduced (Fig. 2A, right panel). We confirmed that these changes were not due to differences in the loading of the total protein using GAPDH as a loading control (Fig. 2B). Thus, we concluded that LPDMs modulated the nature of aSyn species formed in yeast cells, reducing the formation of higher molecular weight oligomeric species of aSyn, which are potentially more toxic12.
A. unedo LPDMs modulate the biochemical nature of aSyn oligomeric species. Yeast cells were treated with 62 μg GAE.mL−1 of LPDMs for 6 h, in galactose medium. (A) Oligomeric aSyn species resolved using sucrose gradients. The resulting fractions were separated on an SDS-PAGE gel followed by immunoblotting for aSyn (left panel); quantification of aSyn in each fraction normalized by total aSyn (right panel). (B) The samples were submitted to the sucrose gradients and evaluated for aSyn expression levels assessed by immunoblot. A representative result is shown, and values represent the mean ± SD of three independent experiments. Significantly different results are indicated between “aSyn” and “aSyn + PDMs” treatment for each fraction. Statistically significant differences between the indicated treatments are shown, *p < 0.05, **p < 0.01, ***p < 0.001.
LPDMs promote the clearance of aSyn by restoring proteasomal and autophagic activities
The impairment of protein clearance pathways is implicated in the pathogenesis of PD, leading to the accumulation of misfolded and aggregated aSyn12,38. Therefore, we next evaluated the effect of LPDMs treatment on aSyn clearance (Fig. 3A). Briefly, 6 h after induction of aSyn expression in galactose medium in the presence of LPDMs (0 h of clearance), cells were switched to glucose-containing medium in order to repress aSyn expression. Cells were then grown in glucose medium for 18 h (18 h clearance), the degradation/clearance of aSyn was assessed by western blotting. Cells treated with LPDMs during the induction period presented lower levels of aSyn after 18 h of clearance (Fig. 3A), suggesting that LPDMs promoted the degradation of aSyn. Thus, we then analyzed the effect of LPDMs on proteostasis pathways.
A. unedo LPDMs increase the clearance of aSyn by promoting autophagy and proteasomal function. Yeast cells were treated with 62 μg GAE.mL−1 of LPDMs for 6 h, in galactose medium. (A) Clearance of aSyn evaluated by western blot at 6 h of aSyn induction (0 h clearance) and after18 h in glucose medium (18 h of clearance) (left panel), and respective quantification normalized to PGK levels used as loading control. (B) Proteasome impairment evaluated by flow cytometry (FCM) using GFPu median fluorescence intensity (MFI), after 6 h of aSyn induction. (C) Autophagy evaluated by GFP-Atg8 processing assay assessed by western blot (left panel), after 6 h of aSyn induction. Atg8 induction quantified by the total GFP signal (GFP-Atg8 and free GFP signal, detected with anti-GFP) (middle panel); autophagic flux quantified by measuring the vacuolar degradation of the Atg8 domain reporter (ratio of free GFP to total GFP signal) (right panel). (D) RPN4 and ATG8 gene relative expression levels assessed by qRT-PCR. (E) SIR2 relative expression levels assessed by qRT-PCR. A representative result is shown, and values represent the mean ± SD of at least three independent experiments. Statistically significant differences between the indicated treatments are shown, *p < 0.05, **p < 0.01, ***p < 0.001. Full-length immunoblots are shown in Supplementary Figure S4.
The clearance of aSyn has been associated with both UPS and autophagy-lysosome pathway18. The effect of LPDMs on proteasome activity was analyzed using an unstable GFP reporter (GFPu) consisting of GFP fused with a constitutive degradation signal (CL-1) that promotes its rapid degradation by the UPS19. FCM analysis revealed that, upon aSyn expression, the levels of GFPu considerably increased (Fig. 3B) indicating that aSyn impaired proteasome function, as previously reported19,24. Interestingly, cells expressing aSyn and treated with LPDMs presented reduced levels of GFPu, indicating an attenuation of the proteasome impairment (Fig. 3B). Treatment with LPDMs had no effect on the levels of GFPu in control cells not expressing aSyn (Fig. 3B), suggesting that this effect was specific for cells expressing aSyn.
Subsequently, we tested whether autophagy was affected by LPDMs using the GFP-Atg8 processing assay as a reporter39,40. Atg8 is one of the key molecules involved in autophagy and its conjugation to the autophagosomal membrane, through an ubiquitin-like conjugation system, is essential for autophagy in eukaryotes40. We observed that aSyn expression led to a significant increase in Atg8 induction and reduced autophagic flux (Fig. 3C), suggesting that aSyn interfered in autophagy function as previously described24. Remarkably, when aSyn-expressing cells were treated with LPDMs the autophagic flux was restored to control levels (Fig. 3C). However, in the control cells not expressing aSyn, both Atg8 induction and autophagic flux remained unchanged when cells were treated with LPDMs (Fig. 3C).
To reinforce the findings described above, we then investigated the involvement of the proteasome and autophagy pathways, by evaluating the mRNA levels of RPN4 and ATG8 genes, since their transcriptional regulation is well characterized. RPN4 encodes for a transcription factor that regulates the expression of proteasome genes41, and is upregulated by the UPR42. We observed that RPN4 transcript levels increased upon aSyn expression (Fig. 3D). In contrast, in cells treated with LPDMs the RPN4 transcript levels did not increase (Fig. 3D). Similarly, aSyn expression induced ATG8 expression, as expected, considering the Atg8 induction observed in the GFP-Atg8 processing assay (Fig. 3C). However, upon LPDMs treatment, ATG8 transcript levels were similar to those in control cells not expressing aSyn (Fig. 3D).
To gain further insight into the regulation of proteostasis by LPDMs treatment, we then analyzed a known inducer of autophagy and modulator of aSyn toxicity, the Sir243. Sir2 is a deacetylase of the sirtuin family and functions as a regulator of autophagy and mitophagy44. Upregulation of Sir2 is linked with the induction of excessive autophagy and a resulting increase in aSyn toxicity, in aged yeast cells43. Consistently, we observed that aSyn expression increased the transcript levels of SIR2 under the conditions tested (Fig. 3E). However, upon treatment with LPDMs, the SIR2 expression levels were similar to the control levels (Fig. 3E). Altogether, these results indicate that the LPDMs might play an important role in regulating autophagy, and that one of the mechanisms involved might be through Sir2 modulation.
LPDMs modulate the unfolded protein response and mitochondrial function
Due to the observed effect of LPDMs on the clearance of aSyn, we then evaluated the effect of LPDMs treatment on the expression levels of a set of UPR related genes. We assessed the mRNA levels of KAR2, LHS1, HSP26 and HRD1. Here, we observed that aSyn expression induced an increase in the expression levels of all the UPR related genes evaluated (Fig. 4A). On the other hand, LPDMs treatment reduced the levels of LHS1, HRD1 and HSP26 transcripts (Fig. 4A), suggesting that LPDMs were able to attenuate the UPR and the associated ER stress.
