Abstract
Bacterial chromosomes harbour a unique origin of bidirectional replication, oriC. They are almost always circular, with replication terminating in a region diametrically opposite to oriC, the terminus. The oriC-terminus organisation is reflected by the orientation of the genes and by the disposition of DNA-binding protein motifs implicated in the coordination of chromosome replication and segregation with cell division. Correspondingly, the E. coli and B. subtilis model bacteria possess a replication fork trap system, Tus/ter and RTP/ter, respectively, which enforces replication termination in the terminus region. Here, we show that tus and rtp are restricted to four clades of bacteria, suggesting that tus was recently domesticated from a plasmid gene. We further demonstrate that there is no replication fork system in Vibrio cholerae, a bacterium closely related to E. coli. Marker frequency analysis showed that replication forks originating from ectopic origins were not blocked in the terminus region of either of the two V. cholerae chromosomes, but progressed normally until they encountered an opposite fork. As expected, termination synchrony of the two chromosomes is disrupted by these ectopic origins. Finally, we show that premature completion of the primary chromosome replication did not modify the choreography of segregation of its terminus region.
Similar content being viewed by others
Introduction
Genome architecture is extremely conserved in bacteria. Bacterial genomes are typically organised on a single chromosome, which rarely exceeds 10 Mbp and which is almost always circular1. Bacterial replication is initiated bidirectionally at a unique origin of replication per chromosome, oriC. Provided that they progress at a similar velocity, two replication forks originating from oriC converge in a zone diametrically opposite to it, which is called the terminus (Ter). Completion of replication at a precise place and time seems to participate in the coordination of the different cellular processes: replication, segregation and cell division2. It was notably illustrated by the observation that the termini of the different circular chromosomes harboured by bacteria with a multipartite genome, such as Vibrio cholerae, are replicated synchronously at mid-cell3,4,5,6,7. The system allowing the synchrony of termination of the two chromosome of V. cholerae is based on the fact that the secondary chromosome (chr2) initiates replication only when crtS, a non-coding locus located 690 kb downstream of oriC1, between VC764 and VC765 genes, on the primary chromosome (chr1) has been replicated8,9,10,11.
Ter is functionally defined via hundreds of small strategically-located DNA motifs2,12. First, FtsK-Orienting Polar Sequences (KOPS) and SpoIIIE-Recogntition Sequences (SRS) are distributed all over the unique chromosome of E. coli and B. subtillis and over the two chromosomes of V. cholerae. KOPS and SRS are skewed octameric motifs directing DNA transaction towards the Ter region, which contributes to the completion of chromosome segregation13,14,15,16,17,18. They notably bring together sister copies of a highly conserved 28 bp motif located in the middle of the Ter region, dif, which contributes to sister chromosome decatenation and enable the resolution of chromosome dimers19,20,21,22,23. Second, E. coli, V. cholerae and B. subtilis contain an inhibitor of cell division, either SlmA or Noc, which prevents the assembly/ stabilisation of a septum ring over the bulk of the nucleoid. SlmA and Noc bind to specific motifs, which are located all over the chromosomes but excluded from the Ter. As a result, septum formation only occurs when chromosomes have been segregated and is precisely located at mid-cell where the Ter is maintained24,25,26,27,28. Third, MatP, which binds to specific DNA motifs in the Ter region, contributes to the coordination of cell division and chromosome segregation by maintaining the two sister copies of the terminus regions together at mid-cell at the time of division in E. coli and in V. cholerae29,30,31. In E. coli, the MatP/matS complexes also stabilises the FtsZ-ring assembly via its direct interaction with the cell division protein ZapB31,32. Hence, the coordination between chromosome segregation and cell division benefits from the completion of DNA synthesis at Ter.
In the absence of replication problems, the two independent replication forks originating from the unique origin of bacterial circular chromosomes naturally converge in the Ter region33,34. However, several challenging situations require a replication fork trap (RFT) system to force termination within the Ter region. Replication initiated at ectopic positions such as those occupied by prophages or those resulting from proposed over-replication are a few examples35. Cells may also benefit from an RFT preventing a replication fork to progress on the illegitimate replichore if the other fork is prematurely halted or inactivated36,37. The drastic loss of viability of mutant cells unable to reactivate replication forks in E. coli suggests that the occurrence of such fork impediments is not rare, especially in rich medium38. Protein-DNA complexes are a major source of replication fork pausing in E. coli39. RFT may also circumvent deleterious head-to-head collisions occurring between replication forks and transcription bubbles by preventing replication forks to proceed replication beyond Ter. Indeed, the transcription of highly expressed genes on each replichore is oriented in the direction of the replication fork, which is supposed to ensure a smooth and processive synthesis of DNA40.
Two unrelated RFT systems were described so far, ter/Tus in E. coli and ter/ RTP in B. subtilis41,42,43. Both systems are based on two types of components and operate similarly. Surrounding the Ter region, several copies of ter-sites, which are non-palindromic sequences, point toward the dif site. The Tus or RTP proteins bind in an oriented manner on these ter-sites in such a way that only the forks progressing from, but not towards, dif are blocked44,45. The efficiency of blockage depends on each ter site46,47 and on the replication fork speed48. In laboratory conditions, inactivation of these systems does not generate strong phenotypes42. However, the fitness is slightly reduced in E. coli tus mutant cells containing an additional ectopic replication origin (oriZ): oriC-oriZ tus cells have a doubling time of 21.6 min compared to the 20.5 min doubling time of oriC–oriZ cells. These observations suggested that completion of replication within the Ter region participated in the processivity of replication fork progression49. The RFT effect on replication fork progression was monitored in E. coli using Marker Frequency Analysis (MFA) in strains harbouring an additional ectopic origin: in presence of an RFT, the convergence between the two forks continued to be in the Ter region, while in its absence (tus mutant), the convergence point was displaced to the midpoint between the two origins36,49. However, no homologous copy of the E. coli tus or the B. subtilis rtp genes was found in its genome with a BLAST-P search, even though V. cholerae contains homologues of all the other elements differentiating Ter from the rest of the chromosome in E. coli, including SlmA-binding sites, matS, KOPS and dif.