Effect of A. unedo LPDMs in the unfolded protein response and oxidative stress. Yeast cells were treated with 62 μg GAE.mL−1 of LPDMs for 6 h, in galactose medium. (A) Transcript expression levels of genes involved in the unfolded protein response, assessed by qRT-PCR. (B) Transcript expression levels of genes involved in oxidative stress response (SOD1, SOD2) and mitochondrial biogenesis (HAP4), assessed by qRT-PCR. (C) Transcript expression levels of genes involved in glutathione metabolism, assessed by qRT-PCR. (D) GSSG/GSH ratio, GSSG and GSH levels assessed by HPLC. Values represent the mean ± SD of three independent experiments. Statistically significant differences between the indicated treatments are shown, *p < 0.05, **p < 0.01, ***p < 0.001.
Oxidative stress and mitochondrial dysfunction are known triggers of protein misfolding and ultimately result in ER stress. SOD1, SOD2 and HAP4 mRNA levels were analysed to gain additional insight into the effect of LPDMs treatment on mitochondrial function (Fig. 4B). Sod1 and Sod2 are superoxide dismutases responsible for detoxifying superoxide radical45. Hap4 is a global regulator of mitochondrial electron transport and activates their expression46. aSyn expressing cells displayed lower levels of SOD1 and SOD2 and, remarkably, higher levels of HAP4, in comparison to the control cells (Fig. 4B). Notably, upon treatment with LPDMs, SOD1 and SOD2 and HAP4 levels were similar to the control cells. For HAP4 its levels in aSyn expressing cells were drastically reduced by LPDMs (Fig. 4B).
We then evaluated the major low-molecular-weight redox buffer, glutathione (GSH), a tripeptide that plays a pivotal role in the protection against oxidative damage, detoxification from xenobiotics and endogenous toxic metabolites47. We assessed both the levels of the key enzymes involved in GSH synthesis and also the ratio of oxidized glutathione to reduced glutathione (GSSG/GSH). Interestingly, the levels of GSH1 and GLR1 were not significantly affected by aSyn expression. However, we observed a slight but significant reduction in GLR1 mRNA levels in cells expressing aSyn treated with LPDMs (Fig. 4C). The cellular glutathione pools were also determined by HPLC, since the GSSG/GSH ratio is a measure of the cell redox state47. We observed that aSyn expression increased the GSSG/GSH ratio, suggesting an increase in oxidative stress. Treatment with LPDMs reduced this ratio, suggesting a reduction of oxidative stress (Fig. 4D).
LPDMs modulate aSyn aggregation, protect from oxidative stress and promote mitochondrial function in human cells
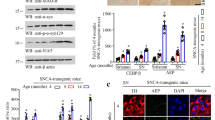
Next, we investigated the effect of the LPDMs on aSyn aggregation in human cells (Fig. 5A, left panel)48,49. Twenty-four hours after co-transfection of cells with plasmids encoding for SynT and Synphilin-1, cells were treated with LPDMs for 16 h. We observed that LPDMs treatment increased the percentage of cells without aSyn inclusions and reduced the percentage of cells presenting more than 10 inclusions (Fig. 5A, middle panel). The mean fluorescence intensity (MFI) of the inclusions in the microscopy images was assessed in cells treated with the LPDMs or vehicle, we did not observe a statistical difference in inclusions MFI between treatments. Importantly, the effect observed was not due to differences in SynT levels due to LPDMs treatment, evaluated by immunoblotting (Fig. 5B).
A. unedo LPDMs modify the distribution of aSyn inclusions, protect from H2O2-induced toxicity and promote mitochondrial function in H4 cells. (A) H4 human neuroglioma cells were transfected with SynT (SynT and Synphilin-1) for 24 h and treated with 2 µg GAE.mL−1 LPDMs for 6 h. Subsequently, cells were subjected to immunofluorescence labeling and the percentage of cells with inclusions was determined by fluorescence microscopy (left panel). Cells were classified by cells without inclusions (□), with less than 10 inclusions ( ), or with 10 or more inclusions (■) (right panel). Scale bar: 10 µm. (B) SynT levels of H4 cells transfected with the empty vector (control), co-transfected with SynT and Synphilin-1 (SynT) or treated with LPDMs (SynT + LPDMs), assessed by western blot. GAPDH was used as loading control. (C) H4 cells were pre-treated with PDMs for 16 h, then subjected to 600 µM of H2O2 for 6 h. Viability was assessed by PI fluorescence versus side scatter (SSC), determined by flow cytometry. PI positive cells are gated, and the respective percentage is presented, statistics were performed in comparison to the control. (D) Mitochondrial membrane potential evaluated using MitoTracker Red by flow cytometry; histogram of MitoTracker Red fluorescence (left panel) and respective median fluorescence intensity (MFI) (right panel) of H4 cells transfected with the empty vector (control), co-transfected with SynT and Synphilin-1 not treated (SynT) or treated with LPDMs (SynT + LPDMs). (E) H4 cells transfected with the empty vector (control), co-transfected with SynT and Synphilin-1 (SynT) or treated with LPDMs (SynT + LPDMs) were immunostained for SynT (green). Mitochondria were visualized using MitoTracker Red (red) while Nuclei were stained with DAPI (blue). Scale bar, 10 µm. A representative result is shown, and values represent the mean ± SD of at least three independent experiments. Statistically significant differences between the indicated treatments are shown, *p < 0.05, **p < 0.01, ***p < 0.001. Full-length immunoblots are shown in Supplementary Figure S4.
), or with 10 or more inclusions (■) (right panel). Scale bar: 10 µm. (B) SynT levels of H4 cells transfected with the empty vector (control), co-transfected with SynT and Synphilin-1 (SynT) or treated with LPDMs (SynT + LPDMs), assessed by western blot. GAPDH was used as loading control. (C) H4 cells were pre-treated with PDMs for 16 h, then subjected to 600 µM of H2O2 for 6 h. Viability was assessed by PI fluorescence versus side scatter (SSC), determined by flow cytometry. PI positive cells are gated, and the respective percentage is presented, statistics were performed in comparison to the control. (D) Mitochondrial membrane potential evaluated using MitoTracker Red by flow cytometry; histogram of MitoTracker Red fluorescence (left panel) and respective median fluorescence intensity (MFI) (right panel) of H4 cells transfected with the empty vector (control), co-transfected with SynT and Synphilin-1 not treated (SynT) or treated with LPDMs (SynT + LPDMs). (E) H4 cells transfected with the empty vector (control), co-transfected with SynT and Synphilin-1 (SynT) or treated with LPDMs (SynT + LPDMs) were immunostained for SynT (green). Mitochondria were visualized using MitoTracker Red (red) while Nuclei were stained with DAPI (blue). Scale bar, 10 µm. A representative result is shown, and values represent the mean ± SD of at least three independent experiments. Statistically significant differences between the indicated treatments are shown, *p < 0.05, **p < 0.01, ***p < 0.001. Full-length immunoblots are shown in Supplementary Figure S4.
The effect of the LPDMs on oxidative injury was analyzed in human H4 cells treated with H2O2 (Fig. 5C). We observed that the pre-treatment with LPDMs for 16 h, prior to stress with 600 µM H2O2 for 6 h, protected cells from oxidative stress-induced cell death, as observed by the reduction of the number of PI positive cells (gated cells) (Fig. 5C). These data show that the LPDMs protected human cells from oxidative stress, validating the results obtained in yeast.
The mitochondria work as generators and targets of ROS13 and its function is impaired by aSyn32,33. Mitochondrial function was studied using MitoTracker Red, a dye that stains mitochondria in live cells and whose accumulation is dependent on mitochondrial membrane potential, a parameter linked to mitochondrial function. In cells expressing SynT, MitoTracker Red fluorescence was lower than in control cells, indicating reduced mitochondrial membrane potential and suggesting impairment of mitochondrial function. On the other hand, treatment with LPDMs of cells expressing SynT increased the MitoTracker Red signal, suggesting a rescue of the mitochondrial function (Fig. 5D). The observation of mitochondria by fluorescence microscopy reinforces the FCM data (Fig. 5E).
Discussion
PD is currently an incurable disorder affecting a growing number of people due to the aging of population11. Therefore, it is urgent to understand the molecular mechanisms underlying PD pathogenesis and to identify novel therapeutic targets and strategies. Phenolic compounds, in food, isolated, or in extracts, are of great interest in nutrition and medicine. They have been associated with the prevention of cancer, chronic and neurodegenerative diseases1,3,4,5,6. Thus, (poly)phenols are attractive compounds in the context of neurodegenerative diseases, since public health policies are focused on the prevention of dementia, through the promotion of a healthy life style, natural nutrition and supplementation1.
In this pioneer study of digested metabolites in PD, we found that PDMs from A. unedo leaves offered significant cytoprotection in established yeast and human cell models of synucleinopathies19,29,48,49. A. unedo possesses several useful biological properties3,27,28,50, nevertheless its neuroprotective potential was not explored. After digestion, the metabolites obtained consisted mainly of myricetin-3-rhamnoside, quercetin-hexoside and kaempferol glycosides. The latter was described as being neuroprotector against ischemic brain injury and neuroinflammation in rats51, while, myricetin and quercetin were described to efficiently inhibit aSyn fibrillation52.
Interestingly, in a previous study we observed that the original undigested fraction of A. unedo leaves was not protective against aSyn-induced toxicity in the conditions tested, measured by growth curves and cells metabolic capacity24. Therefore, the results here obtained indicate that the digestion potentiates the beneficial effects of (poly)phenols in A. unedo, reinforcing their nutritional relevance. Importantly, digestion was also found to potentiate neuroprotective properties of blackberry (poly)phenols53.
PD pathogenesis is associated with proteostasis dysfunction and inefficient protein clearance12,21. Autophagy was shown to be required for the degradation of aSyn aggregates under pathological conditions21,24,32,37,38. Nevertheless, recent studies reported that increased autophagy/mitophagy, promoted by aSyn, has a deleterious effect in aged cells32,43. This appears counterintuitive, as autophagy is generally considered to be a pro-survival process in protein misfolding diseases21. However, autophagy is a complex pathway and, depending on the conditions and models used, it can be either a survival or death mechanism. In the conditions tested herein, aSyn induces the activation of autophagy, as measured by Atg8 protein and mRNA levels. However, this autophagy is dysfunctional, as observed by the reduced autophagic flux. Ultimately, the excessive activation of a dysfunctional autophagy will lead to a loss of selectivity, resulting in the trapping of functional competent organelles in autophagosomes, contributing to the toxicity observed. In fact, LPDMs reduced aSyn toxicity and aggregation, and improved autophagic function, as observed by the increased autophagic flux, in agreement with our previous work where we found that (poly)phenols preferentially affect the autophagic function24, which is impaired by aSyn expression. Nevertheless, it is not clear if the effect of LPDMs on the quality control pathways is direct or a consequence of the modulation of aSyn aggregation.
To ascertain the pathways/molecules associated with aSyn-induced autophagy activation, we investigated the UPR pathway and the levels of ROS. We analysed the expression of ER stress associated genes, where Kar2 and Lhs1 are HSP70 molecular chaperones found in the ER, where they mediate protein folding54,55. Kar2 negatively regulates the UPR via interaction with Ire114,54, a major regulator of the UPR. Lhs1 acts as a nucleotide exchange factor for Kar2 and is upregulated by the UPR55. Hsp26 is a cytosolic chaperone that binds and prevents unfolded proteins from irreversibly forming large protein aggregates56. Hsp26 activity is found only under stress conditions, where it is strongly induced56. Hrd1 is an ubiquitin ligase responsible for recognizing and ubiquitinating misfolded proteins in the ER, for further degradation by the proteasome and is induced in UPR conditions57 (Fig. 6). UPS and autophagy are both mitigators of ER stress16,17,42. Accordingly, increasing evidence suggests the existence of a complex cross-talk between different proteostasis pathways, processes known to play distinct roles in the clearance of specific species of aSyn18,22,23,37.
Schematic representation of the mechanism of aSyn toxicity and protection by A. unedo LPDMs. Accumulation of unfolded proteins (such as aSyn) or oxidative stress leads to endoplasmic reticulum (ER) stress with activation of the unfolded protein response (UPR), leading to oligomerization of the kinase Ire1, and transcription of genes related with protein-folding homeostasis. Kar2 is a negative regulator of UPR via interaction with Ire1. When unfolded proteins accumulate Kar2 is displaced from Ire1, to act as a chaperone aiding proteins conformation. Direct activation of Ire1 by unfolded proteins is an alternative mode for UPR induction. Lhs1, an ER chaperone, mediates protein folding and interacts with Kar2. Cytosolic chaperones also assist aSyn conformation, as Hsp26. Proteins failing to be correctly folded are exported from the ER, a process involving Kar2. Hrd1 is an ubiquitin ligase responsible for recognizing and ubiquitinating misfolded proteins. Afterward ubiquitination, proteins are either degraded in the cytosol by the proteasome or by autophagy, pathways that are both upregulated by the UPR. Aggregated aSyn induces reactive oxygen species (ROS) production and elicits ER stress, with concomitant induction of UPR gene expression. Our findings showed that LPDMs reduced aSyn aggregation and the induction of UPR, while promoting autophagy and proteasome function, pathways involved in aSyn degradation. Moreover, LPDMs reduced oxidative stress and mitochondrial dysfunction, by modulation of redox homeostasis. Overall, LPDMs increased cell viability by a pleiotropic protective role.
Our findings demonstrate that aSyn-elicited ER stress and the concomitant activation of the UPR, might underlie the observed induction of genes related with autophagy and proteasomal function. LPDMs reduced both autophagy activation and the induction of UPR genes, while promoting autophagy and proteasome function (Fig. 6). In line with this observation, it is known that Rpn4, a transcription factor that stimulates the expression of proteasome genes, is regulated by the 26S proteasome in a negative feedback control mechanism41.