Here, we investigated the possible existence of a replication fork trap (RFT) in V. cholerae. We confirmed using a Hidden-Markov Model (HMM) approach that V. cholerae possessed neither tus nor rtp. Our phylogenetic observations further revealed that “resident” tus and rtp are limited to a narrow range of bacteria, which suggests that tus was recently domesticated from plasmid genes. To verify whether V. cholerae uses another yet unknown RFT system or not, we directly analysed the positions where forks converged in strains harbouring ectopic origins of replication. Our results demonstrate that there is no RFT on either of the two V. cholerae chromosomes. Finally, we show that the coordination between the segregation of the primary chromosome and cell division is not significantly impaired when Ter is not the last region to be replicated.
Results
tus and rtp are absent in the Vibrionales
We searched the UniProt database for the presence of protein sequences containing the Tus (PF05472) or the RTP (PF02334) HMM profiles. No RTP-related sequences were found outside of a subgroup of Bacillales (data not shown). Similarly, we identified tus genes only among γ-proteobacteriales strains (Fig. 1). Some tus genes were detected in the Vibrionales, like V. cholerae. A fraction is plasmid-borne while other, located on the chromosome, appears to be distributed erratically among the Vibrionales. This led us to define which among the tus genes are “resident” or “mobile” (see Supp. Information).
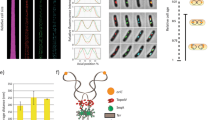
Tus-related homologs in bacteria. Phylogenetic tree of the proteins containing the Tus HHM profile (PF05472) found in complete genome of UniProt database and named as the sequenced species. Only three monophyletic groups of proteins were considered as “resident” as they present a distribution congruent with that of the species containing these Tus proteins established with a DnaABEX-based phylogenetic tree. Most Enterobacteriales (only a subset is indicated) highlighted in purple ①, the Pseudoalteromonas in which tus is present on the second chromosome highlighted in blue ②, and all the Aeromonadales, except Tolumonas aurensis TA4, highlighted in green ③. By opposition, the other tus-related genes, which do not meet the residency criteria explained in the main text, are considered as “mobile” genes. Some are plasmid-borne (highlighted in yellow) and other are located on chromosomes (not highlighted). Scale bar represents 0.1 substitutions per site; Bootstrap scores of domestication branches are indicated on the figure.
We identified three monophyletic groups of Tus proteins. The distribution of the Tus protein within each of these three groups is congruent with the phylogenetic tree based on the DnaABEX proteins of the species containing these Tus proteins. In addition, the genomic context of theses tus gene is conserved among each group. From these two criteria, we define these tus gene as resident. The three groups with a resident tus are: most Enterobacteriales (only a subset is indicated) highlighted in purple ①, the Pseudoalteromonas in which tus is present on the second chromosome highlighted in blue ②, and all the Aeromonadales, except Tolumonas aurensis TA4, highlighted in green ③ (Fig. 1). The other identified tus genes do not fulfil these criteria and were then defined as “mobile”. Their genomic location is not conserved and their distribution is not congruent with that of the species in which they were found. Indeed, they are frequently plasmid-borne (highlighted in yellow, Fig. 1).
The proposed position of tus domestication in the three distinct clades was deduced from the distribution of resident tus. The vast majority of Enterobacteriales contains tus (highlighted in purple, Fig. S1) and an analysis of the tus genomic context reveals that it is strictly conserved (data not shown). A monophyletic group of species, containing mostly endosymbionts, might have lost tus in the course of evolution. Finally, we noticed that tus is absent in Plesiomonas shigelloides, a bacterium that split early in the Enterobacteriales (Fig. S1). Altogether, these observations suggest that tus was acquired and domesticated shortly after the origination of the Enterobacteriales (①, Figs 1, S1). The second clade containing tus regroups the Pseudoaleromonadacae (②, Figs 1, S1). These species are atypical Alteromonadales since they contain a second chromosome of plasmid origin. Within the Pseudoaleromonadacae, tus is strictly and systematically associated with the secondary chromosome, suggesting that the gene became resident during the domestication of the plasmid into a chromosome. Interestingly, in the Pseudoaleromonadacae tus is systematically located next to the origin of replication. Additionally, tus was identified within all Aeromonadales except Tolumonas aurensis TA4 where it might have been lost (③, Figs 1, S1). We conclude from this phylogenomic analysis that there is neither resident tus nor rtp in Vibrionales and in most bacterial orders.