The reduction in the levels of LHS1 and RPN4 mRNA by LPDMs is in agreement with an extensive screen for small molecules capable of attenuating aSyn toxicity58. In this study, aSyn expression induced the upregulation of LHS1 and RPN4, among other genes, and the protective compounds markedly reversed the transcriptional changes induced by high levels of aSyn58, as observed here for the LPDMs under study.
ROS, particularly superoxide anions, are crucial signaling molecules implicated in the control and regulation of autophagy and aging59. Sod1 and Sod2 are superoxide dismutase proteins responsible for detoxifying superoxide radical, which are tightly regulated at transcription level in response to oxidative stress60. Sod1 is mainly cytosolic while Sod2 is found in the mitochondria, both are strongly implicated in the aging of yeast cells, and their deletion dramatically reduces the chronological and replicative life span of yeast59. Moreover, the efficiency of the protein folding status of the ER depends on specific redox condition in its lumen14. Our data suggest that the upregulation of SOD1 and SOD2 might be the underlying mechanism behind LPDMs-mediated decreased in superoxide anion levels, a ROS involved in mitochondrial functionality13. These observations are further substantiated by recent studies of the activity of antioxidant enzymes in PD models. Induction of PD by the expression of mutant aSyn61 or DJ-162 in flies, and MPP+ exposure in a human cell line63, resulted in reduced SOD activity and increased production of superoxide. It was proposed that the increase in ROS levels observed in the PD models was due to a deficient activity of the antioxidant enzymes. In addition, treatment with protective compounds increased SOD activity and rescued the PD phenotype61,62,63. Moreover, treatment with (poly)phenols reduced the ratio of oxidized/reduced glutathione (GSSG/GSH), supporting their role against oxidative stress damage. In our study, the evidence that LPDMs improved oxidative homeostasis and mitochondrial function was further confirmed in human cells, as treatment with LPDMs increased mitochondrial membrane potential in cells expressing aSyn.
Mammalian SIRT1 was recently described as an inducer of autophagy under starvation conditions, and is required to sustain autophagy through deacetylation of autophagic regulators, including Atg5, Atg7, Atg8 and Atg1264. Here, we also investigated the effect of LPDMs on Sir2, the yeast orthologous of SIRT1. Notably, SIR2 overexpression increases aSyn toxicity by regulating ATG8 and, particularly, ATG32 mRNA, leading to excessive mitophagy43. Our findings revealed that aSyn expression induced SIR2 and, impressively, LPDMs treatment maintained SIR2 expression at basal levels, in line with the autophagy induction results, and the described effect of acetylation on autophagy regulation64. Altogether, the effect of LPDMs on SIR2 expression, autophagy flux and aSyn toxicity protection are in agreement with previous studies where it was observed that deletion of the SIR2 gene alleviated aSyn toxicity and drastically inhibited autophagy induction43.
Overall, LPDMs reduced aSyn toxicity and aggregation, promoting the formation of smaller oligomeric species. LPDMs also enhanced the efficiency of the UPS and autophagy and reduced oxidative stress. In total, our study provides important hints for the development of nutraceutical/therapeutic strategies against PD and other synucleinopathies.
Materials and Methods
Plant material and extraction procedure
Fruits and leaves of A. unedo L. were collected by random sampling in an extensive area of Arrábida Natural Park. Plant material was harvested, frozen, freeze-dried and ground in an IKA M20 mill to pass a 0.5 mm sieve and stored at −80 °C. Extracts were prepared using 1 g of lyophilized powder to 12 mL of 50% (v/v) hydroethanolic solution as described53.
In vitro digestion
In vitro digestion was performed as previously described25. Briefly, pH of the extract was adjusted to 1.7. Afterwards 315 units.mL−1 of porcine pepsin (Sigma ref P6887) was added and the solution was incubated at 37 °C, 2 h, 100 rpm. Afterwards 4 mg.mL−1 pancreatin from porcine pancreas (Sigma ref P7545) and 25 mg.mL−1 bile salts mixture were added. A segment of cellulose dialysis tubing (cut-off 12 kDa) containing 0.1 M NaHCO3 to neutralize titratable acidity, was placed inside the beaker containing the solution. After 2 h at 37 °C, the solution inside and outside the dialysis tubing were collected. The soluble materials were applied to C18 solid phase extraction columns (GIGA tubes, 1000 mg capacity, Phenomenex Ltd.), as described in53,65. The (poly)phenol-digested metabolites (PDMs) obtained were concentrated and freeze-dried to suitable phenol concentrations determined by the Folin-Ciocalteau method53.
Liquid chromatography mass spectrometric (LC-MS) analysis of phenolic profile
The samples were dissolved in 1.25 mL water/acetonitrile (80:20, v/v) and 0.4 mL was transferred to a 0.45 mm filter vial. Samples were injected via an autosampler onto a Hypersil Gold (50 mm × 2.1 mm; 1.9 µm, Thermo Scientific) reverse-phase UPLC column at 30 °C. The HPLC system consisted of an Accella 600 quaternary pump and an Accella PDA detector, coupled to an LTQ Orbitrap mass spectrometer (Thermo Fisher Scientific, Stafford House, Boundary Way, Hemel Hempstead, UK). The mobile phases were solution A (0.1% (v/v) aqueous formic acid) and solution B (0.1% (v/v) formic acid in acetonitrile/water (50:50, v/v). The flow rate was 450 µL min−1, and the gradient was 0 min; 97% A, 0–3 min; 85% A, 3–7 min; 75% A, 7–10 min; 50% A, 10–13 min; 50% A, 13–14 min 0% A, 14–16 min 0% A, 16–17 min 97% A, 17–20 min 97% A). The PDA detector range was 200–600 nm. Mass spectrometer mass range was m/z 80–2000 with alternative full scan MS and MS/MS data-dependent scans in negative mode66. Exact mass data was used to predict the molecular formula using the resident software Xcalibur™ (errors <2 ppm). Components were identified using PDA maxima, m/z and MS2 high-resolution fragmentation data with study of the suggested formula for the fragments, and comparison with the values available in the databases.
Liquid chromatography UPLC-MS-NMR analysis of (poly)phenolic profile
The samples were dissolved in 200 μL methanol, were filtered with a 0.22 µm filter and transferred to HPLC vials. Samples were injected via an autosampler reverse-phase UPLC column at 30 °C. The UPLC system consisted of a Waters Acquity UPLC with a BEH C18 Waters (2.1 × 50 mm, 1.7 µm) and a Quadrupole–TOF mass spectrometer (QTOF maXis impact, Bruker, Daltonics, Bremen, Germany), using an orthogonal Z-spray-ESI interface operating in negative ion mode detector, coupled to an NMR. This setup was coupled to a Bruker Avance III HD spectrometer equipped with Ascend TM magnet, 11.7 T (1H operating frequency 500 MHz) and a cryoprobe (Cryoplatform Prodigy BBO) for increased sensitivity. The mobile phases used were solution A (0.1% (v/v) aqueous formic acid) and solution B (0.1% (v/v) formic acid in acetonitrile/water (50:50, v/v)). The flow rate was 5 µL min-1, and the gradient was 0 min; 97% A, 0–3 min; 85% A, 3–7 min; 75% A, 7–10 min; 50% A, 10–13 min; 50% A, 13–14 min 0% A, 14–16 min 0% A, 16–17 min 97% A, 17–20 min 97% A. The PDA detector range was 280–350 nm. Mass spectrometer mass range was m/z 80–2000 with alternative full scan MS and MS/MS data-dependent scans in negative mode. 1H NMR spectra were obtained at 500 MHz in CD3OD with chemical shift values (δ) in ppm.