A marker frequency analysis method to define fork convergence points
In exponentially growing cells, forks originating from the same origin of replication progress at similar speed and converge in a region precisely opposite to the origin. In other word, the fork convergence point (fcp) corresponds to the midpoint of the fragment replicated (mp). It is noteworhy that, using replication-synchronised conditions, the two sides of the E. coli oriC or of the V. cholerae oriC1 origins were shown to present an asymmetry of replication start leading to displacement of the expected fcp from the mp7,33. However, we could not observe any displacement in our data of WT situations (Figs 2, 3). In contrast, because of the presence of the RFT in E. coli, the fcp may significantly differ from the mp when the origin of replication is displaced or when an extra origin of replication is added36,49,50. Hence, this fcp, displaced from mp, would indicate the position of the RFT. Therefore, we decided to check whether the addition of extra-origins of replication could displace the fcp away from the mp on the two V. cholerae chromosomes. Without RFT, the coincidence between mp (midpoint between the two origins) and fcp is expected if the ectopic origins initiate replication synchronously with the endogenous origin. To this end, we developed an MFA method to quantify the deviation of the fcp of any two converging forks from the expected mp. For all the strains we studied, the marker frequencies decreased exponentially between the replication origins and their cognate fcp, as expected from the Cooper-Helmstetter model of replication (see Figs 2, 3;51). We could thus calculate the fcp between any two origins as the position that minimised the error between the log of the experimental marker frequency values and the theoretic values given by their linear regression (see Supp. Methods). In the case of cells carrying ectopic origins of replication, we noticed a slight deviation of the log of the experimental marker frequencies from the linear regression model around the calculated fcp. Since the fcp derived from MFA is the average position where replication forks converged in a population of exponentially growing cells, we corrected the model by considering that the frequency of the cells in which forks converged at any specific position formed a gaussian centred on fcp. The width of the gaussian minimising the error between the experimental data and the gaussian-corrected model indicates the size of the region centred on the fcp within which 95% of the forks converged, which we termed S95 (see Supp. Methods). In order to validate our MFA method, we used it to define the origin regions of our WT strains. In both replicates and using different sliding window sizes, we could localise the position of “origin” at less than 0.5% from the real oriC1 (see Supp. Methods).
MFA of the two-chromosome strains: V. cholerae N16961 (EPV50; A) and derivatives containing ectopic oriC1 ((EGV140; (B) and EGV111; C)). Marker frequencies (grey dots after trimming (see Supp. Methods) and normalisation on the total number of reads) are represented in Log2 as a function of the genome position. The oriC1 or oriC2 of chr1 or chr2, respectively, are indicated at each extremity. Position of dif1, dif2, ectopic origin if applicable (oriL3 or oriR4), crtS, the different mp (origins mid-point) are indicated. The lowest point on chr1 was set to “1” in such a way that log2(1) = “0” and all data were normalized to this point. The curve fitting the marker frequency data (see Supp. Methods for MFA method) are indicated by either a blue or a red line for chr1 and chr2, respectively. They define the forks convergence points (fcp), indicated under the data. On the left side of the marker frequency data, a scheme representing the program of replication of chr1 is indicated on the circular map of the different strains. The program of replication of chr2 is represented only for EPV50 as it is not modified in EGV140 and EGV111. The program of replication deduced from the MFA is as follow: plain grey line corresponds to the wild-type direction of fork progression and the dashed grey line to the reverse direction of fork progression. The distance between fcp and its mp (noted fcp-mp) is indicated in % of the replicon fraction, oriented from the first origin encounters in the clock-wise direction.
MFA of the mono-chromosome strains: V. cholerae MCH1 (A) and derivatives containing ectopic oriC1 (EGV369; (B) and EGV366; (C)). See Fig. 2 legend, adapted for the fusion between chr1 and chr2 (deleting dif1 region and oriC2 region).
V. cholerae chr1 lacks an RFT
We first applied our MFA method to EPV50, an El Tor N16961 V. cholerae strain, to EGV140, a derivative harbouring an additional copy of the origin of replication of chr1 (oriC1) at 0.650 Mbp (position called L3 previously52) from oriC1 on the left replichore of chr1 (oriL3) and to EGV111, a derivative harbouring an additional copy of oriC1 at 1.190 Mpb (position called R4 previously52) from oriC1 on the right replichore (oriR4). The presence of the ectopic origins did not affect considerably the generation time in rich and minimal medium (Table S2).
The MFA of exponentially growing EPV50 cells in minimal medium showed that the fcp of chr1, fcp1, was only ~45 kb away from dif1 and deviated from the mp of the replicon, mp1, by only 0.1% of the replicon length (Fig. 2A and Table S4). The MFA of exponentially growing EGV140 and EGV111 cells in minimal medium presented a drastically modified chr1 profile (Fig. 2B,C). Consistent with synchronous initiation of replication at the two origins of chr1, the marker frequencies of the oriC1 and oriL3 loci were the highest and were roughly identical (Fig. 2B and Table S2). The fcp of the right replicon (fcpR) was moved ~320 kb away from fcp1 (Fig. 2B and Table S4). It was only at 14 kbp from the midpoint of the right replicon, mpR, which corresponds to a deviation of 0.6% of the replicon length (Fig. 2B and Table S4). The fcp observed on the left replicon, fcpL, was about 9 kb from the midpoint of the replicon, mpL, corresponding to a 1.6% deviation (Fig. 2B and Table S4). Similarly, replication was initiated synchronously at the two chr1 origins of EGV111, oriC1 and oriR4 (Fig. 2C and Table S2). The fcp of the right replicon, fcpR, was only at 1 kbp from the midpoint of this replicon, mpR, corresponding to a 0.2% deviation (Fig. 2C and Table S4). The fcp of the left replicon, fcpL, was at 9 kbp from the midpoint of the replicon, mpL, corresponding to a 0.8% deviation (Fig. 2C and Table S4). Thus, a replication fork originating from oriR4 can progress normally past fcp1 and dif1 over 585 kb up to fcpL of EGV111, corresponding to ~40% of the left replichore. A replication fork can also progress over 810 kb without perturbation from oriR4 toward the fcpR of EGV111. With the addition to the 320 kb already tested in EGV140, it corresponds to a total of ~60% of the right replichore (Fig. 2B,C). Taken together, these results demonstrate that there is no RFT on chr1.
Additional origins of replication affect termination synchrony of chr1 and chr2
The MFA of exponentially growing EPV50 cells showed that the fcp of chr2, fcp2, was only ~30 kbp away from dif2 and deviated from the mp of the replicon, mp2, by only 0.04% of the replicon length (Fig. 2A and Table S4). The presence of oriL3 and oriR4 on chr1 did not affect significantly the fcp on chr2 (Fig. 2B,C and Table S4). However, it perturbed the synchrony of termination of chr1 and chr2. The synchrony of termination of the two chromosomes is due to a time-delay between the initiation of chr1 and chr2 replication, chr2 replication initiation requiring duplication of a specific DNA motif adequately located on chr1, crtS7,8,9,53.