Yeast transformation and plasmids
BY4741 (MATa; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0) cells were transformed using lithium acetate standard method67. The empty plasmids pRS426GAL, p413-GPD or p415 were used as control68. The pRS316-GFP-ATG8 plasmid was a gift from Prof Yoshinori Oshumi (National Institute for Basic Biology, Okazaki, Japan) and was used to sub-clone GFP-ATG8 as well as the endogenous ATG8 promoter into the SacI - XhoI sites of p415 plasmid39. Other plasmids used were previously described: p426GAL-aSyn-GFP and p426GAL-aSyn carrying the human gene of aSyn with or without a C-terminal fusion to GFP, under the regulation of GAL1 inducible promoter19; p413-GPD-GFPu expressing an unstable GFP under the regulation of GPD constitutive promoter19.
Yeast growth and compound testing
Cells in log growth phase were obtained using synthetic complete (SC) medium [0.67% (w/v) yeast nitrogen base without amino acids (Difco), 1% (w/v) raffinose and the appropriate auxotrophy supplement mixture (QBiogene)], 200 rpm at 30 °C24. aSyn expression was induced by growing cells (OD600 nm 0.2) in SC selective medium 1% (w/v) galactose (aSyn ON) not supplemented or with 62 μg GAE.mL−1 leaf or fruit PDMs, for 6 h at 30 °C, 200 rpm. For spot assays D600 nm of treated cells was set to 0.1 ± 0.005 and 1:10 serially dilutions were prepared24. Then, 4 µL of each dilution was spotted in solid SC solid selective medium 1% (w/v) galactose and incubated at 30 °C, 42 h. Images were acquired using ChemidocTM XRS and Image Lab software. For clearance experiments, after 6 h of aSyn expression induction (with or without LPDMs treatment), cells were centrifuged, washed in PBS, resuspended in 2% (w/v) glucose SC liquid medium (aSyn expression OFF) and incubated at 30 °C, 200 rpm, 18 h. aSyn levels were determined by western blotting at 6 h of induction (corresponding to 0 h of clearance) and 18 h of clearance.
H4 cell culture and transfections
Human H4 neuroglioma cells (gift from Dr. Bradley T. Hyman, Harvard Medical School) were maintained at 37 °C in OPTI-MEM I (Gibco, Invitrogen, Barcelona, Spain) supplemented with 10% (v/v) fetal bovine serum and seeded at 80,000 cells.cm−2 density 24 h prior to transfection. Cells were transfected as previously described with pcDNA3.1-aSynT and pcDNA3.1-Synphilin-148,49. For H2O2 toxicity assay cells were treated with 2 μg GAE. mL−1 A. unedo LPDMs for 16 h, the medium was removed and cells were treated with 600 µM H2O2 for 6 h, viability was assessed using PI and the percentage of cells labeled with PI is presented (gated cells). To evaluate the effect of LPDMs in aSyn aggregation, 24 h after transfection cells were treated with 2 μg GAE. mL−1 LPDMs for 16 h.
Fluorescence microscopy
To determine the percentage of yeast cells with aSyn inclusions, cells were grown as described above and GFP fluorescence was visualized, the percentage of cells presenting aSyn inclusions was determined by counting at least 800 cells for each treatment. Transfected H4 cells were fixed and permeabilized with methanol and blocked in 1.5% (v/v) normal goat serum in PBS for 1 h. Cells were incubated with primary antibody overnight at 4 °C (mouse anti-aSyn; BD Transduction Laboratories, San Jose, CA, USA) followed by secondary antibody incubation for 1 h (goat anti-mouse IgG-Alexa488, Invitrogen Corporation, Carlsbad, CA, USA). For mitochondria assays, cells were labeled with 200 nM MitoTracker Red FM (Invitrogen Corporation, Carlsbad, CA, USA), for 30 min at 37 °C protected from light. To stain the nucleus 4 µg. mL−1 of DAPI (Sigma-Aldrich, St. Louis, MO, USA) was used. The proportion of cells with aSyn inclusions within the population was then determined by counting at least 100 cells per condition. Slides were subjected to fluorescence microscopy with a Zeiss Axiovert 200 M Widefield Fluorescence microscope and the counting was performed using ImageJ software.
Western blotting
For aSyn and GFP-Atg8 quantification total yeast protein extraction was performed using the method described69. Atg8 induction was quantified by the determination of the fold increase of total GFP signal (GFP-Atg8 and free GFP signal, detected with anti-GFP) normalized to PGK; autophagic flux was quantified by measuring the vacuolar degradation of the Atg8 domain reporter (ratio of free GFP to total GFP signal)40. For aSyn quantification in H4 cells, cells were lysed with NP-40 lysis buffer in the presence of protease and phosphatase inhibitor cocktail (Roche, Mannheim, Germany), western blot was performed following standard procedures. Antibodies used: aSyn (BD Transduction Laboratories, San Jose, CA, USA), GAPDH (Ambion, Cambridgeshire, UK), GFP (Antibodies Incorporated, Davis, CA, USA), PGK (Life Technologies, Paisley, UK). The western blots shown are representative of three independent experiments with similar outcomes, densitometry is measured from the western blot panel shown. The protein bands were quantified using ImageJ software for statistical analysis.
Sucrose gradients
Total protein was obtained from cells expressing aSyn and applied on a 5 to 30% (w/v) sucrose gradient as described before37. Fractions were collected, precipitated for 4 h at 4 °C in trichloroacetic acid, washed in acetone three times and suspended in protein sample buffer (0.5 M Tris-HCl, pH 6.8, 10% (w/v) glycerol, 0.1% (w/v) SDS, 6 mM bromophenol blue). Proteins were resolved by SDS-PAGE, the estimation of the molecular sizes for each fraction was previously described70. The western blots shown are representative of three independent experiments with similar outcomes, densitometry is measured from the western blot panel shown. The protein bands were quantified using ImageJ software for statistical analysis.
Flow cytometry (FCM)
FCM was performed in a FACS BD LSR Fortessa, equipped with the 695/40 BP and the 685 LP, as previously described24. ROS was determined in yeast cells transformed with the aSyn encoding plasmid or the empty plasmid, incubated with 30 µM DHE (dihydroethidium), for 15 min at 30 °C, 200 rpm. For cell viability cells transformed with the aSyn-GFP encoding plasmid or the empty plasmid were incubated with PI 5 µg.mL−1, for 15 min at 30 °C, 200 rpm. To study the proteasome, yeast cells were transformed with the aSyn and GFPu encoding plasmid or the respective empty plasmid. For mitochondria assays in H4 cells, cells were labeled with 200 nM MitoTracker Red FM, for 30 min at 37 °C. Data analysis was performed using FlowJo software. A minimum of 10000 events were collected for each experiment. Results were expressed as median fluorescence intensity (MFI) of a molecule. Image J was used for cropping and quantification of bands.
Real-time PCR
Quantitative real-time PCR (qRT-PCR) analyzes was performed as described in43. The oligonucleotides listed in Supplementary Table 1 were used to evaluate expression of SOD1, SOD2, HAP4, LHS1, KAR2, HRD1, GSH1, GLR1, RPN4, SIR2, and ATG8 genes. Relative standard curves were constructed for each gene, using triplicate serial dilutions of cDNA. The relative expression of the genes was calculated by the relative quantification method with efficiency correction, using Applied Biosystems® ViiA™ 7 software. ACT1, PDA1 and PGK were used as internal standards and for the normalization of mRNA expression levels43. The results were expressed as fold change mRNA levels relative to the control condition and normalized to the reference genes (relative expression).
Glutathione (GSH) and glutathione disulphide (GSSG) quantification
Yeast cells were grown as previously described and GSH and GSSG were quantified by HPLC after sample derivatization, as described65.
Statistical analysis
The results reported in this work are the average of at least three independent biological replicates and are represented as the mean ± SD. Differences among treatments were assessed by analysis of variance with Tuckey HSD (Honestly Significant Difference) multiple comparison test (a = 0.05) using SigmaStat 3.10.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Mecocci, P., Tinarelli, C., Schulz, R. J. & Polidori, M. C. Nutraceuticals in cognitive impairment and Alzheimer’s disease. Frontiers in Pharmacology 5, 147, https://doi.org/10.3389/fphar.2014.00147 (2014).
Del Rio, D. et al. Dietary (poly)phenolics in human health: structures, bioavailability, and evidence of protective effects against chronic diseases. Antioxid Redox Signal 18, 1818–1892, https://doi.org/10.1089/ars.2012.4581 (2013).
Afkir, S. et al. Arbutus unedo prevents cardiovascular and morphological alterations in L-NAME-induced hypertensive rats - Part 1: Cardiovascular and renal hemodynamic effects of Arbutus unedo in L-NAME-induced hypertensive rats. Journal of Ethnopharmacology 116, 288–295, https://doi.org/10.1016/j.jep.2007.11.029 (2008).
Carcache-Blanco, E. J. et al. Potential cancer chemopreventive agents from Arbutus unedo. Natural Product Research 20, 327–334, https://doi.org/10.1080/14786410500161205 (2006).
DeFeudis, F. V. & Drieu, K. Ginkgo biloba extract (EGb 761) and CNS functions: basic studies and clinical applications. Current Drug Targets 1, 25–58, https://doi.org/10.2174/1389450003349380 (2000).
Mandel, S. & Youdim, M. B. Catechin polyphenols: neurodegeneration and neuroprotection in neurodegenerative diseases. Free Radical Biology and Medicine 37, 304–317, doi:S0891584904003168 (2004).
Alminger, M. et al. In vitro models for studying secondary plant metabolite digestion and bioaccessibility. Comprehensive Reviews in Food Science and Food Safety 13, 413–436, https://doi.org/10.1111/1541-4337.12081 (2014).
Janle, E. M. et al. Pharmacokinetics and tissue distribution of 14C-labeled grape polyphenols in the periphery and the central nervous system following oral administration. J Med Food 13, 926–933, https://doi.org/10.1089/jmf.2009.0157 (2010).
Andres-Lacueva, C. et al. Anthocyanins in aged blueberry-fed rats are found centrally and may enhance memory. Nutr Neurosci 8, 111–120, https://doi.org/10.1080/10284150500078117 (2005).
Joseph, J. A. et al. Reversals of age-related declines in neuronal signal transduction, cognitive, and motor behavioral deficits with blueberry, spinach, or strawberry dietary supplementation. J Neurosci 19, 8114–8121 (1999).
von Campenhausen, S. et al. Prevalence and incidence of Parkinson’s disease in Europe. European neuropsychopharmacology: the journal of the European College of Neuropsychopharmacology 15, 473–490, https://doi.org/10.1016/j.euroneuro.2005.04.007 (2005).
Lashuel, H. A., Overk, C. R., Oueslati, A. & Masliah, E. The many faces of alpha-synuclein: from structure and toxicity to therapeutic target. Nat Rev Neurosci 14, 38–48, https://doi.org/10.1038/nrn3406 (2013).
Lee, J., Giordano, S. & Zhang, J. Autophagy, mitochondria and oxidative stress: cross-talk and redox signalling. Biochemical Journal 441, 523–540, https://doi.org/10.1042/BJ20111451 (2012).
Hetz, C., Chevet, E. & Harding, H. P. Targeting the unfolded protein response in disease. Nature Reviews Drug Discovery 12, 703–719, https://doi.org/10.1038/Nrd3976 (2013).
Kozutsumi, Y., Segal, M., Normington, K., Gething, M. J. & Sambrook, J. The Presence of Malfolded Proteins in the Endoplasmic-Reticulum Signals the Induction of Glucose-Regulated Proteins. Nature 332, 462–464, https://doi.org/10.1038/332462a0 (1988).
Ding, W. X. & Yin, X. M. Sorting, recognition and activation of the misfolded protein degradation pathways through macroautophagy and the proteasome. Autophagy 4, 141–150, https://doi.org/10.4161/auto.5190 (2008).
Yorimitsu, T., Nair, U., Yang, Z. & Klionsky, D. J. Endoplasmic reticulum stress triggers autophagy. J Biol Chem 281, 30299–30304, https://doi.org/10.1074/jbc.M607007200 (2006).
Engelender, S. alpha-synuclein fate: Proteasome or autophagy? Autophagy 8, 418–420, https://doi.org/10.4161/auto.19085 (2012).
Outeiro, T. F. & Lindquist, S. Yeast cells provide insight into alpha-synuclein biology and pathobiology. Science 302, 1772–1775, https://doi.org/10.1126/science.1090439302/5651/1772 (2003).
Cuervo, A. M., Stefanis, L., Fredenburg, R., Lansbury, P. T. & Sulzer, D. Impaired degradation of mutant alpha-synuclein by chaperone-mediated autophagy. Science 305, 1292–1295, https://doi.org/10.1126/science.1101738305/5688/1292 (2004).
Chu, C. T. Diversity in the regulation of autophagy and mitophagy: lessons from Parkinson’s disease. Parkinson’s Disease 2011, 789431, https://doi.org/10.4061/2011/789431 (2011).
Petroi, D. et al. Aggregate clearance of alpha-synuclein in Saccharomyces cerevisiae depends more on autophagosome and vacuole function than on the proteasome. The Journal of biological chemistry 287, 27567–27579, https://doi.org/10.1074/jbc.M112.361865 (2012).