In EGV140, the crtS locus is located at about 45 kb from oriL3. It is replicated soon after chr1 replication initiation and in agreement with the function of crtS, chr2 replication is initiated early and the marker frequency of oriC2 is quite similar to the marker frequency oriL3 (Fig. 2B and Table S2). As a result, the marker frequency of fcp2 is significantly higher than the marker frequency of the fcpR indicating that chr2 replication terminates ahead of chr1 replication (Figs 2B, S4 and Table S2). Reciprocally, in the strain containing oriR4, the replication of crtS is delayed since the sequence is located close to fcpL, the last region of chr1 to be replicated, and the marker frequency of oriC2 is similar to the marker frequency of fcpL (Fig. 2C and Table S2). As result, chr2 replication terminates well after chr1 has been replicated (Figs 2C, S4 and Table S2).
Progression on an illegitimate replichore slightly perturbs replication
In EGV140, the fcp of the left replicon, fcpL, was shifted 9 kbp closer to the ectopic origin than expected at position mpL. In addition, we noticed that the marker frequencies immediately on the left and right of oriL3 and the slopes of the oriL3-fcpL and oriL3-fcpR regression lines were markedly different (Fig. 2B and Table S2). A likely explanation for these observations was the presence of a ribosomal RNA genes (rrn) operon in oriL3-fcpL replichore, which is transcribed in a direction opposite to that of the replication (Fig. 2B, double arrow head). Indeed, previous work indicated that head-on collisions between replication forks and transcription bubbles could result in fork stalling and/or DNA degradation-dependent repair37. In agreement with this hypothesis, marker frequencies were significantly lower on the right of oriL3 than its left in fast growing cells where the transcription of the rrn operon is expected to be stronger (Figs S5, S6 and Table S2). Moreover, the fcpL was closer to oriL3, now deviating from mpL by 11.4% (Fig. S6 and Table S4). No such dramatic deviation of the fcp from the mp was observed for the right replicon of EGV140 and the two replicons of EGV111 (Fig. 2B,C, Table S4). However, under fast growth conditions, the fcp was always situated on the side of the mp that minimised replication on illegitimate replichores (Figs S5–S7 and Table S4). In addition, S95, the length of the region centred on the fcp within which 95% of forks converged, increased from 4 kbp for the fcp1 of EPV50 to 128 kbp for the fcpL of EGV140, 628 kpb for the fcpR of EGV140, 396 kbp for the fcpL of EGV111 and 176 kbp for the fcpR of EGV111 (Fig. 2 and Table S4). Together, these results suggest that replication is slightly perturbed when ectopic origins were added.
V. cholerae chr2 lacks an RFT
The Ter of chr2 (Ter2) appears to contain all the attributes of Ter1 such as the underrepresentation of the SlmA-binding sites, the presence of matS sites, the convergence of the KOPS toward a dif site20,24,29,30. To investigate the presence of an RFT on chr2, we added an additional ectopic origin at two positions in a mono-chromosomal V. cholerae strain, MCH154, in which the two replicons are fused to each other through the replacement of the dif1 region of chr1 by the entire sequence of chr2, excepting the oriC2 and partition machinery region (Fig. 3A). The MFA of MCH1 showed that replication initiated at oriC1 and that replication forks progressed across chr1 and chr2 DNA to converge close to dif2. The fcp was at 13 kb away from the midpoint, corresponding to a deviation of ~0.3% of the replicated DNA segment. Marker frequency slopes decreased exponentially at the same rate on each replication arm (Fig. 3A).
The marker frequency profiles of the MCH1 strains containing oriL3 (EGV369) or oriR4 (EGV366) indicated that replication initiated at both the native and the ectopic origin. In EGV369, we noticed a reduction in marker frequency associated with the right replication fork, emerging from oriL3, which is probably caused by the transcription of the rrnE operon. The marker frequencies were noisy and we did not trust any fitting over the corresponding replicon. However, it did not prevent the analysis of the other replicon of EGV369. Likewise, the unexpectedly lower marker frequency at oriC1 than at oriR4 in EGV366 did not prevent the analysis of the fcp of the two replicons. In EGV369, the fcpR was located 360 kb away from dif2 (Fig. 3B) and in EGV366, the fcpL was located 640 kb away from it (Fig. 3C). Both fcp were located close to their corresponding midpoint, at less than 1.5% of each replicated DNA segment (Fig. 3B,C, Table S5). These results show that replication forks may progress beyond dif2 over ~67% of the left replichore of chr2 and over the entire right replichore of chr2. Taken together, these data led us to conclude that no RFT restricts the completion of replication within Ter1 or Ter2.
Impacts of a premature replication of Ter
Because of the presence of oriR4, dif1 is replicated 600 kbp before the fcpL in EGV111. Similarly, because of oriL3, dif1 is replicated about 350 kbp before the fcpR in EGV140. Using strains with fluorescent tags allowing to visualise dif1 and dif2 foci, the fraction of cells with duplicated dif1 and the localisation of dif1 as a function of cell length were analysed in oriL3- and oriR4-containing strains (EGV362 and EGV361, respectively) and compared to the corresponding WT strain, EGV360 (Fig. 4). The cell length distribution of the 3 strains were similar (Fig. 4A inset). We observed a higher proportion of cells with two dif1 foci in the 2.5 to 4.2 µm sizes of strains EGV361 and EGV362 than in the WT strain EGV360 (Figs 4A, S4). In agreement with the marker frequency data, it suggested that dif1 replication occurred earlier in the cell cycle in EGV361 and EGV362 cells than it did in WT EGV360 cells. More importantly, dif1 loci still globally behaved like genuine terminus DNA in EGV362 and EGV361 strains: in most of the cases, dif1 foci only duplicated after the mobilisation of the focus to mid-cell (Fig. 4B,52). It is noteworthy that duplicated dif1 copies remained close to each other, near mid-cell in EGV361 and EGV362 cells, even if duplication occurred an earlier stage of the cell cycle (i.e. in shorter cells) than in EGV360. This likely explains why strains EGV361 and EGV362 exhibited no obvious defect in the coordination of chromosome segregation and cell division. In particular, we observed no anucleated nor filamentous cells.
Duplication and choreography of dif1 locus in WT and ectopic oriC1-containing derivatives. (A) Proportion of cells with 2 dif1-foci according to the cell length (cell size intervals of 0.1 µm). Population cell size is represented in the inset. (B) Choreography of dif1 in EGV360 (top panel), in EGV362 (middle panel) and in EGV361 (bottom panel). The relative positioning of dif1 along the cell length axis is in function of the cell length (cell size intervals of 0.1 µm); For each size interval, the median positioning of dif1 among the cells with 1 dif1- focus (plain dot) and the pair of median dif1 positioning among the cells with 2 dif1-foci (bi-coloured dot) are indicated (when proportion exceed 5% as shown by the dashed line in (A). The size of the dots reflects qualitatively the proportion between the 1- and the 2-dif1 cells among each size interval. (0: new pole; 1: old pole). For each strain, 2500 cells minimum were analysed.
Discussion
Our results unambiguously demonstrate that no replication fork trap (RFT) exists in V. cholerae. The addition of an extra origin of replication on the chromosome of V. cholerae resulted in a modification of the DNA synthesis program such that replication terminated about halfway from the origin(s), irrespective of the chromosomal region, not necessarily around the dif site nor within Ter, for both chr1 or chr2.
The absence of RFT on chr2 was already suggested in V. cholerae55. A strain in which the origin of replication of chr2 is inactivated may propagate, granted that chr2 fuses with chr1. In one such strain, the two chromosomes fused through their dif sites sequences. As a consequence, the orientation of the chr2 sequence was inverted with respect to that of its replication when autonomous and any functional replication fork trap would have forbidden the completion of replication, which would have prevented the propagation of the strain. However, it was not possible to exclude that the strain, obtained at low frequency, had acquired a secondary mutation that prevented the conjectural RFT.
In E. coli, initiation of replication at ectopic origins may be silenced, depending on their genomic location. When functional, their initiation frequency appears similar to that of the origin on its endogenous context, independently of its position or the length of the origin fragment used (from 449-bp to 5-kbp)36,50. We observed in some of our V. cholerae MFA that the copy number of the 384-bp ectopic origin (oriR4 in Fig. 3) is higher than that of oriC1 at its endogenous locus. This could be explained by the absence of a negative cis-regulation of the initiation of replication on the 384-bp fragment. The absence of the negative regulation may be masked by the perturbation created by the transcription of rrnE (oriL3) and by the different growth conditions used in our MFA, as judged by the different level of oriC1/fcpR ratio between the replicates (oriR4: 1.21 and 1.14).
Our results further showed that a fork progressing in the opposite direction of a rrn operon transcription was impeded (Figs 2, 3 and S6). Transcription rrn increases as a function of the richness of the medium, likely explaining the higher level of replication perturbation when the strain was grown in rich medium. These transcription-replication conflicts may explain the lower level of MF observed at the oriL3 and crtS loci because of local DNA degradation in the rrnE region. Consequently, the copy number of chr2, whose replication initiation depends on the duplication of the crtS locus located on chr1, was lower in the strain grown in the rich medium than in the strain grown in the medium (Figs 2 and S6). However, no dramatic perturbation was observed when forks progressed on an illegitimate replichore around the Ter regions. Furthermore, our results indicated that replication completion at Ter was not required to ensure the proper coordination between chromosome segregation and cell division (Figs 2, 4).
The MFA showed that replication terminates very accurately near dif in cultures of WT V. cholerae (Fig. 2). In contrast, the deviation of the fork convergence points (fcp) from the midpoints between two origins (mp) and the increased zone of termination (S95 value) indicated that the robustness of the process was significantly reduced in strains in which an additional origin of replication was integrated elsewhere on the chromosome (Table S4 and S5). It suggests that the progression of replication forks is perturbed on illegitimate replichores. In bacterial genomes, strongly expressed genes such as those contained in ribosomal operons (rrn) are usually transcribed in the same direction as the progression of the replication forks. Hence, the difference in terms of precision of replication completion between the WT and the two mutant strains could reflect the deleterious consequences of opposite transcription and replication orientations. In EGV140, containing an ectopic oriC1 close to rrnE, the distance between fcpL and mpL was probably increased linked to the rrnE perturbation; in EGV111, the probability to terminate far from fcpR or fcpL (S95 parameter) was probably increased by perturbations on each side of fcpR or fcpL. Head-to-head collisions between transcription bubbles and replication forks were shown to lead frequently to replication fork inactivation in fast growth conditions in E. coli to the point that recA became essential in strains in which the orientation of rrn operons were inverted37. Similarly, doubling time was reduced by a combination of ectopic origins that would rebalance the replichore size36. These replication hazards had also an effect on growth rate in V. cholerae, although to a lesser extent (Table S2).