Tanik, S. A., Schultheiss, C. E., Volpicelli-Daley, L. A., Brunden, K. R. & Lee, V. M. Lewy body-like alpha-synuclein aggregates resist degradation and impair macroautophagy. The Journal of biological chemistry 288, 15194–15210, https://doi.org/10.1074/jbc.M113.457408 (2013).
Macedo, D. et al. (Poly)phenols protect from alpha-synuclein toxicity by reducing oxidative stress and promoting autophagy. Hum Mol Genet 24, 1717–1732, https://doi.org/10.1093/hmg/ddu585 (2015).
McDougall, G. J., Fyffe, S., Dobson, P. & Stewart, D. Anthocyanins from red wine - Their stability under simulated gastrointestinal digestion. Phytochemistry 66, 2540–2548, https://doi.org/10.1016/j.phytochem.2005.09.003 (2005).
Jardim, C. et al. (Poly)phenol metabolites from Arbutus unedo leaves protect yeast from oxidative injury by activation of antioxidant and protein clearance pathways. J Funct Foods 32, 333–346, https://doi.org/10.1016/j.jff.2017.03.010 (2017).
El Haouari, M., Lopez, J. J., Mekhfi, H., Rosado, J. A. & Salido, G. M. Antiaggregant effects of Arbutus unedo extracts in human platelets. Journal of Ethnopharmacology 113, 325–331, https://doi.org/10.1016/j.jep.2007.06.016 (2007).
Mariotto, S. et al. Aqueous extract of Arbutus unedo inhibits STAT1 activation in human breast cancer cell line MDA-MB-231 and human fibroblasts through SHP2 activation. Medicinal Chemistry 4, 219–228, https://doi.org/10.2174/157340608784325179 (2008).
Tenreiro, S., Munder, M. C., Alberti, S. & Outeiro, T. F. Harnessing the power of yeast to unravel the molecular basis of neurodegeneration. J Neurochem 127, 438–452, https://doi.org/10.1111/jnc.12271 (2013).
Giasson, B. I. et al. Oxidative damage linked to neurodegeneration by selective alpha-synuclein nitration in synucleinopathy lesions. Science 290, 985–989, doi:8948 (2000).
Flower, T. R., Chesnokova, L. S., Froelich, C. A., Dixon, C. & Witt, S. N. Heat shock prevents alpha-synuclein-induced apoptosis in a yeast model of Parkinson’s disease. J Mol Biol 351, 1081–1100, https://doi.org/10.1016/j.jmb.2005.06.060 (2005).
Arduino, D. M. et al. Mitochondrial metabolism in Parkinson’s disease impairs quality control autophagy by hampering microtubule-dependent traffic. Human molecular genetics 21, 4680–4702, https://doi.org/10.1093/hmg/dds309 (2012).
Hsu, L. J. et al. Alpha-synuclein promotes mitochondrial deficit and oxidative stress. The American journal of pathology 157, 401–410 (2000).
Liu, C. et al. Rehmanniae Radix in osteoporosis: A review of traditional Chinese medicinal uses, phytochemistry, pharmacokinetics and pharmacology. J Ethnopharmacol 198, 351–362, https://doi.org/10.1016/j.jep.2017.01.021 (2017).
Mamadalieva, N. Z., Sharopov, F., Girault, J. P., Wink, M. & Lafont, R. Phytochemical analysis and bioactivity of the aerial parts of Abutilon theophrasti (Malvaceae), a medicinal weed. Nat Prod Res 28, 1777–1779, https://doi.org/10.1080/14786419.2014.939080 (2014).
Fiorentino, A. et al. Polyphenols from the hydroalcoholic extract of Arbutus unedo living in a monospecific Mediterranean woodland. Biochemical Systematics and Ecology 35, 809–811, https://doi.org/10.1016/j.bse.2007.04.005 (2007).
Tenreiro, S. et al. Phosphorylation modulates clearance of alpha-synuclein inclusions in a yeast model of Parkinson’s disease. PLoS Genetics 10, e1004302, https://doi.org/10.1371/journal.pgen.1004302 (2014).
McNaught, K. S. & Jenner, P. Proteasomal function is impaired in substantia nigra in Parkinson’s disease. Neuroscience Letters 297, 191–194, https://doi.org/10.1016/S0304-3940(00)01701-8 (2001).
Suzuki, K. et al. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO Journal 20, 5971–5981, https://doi.org/10.1093/emboj/20.21.5971 (2001).
Klionsky, D. J. et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 12, 1–222, https://doi.org/10.1080/15548627.2015.1100356 (2016).
Xie, Y. M. & Varshavsky, A. RPN4 is a ligand, substrate, and transcriptional regulator of the 26S proteasome: A negative feedback circuit. Proceedings of the National Academy of Sciences of the United States of America 98, 3056–3061, https://doi.org/10.1073/pnas.071022298 (2001).
Ng, D. T. W., Spear, E. D. & Walter, P. The unfolded protein response regulates multiple aspects of secretory and membrane protein biogenesis and endoplasmic reticulum quality control. Journal of Cell Biology 150, 77–88, https://doi.org/10.1083/Jcb.150.1.77 (2000).
Sampaio-Marques, B. et al. SNCA (alpha-synuclein)-induced toxicity in yeast cells is dependent on sirtuin 2 (Sir2)-mediated mitophagy. Autophagy 8, 1494–1509, https://doi.org/10.4161/auto.21275 (2012).
Ivy, J. M., Klar, A. J. S. & Hicks, J. B. Cloning and characterization of 4 Sir genes of Saccharomyces cerevisiae. Molecular and Cellular Biology 6, 688–702 (1986).
Subramaniam, S. R. & Chesselet, M. F. Mitochondrial dysfunction and oxidative stress in Parkinson’s disease. Progress in Neurobiology 106, 17–32, https://doi.org/10.1016/j.pneurobio.2013.04.004 (2013).
Forsburg, S. L. & Guarente, L. Identification and Characterization of Hap4 - a 3rd Component of the Ccaat-Bound Hap2 Hap3 Heteromer. Genes & Development 3, 1166–1178, https://doi.org/10.1101/Gad.3.8.1166 (1989).
Grant, C. M. Role of the glutathione/glutaredoxin and thioredoxin systems in yeast growth and response to stress conditions. Molecular Microbiology 39, 533–541, https://doi.org/10.1046/j.1365-2958.2001.02283.x (2001).
McLean, P. J., Kawamata, H. & Hyman, B. T. Alpha-synuclein-enhanced green fluorescent protein fusion proteins form proteasome sensitive inclusions in primary neurons. Neuroscience 104, 901–912, https://doi.org/10.1016/S0306-4522(01)00113-0 (2001).
Klucken, J. et al. Alpha-synuclein aggregation involves a bafilomycin A 1-sensitive autophagy pathway. Autophagy 8, 754–766, https://doi.org/10.4161/auto.19371 (2012).
Miguel, M. G., Faleiro, M. L., Guerreiro, A. C. & Antunes, M. D. Arbutus unedo L.: chemical and biological properties. Molecules 19, 15799–15823, https://doi.org/10.3390/molecules191015799 (2014).