We initiated this work with the idea that in E. coli RFT might participate in the positioning of Ter at mid-cell at the time of cell division by forcing termination in this region56,57. However, our data demonstrate that whatever the replication timing, dif1 sister loci remain colocalised until their positioning at mid-cell at the time of division in V. cholerae (Fig. 4A). Hence, we envision that the spatial arrangement of the chromosome during the cell cycle and the associated coordination between chromosome segregation with cell division could be the rationale behind the presence of an RFT system in some organisms and not in others. In contrast to V. cholerae, the chromosome of E. coli follows a Left-Right transversal arrangement, implying that merely the region where replication is completed, and therefore not necessarily Ter, is located at mid-cell. Indeed, modifications of the RFT zone or perturbations of one of the two replication forks progression have important consequences on the spatial positioning of the Ter region in E. coli58,59. Assuming that the localization of Ter is critical for the formation of the septal ring at mid-cell, the Left-Right arrangement of the chromosome would benefit from the assistance of an RFT to force the localisation of Ter at mid-cell when replication is completed. Given that only a few species have a Left-Right chromosome arrangement, it is likely that RFT acquisition followed the loss of the ori-ter arrangement to circumvent problems associated with Ter mispositioning.
Only a few bacteria encode an RTF system. We observed that the ter/Tus system is carried by plasmids that proliferate in γ-proteobacteria and we proposed that, in the course of evolution, tus was directly or indirectly domesticated only thrice and relatively recently within this bacterial class. The absence of such a system in Plesiomonas shigelloides, an atypical enterobacteria that naturally lives in brackish waters and that diverged early during the evolution of the Enterobacteriales, suggests that the domestication occurred simultaneously with or in response to the switch to another ecological niche in that order. Finally, the absence of RFT in V. cholerae, and in most bacteria, strongly suggests that the domestication of the ter/Tus system was not achieved at the expenses of an ancestral system, but rather as a gain of function, possibly to ensure an improved proliferation rate of the cells.
Methods
Plasmids and strains
Bacterial strains and plasmids used in this study are listed in Table S1. All V. cholerae mutants were constructed by integration-excision or natural transformation52. To this end, derivatives of the El Tor V. cholerae N16961 and MCH124 were rendered competent by the insertion of hapR by specific transposition60. Description of strain construction is included in Sup MM.
Genomic DNA bank preparation for MFA
Cells were grown in M9 minimal medium supplemented with 0.4% fructose or in LB rich medium to exponential phase (0.05 and 0.2 OD at 650 nm in M9 and LB, respectively). Chromosomal DNA was extracted using the Sigma GenElute® bacterial genomic DNA kit to generate a genomic library according to Illumina’s protocol. The libraries and the sequencing were performed by the High-throughput Sequencing facility of the I2BC (http://www.i2bc.paris-saclay.fr/spip.php?article399&lang=en, CNRS, Gif-sur-Yvette, France). Genomic DNA libraries were made with the ‘Nextera DNA library preparation kit’ (Illumina) following the manufacturer’s recommendations. Library quality was assessed on an Agilent Bioanalyzer 2100, using an Agilent High Sensitivity DNA Kit (Agilent technologies). Libraries were pooled in equimolar proportions. 75 bp single reads were generated on an Illumina MiSeq instrument, using a MiSeq Reagent kit V2 (500 cycles) (Illumina), with an expected depth of 217X.
Marker frequency analysis
An in-lab written MATLAB-based script (available on demand) was used to perform marker frequency analysis. The determination of the termination parameters (fork convergence point (fcp) and S95) are described in Supp. Methods. The MFA data have been submitted to the ArrayExpress repository. The access number for these data is E-MTAB-7193.
Fluorescence microscopy
Cells were grown in M9 minimal medium supplemented with 0.4% fructose to exponential phase and spread on a 1% (wt/vol) agarose pad for analysis. Cell images were acquired using a DM6000-B (Leica) microscope with MetaMorph software (Version 7.8.8.0, Molecular Devices). Cell outlines and spot positions were determined using Microbetracker61. More than 2500 cells were analysed in each experiment. Orientation of the cells containing only one dif1 focus was arbitrary: the pole closer to dif1 was called pole 0.
References
Casjens, S. The diverse and dynamic structure of bacterial genomes. Annu. Rev. Genet. 32, 339–377 (1998).
Kleckner, N. E., Chatzi, K., White, M. A., Fisher, J. K. & Stouf, M. Coordination of Growth, Chromosome Replication/Segregation, and Cell Division in E. coli. Front. Microbiol. 9, 1469 (2018).
Rasmussen, T., Jensen, R. B. & Skovgaard, O. The two chromosomes of Vibrio cholerae are initiated at different time points in the cell cycle. EMBO J. 26, 3124–3131 (2007).
Srivastava, P., Fekete, R. A. & Chattoraj, D. K. Segregation of the replication terminus of the two Vibrio cholerae chromosomes. J. Bacteriol. 188, 1060–1070 (2006).
Val, M.-E., Soler-Bistué, A., Bland, M. J. & Mazel, D. Management of multipartite genomes: the Vibrio cholerae model. Curr. Opin. Microbiol. 22, 120–126 (2014).
Du, W.-L. et al. Orderly Replication and Segregation of the Four Replicons of Burkholderia cenocepacia J2315. PLoS Genet. 12, e1006172 (2016).
Kemter, F. S. et al. Synchronous termination of replication of the two chromosomes is an evolutionary selected feature in Vibrionaceae. PLoS Genet. 14, e1007251 (2018).
Val, M.-E. et al. A checkpoint control orchestrates the replication of the two chromosomes of Vibrio cholerae. Sci. Adv. 2, e1501914 (2016).
Ramachandran, R., Ciaccia, P. N., Filsuf, T. A., Jha, J. K. & Chattoraj, D. K. Chromosome 1 licenses chromosome 2 replication in Vibrio cholerae by doubling the crtS gene dosage. PLoS Genet. 14, e1007426 (2018).
de Lemos Martins, F., Fournes, F., Mazzuoli, M.-V., Mazel, D. & Val, M.-E. Vibrio cholerae chromosome 2 copy number is controlled by the methylation-independent binding of its monomeric initiator to the chromosome 1 crtS site. Nucleic Acids Res. 46, 10145–10156 (2018).
Bruhn, M. et al. Functionality of Two Origins of Replication in Vibrio cholerae Strains With a Single Chromosome. Front. Microbiol. 9, 2932 (2018).
Touchon, M. & Rocha, E. P. C. Coevolution of the Organization and Structure of Prokaryotic Genomes. Cold Spring Harb. Perspect. Biol. 8, a018168 (2016).
Ptacin, J. L. et al. Sequence-directed DNA export guides chromosome translocation during sporulation in Bacillus subtilis. Nat. Struct. Mol. Biol. 15, 485–493 (2008).
Stouf, M., Meile, J.-C. & Cornet, F. FtsK actively segregates sister chromosomes in Escherichia coli. Proc. Natl. Acad. Sci. USA 110, 11157–11162 (2013).
Galli, E., Midonet, C., Paly, E. & Barre, F.-X. Fast growth conditions uncouple the final stages of chromosome segregation and cell division in Escherichia coli. PLoS Genet. 13, e1006702 (2017).
Biller, S. J. & Burkholder, W. F. The Bacillus subtilis SftA (YtpS) and SpoIIIE DNA translocases play distinct roles in growing cells to ensure faithful chromosome partitioning. Mol. Microbiol. 74, 790–809 (2009).
Bigot, S. et al. KOPS: DNA motifs that control E. coli chromosome segregation by orienting the FtsK translocase. EMBO J. 24, 3770–3780 (2005).
Aussel, L. et al. FtsK Is a DNA motor protein that activates chromosome dimer resolution by switching the catalytic state of the XerC and XerD recombinases. Cell 108, 195–205 (2002).
Ip, S. C. Y., Bregu, M., Barre, F.-X. & Sherratt, D. J. Decatenation of DNA circles by FtsK-dependent Xer site-specific recombination. EMBO J. 22, 6399–6407 (2003).
Val, M.-E. et al. FtsK-dependent dimer resolution on multiple chromosomes in the pathogen Vibrio cholerae. PLoS Genet. 4, e1000201 (2008).
Kennedy, S. P., Chevalier, F. & Barre, F.-X. Delayed activation of Xer recombination at dif by FtsK during septum assembly in Escherichia coli. Mol. Microbiol. 68, 1018–1028 (2008).
Dubarry, N., Possoz, C. & Barre, F.-X. Multiple regions along the Escherichia coli FtsK protein are implicated in cell division. Mol. Microbiol. 78, 1088–1100 (2010).
Castillo, F., Benmohamed, A. & Szatmari, G. Xer Site Specific Recombination: Double and Single Recombinase Systems. Front. Microbiol. 8, 453 (2017).
Galli, E. et al. Cell division licensing in the multi-chromosomal Vibrio cholerae bacterium. Nat. Microbiol. 1, 16094 (2016).
Bernhardt, T. G. & de Boer, P. A. J. SlmA, a nucleoid-associated, FtsZ binding protein required for blocking septal ring assembly over Chromosomes in E. coli. Mol. Cell 18, 555–564 (2005).
Cho, H., McManus, H. R., Dove, S. L. & Bernhardt, T. G. Nucleoid occlusion factor SlmA is a DNA-activated FtsZ polymerization antagonist. Proc. Natl. Acad. Sci. USA 108, 3773–3778 (2011).
Wu, L. J. et al. Noc protein binds to specific DNA sequences to coordinate cell division with chromosome segregation. EMBO J. 28, 1940–1952 (2009).
Wu, L. J. & Errington, J. Coordination of cell division and chromosome segregation by a nucleoid occlusion protein in Bacillus subtilis. Cell 117, 915–925 (2004).
Mercier, R. et al. The MatP/matS site-specific system organizes the terminus region of the E. coli chromosome into a macrodomain. Cell 135, 475–485 (2008).
Demarre, G. et al. Differential management of the replication terminus regions of the two Vibrio cholerae chromosomes during cell division. PLoS Genet. 10, e1004557 (2014).
Espéli, O. et al. A MatP-divisome interaction coordinates chromosome segregation with cell division in E. coli. EMBO J. 31, 3198–3211 (2012).
Buss, J. A., Peters, N. T., Xiao, J. & Bernhardt, T. G. ZapA and ZapB form an FtsZ-independent structure at midcell. Mol. Microbiol. 104, 652–663 (2017).
Breier, A. M., Weier, H.-U. G. & Cozzarelli, N. R. Independence of replisomes in Escherichia coli chromosomal replication. Proc. Natl. Acad. Sci. USA 102, 3942–3947 (2005).
Reyes-Lamothe, R., Possoz, C., Danilova, O. & Sherratt, D. J. Independent positioning and action of Escherichia coli replisomes in live cells. Cell 133, 90–102 (2008).
Lloyd, R. G. & Rudolph, C. J. 25 years on and no end in sight: a perspective on the role of RecG protein. Curr. Genet. 62, 827–840 (2016).
Dimude, J. U. et al. Origins Left, Right, and Centre: Increasing the Number of Initiation Sites in the Escherichia coli Chromosome. Genes 9 (2018).
De Septenville, A. L., Duigou, S., Boubakri, H. & Michel, B. Replication fork reversal after replication-transcription collision. PLoS Genet. 8, e1002622 (2012).
Michel, B. & Sandler, S. J. Replication Restart in Bacteria. J. Bacteriol. 199 (2017).
Gupta, M. K. et al. Protein–DNA complexes are the primary sources of replication fork pausing in Escherichia coli. Proc. Natl. Acad. Sci. 110, 7252–7257 (2013).
Rocha, E. P. C. The organization of the bacterial genome. Annu. Rev. Genet. 42, 211–233 (2008).