Yu, L. et al. Neuroprotective effect of kaempferol glycosides against brain injury and neuroinflammation by inhibiting the activation of NF-kappaB and STAT3 in transient focal stroke. PLoS One 8, e55839, https://doi.org/10.1371/journal.pone.0055839 (2013).
Masuda, M. et al. Small molecule inhibitors of alpha-synuclein filament assembly. Biochemistry 45, 6085–6094, https://doi.org/10.1021/bi0600749 (2006).
Tavares, L. et al. Neuroprotective effect of blackberry (Rubus sp.) polyphenols is potentiated after simulated gastrointestinal digestion. Food Chemistry 131, 1443–1452, https://doi.org/10.1016/j.foodchem.2011.10.025 (2012).
Okamura, K., Kimata, Y., Higashio, H., Tsuru, A. & Kohno, K. Dissociation of Kar2p/BiP from an ER sensory molecule, Ire1p, triggers the unfolded protein response in yeast. Biochemical and Biophysical Research Communications 279, 445–450, https://doi.org/10.1006/bbrc.2000.3987 (2000).
Baxter, B. K., James, P., Evans, T. & Craig, E. A. SSI1 encodes a novel Hsp70 of the Saccharomyces cerevisiae endoplasmic reticulum. Molecular and Cellular Biology 16, 6444–6456 (1996).
Burnie, J. R., Carter, T. L., Hodgetts, S. J. & Matthews, R. C. Fungal heat-shock proteins in human disease. Fems Microbiology Reviews 30, 53–88, https://doi.org/10.1111/j.1574-6976.2005.00001.x (2006).
Kaneko, M., Ishiguro, M., Niinuma, Y., Uesugi, M. & Nomura, Y. Human HRD1 protects against ER stress-induced apoptosis through ER-associated degradation. FEBS Letters 532, 147–152, https://doi.org/10.1016/S0014-5793(02)03660-8 (2002).
Su, L. J. et al. Compounds from an unbiased chemical screen reverse both ER-to-Golgi trafficking defects and mitochondrial dysfunction in Parkinson’s disease models. Disease Models and Mechanisms 3, 194–208, https://doi.org/10.1242/dmm.004267 (2010).
Longo, V. D., Gralla, E. B. & Valentine, J. S. Superoxide dismutase activity is essential for stationary phase survival in Saccharomyces cerevisiae - Mitochondrial production of toxic oxygen species in vivo. Journal of Biological Chemistry 271, 12275–12280, https://doi.org/10.1074/jbc.271.21.12275 (1996).
Miao, L. & St Clair, D. K. Regulation of superoxide dismutase genes: implications in disease. Free Radic Biol Med 47, 344–356, https://doi.org/10.1016/j.freeradbiomed.2009.05.018 (2009).
Jahromi, S. R., Haddadi, M., Shivanandappa, T. & Ramesh, S. R. Attenuation of neuromotor deficits by natural antioxidants of Decalepis hamiltonii in transgenic Drosophila model of Parkinson’s disease. Neuroscience 293, 136–150, https://doi.org/10.1016/j.neuroscience.2015.02.048 (2015).
Casani, S., Gomez-Pastor, R., Matallana, E. & Paricio, N. Antioxidant compound supplementation prevents oxidative damage in a Drosophila model of Parkinson’s disease. Free Radical Biology and Medicine 61, 151–160, https://doi.org/10.1016/j.freeradbiomed.2013.03.021 (2013).
Lee, D. H., Kim, C. S. & Lee, Y. J. Astaxanthin protects against MPTP/MPP plus -induced mitochondrial dysfunction and ROS production in vivo and in vitro. Food and Chemical Toxicology 49, 271–280, https://doi.org/10.1016/j.fct.2010.10.029 (2011).
He, C. C. & Klionsky, D. J. Regulation Mechanisms and Signaling Pathways of Autophagy. Annual Review of Genetics 43, 67–93, https://doi.org/10.1146/annurev-genet-102808-114910 (2009).
Tavares, L. et al. Neuroprotective effects of digested polyphenols from wild blackberry species. European Journal of Nutrition 52, 225–236, https://doi.org/10.1007/s00394-012-0307-7 (2013).
McDougall, G. J. et al. Tracking (Poly)phenol components from raspberries in ileal fluid. J Agric Food Chem 62, 7631–7641, https://doi.org/10.1021/jf502259j (2014).
Gietz, D., St Jean, A., Woods, R. A. & Schiestl, R. H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Research 20, 1425 (1992).
Mumberg, D., Muller, R. & Funk, M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156, 119–122, doi:0378111995000377 (1995).
Cheong, H. et al. Atg17 regulates the magnitude of the autophagic response. Molecular Biology of the Cell 16, 3438–3453, https://doi.org/10.1091/mbc.E04-10-0894 (2005).
Tanese, N. Small-scale density gradient sedimentation to separate and analyze multiprotein complexes. Methods-A Companion to Methods in Enzymology 12, 224–234, https://doi.org/10.1006/meth.1997.0475 (1997).
Acknowledgements
We acknowledge Rita Ramos for support with qRT-PCR and Regina Menezes for the selection of primers; António Temudo and Ana M. Nascimento for imaging support; The IMM-JLA Flow Cytometry Facility. We also thank Prof. Kuninori Suzuki (Tokyo Institute of Technology, Yokohama, Japan) for the 2xmCherry-ATG8 plasmid. TFO is supported by the DFG Center for Nanoscale Microscopy and Molecular Physiology of the Brain. This work was supported by Fundação para a Ciência e Tecnologia [iNOVA4Health: UID/Multi/04462/2013, SFRH/BD/73429/2010 and IMM/BI/78-2017 to DM, SFRH/BD/86584/2012 to IF, IF/01097/2013 to CNS, SFRH/BPD/35767/2007 and SFRH/BPD/101646/2014 to ST]. BacHBerry FP7 KBBE-2013-7 613793 to CNS, DM and CJ, Marie Curie International Reintegration Grant and an EMBO Installation Grant to TFO. TFO is supported by the DFG Center for Nanoscale Microscopy and Molecular Physiology of the Brain. The author(s) would like to acknowledge the STSM to AFA and networking support by the COST Action FA 1403 POSITIVe (Interindividual variation in responseto consumption of plant food bioactives and determinants involved), supported by COST (European Cooperation in Science and Technology).
Author information
Authors and Affiliations
Contributions
D.M. preformed the majority of experiments; C.J. determined GSSG and GSH levels; I.F., G.J.M. and D.S. performed and analysed the LC-MS characterization of A. unedo; A.F.A., J.E.Y. and F.T.B. performed the UPLC-MS-NMR analysis and interpretation; D.M., S.T., T.F.O., C.N.S. designed the study and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Macedo, D., Jardim, C., Figueira, I. et al. (Poly)phenol-digested metabolites modulate alpha-synuclein toxicity by regulating proteostasis. Sci Rep 8, 6965 (2018). https://doi.org/10.1038/s41598-018-25118-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-25118-z
This article is cited by
-
Natural Phenolic Compounds with Neuroprotective Effects
Neurochemical Research (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.