Hill, T. M., Pelletier, A. J., Tecklenburg, M. L. & Kuempel, P. L. Identification of the DNA sequence from the E. coli terminus region that halts replication forks. Cell 55, 459–466 (1988).
Hill, T. M., Tecklenburg, M. L., Pelletier, A. J. & Kuempel, P. L. tus, the trans-acting gene required for termination of DNA replication in Escherichia coli, encodes a DNA-binding protein. Proc. Natl. Acad. Sci. USA 86, 1593–1597 (1989).
Smith, M. T. & Wake, R. G. Definition and polarity of action of DNA replication terminators in Bacillus subtilis. J. Mol. Biol. 227, 648–657 (1992).
Kamada, K., Horiuchi, T., Ohsumi, K., Shimamoto, N. & Morikawa, K. Structure of a replication-terminator protein complexed with DNA. Nature 383, 598–603 (1996).
Vivian, J. P., Porter, C. J., Wilce, J. A. & Wilce, M. C. J. An asymmetric structure of the Bacillus subtilis replication terminator protein in complex with DNA. J. Mol. Biol. 370, 481–491 (2007).
Duggin, I. G. & Bell, S. D. Termination structures in the Escherichia coli chromosome replication fork trap. J. Mol. Biol. 387, 532–539 (2009).
Moreau, M. J. J. & Schaeffer, P. M. Differential Tus-Ter binding and lock formation: implications for DNA replication termination in Escherichia coli. Mol. Biosyst. 8, 2783–2791 (2012).
Elshenawy, M. M. et al. Replisome speed determines the efficiency of the Tus-Ter replication termination barrier. Nature 525, 394–398 (2015).
Ivanova, D. et al. Shaping the landscape of the Escherichia coli chromosome: replication-transcription encounters in cells with an ectopic replication origin. Nucleic Acids Res. 43, 7865–7877 (2015).
Milbredt, S., Farmani, N., Sobetzko, P. & Waldminghaus, T. DNA Replication in Engineered Escherichia coli Genomes with Extra Replication Origins. ACS Synth. Biol. 5, 1167–1176 (2016).
Bremer, H. & Churchward, G. An examination of the Cooper-Helmstetter theory of DNA replication in bacteria and its underlying assumptions. J. Theor. Biol. 69, 645–654 (1977).
David, A. et al. The two Cis-acting sites, parS1 and oriC1, contribute to the longitudinal organisation of Vibrio cholerae chromosome I. PLoS Genet. 10, e1004448 (2014).
Baek, J. H. & Chattoraj, D. K. Chromosome I controls chromosome II replication in Vibrio cholerae. PLoS Genet. 10, e1004184 (2014).
Val, M.-E., Skovgaard, O., Ducos-Galand, M., Bland, M. J. & Mazel, D. Genome engineering in Vibrio cholerae: a feasible approach to address biological issues. PLoS Genet. 8, e1002472 (2012).
Val, M.-E. et al. Fuse or die: how to survive the loss of Dam in Vibrio cholerae. Mol. Microbiol. 91, 665–678 (2014).
Esnault, E., Valens, M., Espéli, O. & Boccard, F. Chromosome structuring limits genome plasticity in Escherichia coli. PLoS Genet. 3, e226 (2007).
Lesterlin, C., Pages, C., Dubarry, N., Dasgupta, S. & Cornet, F. Asymmetry of chromosome Replichores renders the DNA translocase activity of FtsK essential for cell division and cell shape maintenance in Escherichia coli. PLoS Genet. 4, e1000288 (2008).
Wang, X., Possoz, C. & Sherratt, D. J. Dancing around the divisome: asymmetric chromosome segregation in Escherichia coli. Genes Dev. 19, 2367–2377 (2005).
Liu, X., Wang, X., Reyes-Lamothe, R. & Sherratt, D. Replication-directed sister chromosome alignment in Escherichia coli. Mol. Microbiol. 75, 1090–1097 (2010).
Nielsen, A. T. et al. RpoS controls the Vibrio cholerae mucosal escape response. PLoS Pathog. 2, e109 (2006).
Sliusarenko, O., Heinritz, J., Emonet, T. & Jacobs-Wagner, C. High-throughput, subpixel precision analysis of bacterial morphogenesis and intracellular spatio-temporal dynamics. Mol. Microbiol. 80, 612–627 (2011).
Acknowledgements
We thank F. Alberge for helpful discussions on MFA, N. Benita for pNB1 plasmid construction, and reviewer’ suggestions to validate our MFA method. We are very grateful to the High-throughput Sequencing facility of the I2BC (http://www.i2bc.paris-saclay.fr/spip.php?article399&lang=en, CNRS, Gif-sur-Yvette, France) for the realisation of libraries and sequencing for MFA studies. This work was supported by the European Research Council under the European Community’s Seventh Framework Programme [FP7/2007–2013 Grant Agreement no. 281590] and the ANR [PhenX/16-CE12-0030-01].
Author information
Authors and Affiliations
Contributions
Christophe Possoz and François-Xavier Barre and Marie-Eve Val conceived and designed the experiments; Elisa Galli constructed the strains, realised the marker frequency and the microscopy experiments; Jean-Michel Desfontaines, François-Xavier Barre, Christophe Possoz and Ole Skovgaard analysed the marker frequency experiments; Jean-Luc Ferat performed the phylogenetic analysis. Christophe Possoz, Jean-Luc Ferat and François-Xavier Barre wrote the manuscript. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Galli, E., Ferat, JL., Desfontaines, JM. et al. Replication termination without a replication fork trap. Sci Rep 9, 8315 (2019). https://doi.org/10.1038/s41598-019-43795-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-43795-2
This article is cited by
-
The in vivo measurement of replication fork velocity and pausing by lag-time analysis
Nature Communications (2023)
-
Computational analysis of LexA regulons in Proteus species
3 Biotech (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.