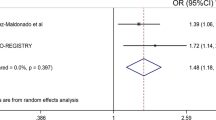
Abstract
Here, we assessed whether 41 SNPs within steroid hormone genes associated with erosive disease. The most relevant finding was the rheumatoid factor (RF)-specific effect of the CYP1B1, CYP2C9, ESR2, FcγR3A, and SHBG SNPs to modulate the risk of bone erosions (P = 0.004, 0.0007, 0.0002, 0.013 and 0.015) that was confirmed through meta-analysis of our data with those from the DREAM registry (P = 0.000081, 0.0022, 0.00074, 0.0067 and 0.0087, respectively). Mechanistically, we also found a gender-specific correlation of the CYP2C9rs1799853T/T genotype with serum vitamin D3 levels (P = 0.00085) and a modest effect on IL1β levels after stimulation of PBMCs or blood with LPS and PHA (P = 0.0057 and P = 0.0058). An overall haplotype analysis also showed an association of 3 ESR1 haplotypes with a reduced risk of erosive arthritis (P = 0.009, P = 0.002, and P = 0.002). Furthermore, we observed that the ESR2, ESR1 and FcγR3A SNPs influenced the immune response after stimulation of PBMCs or macrophages with LPS or Pam3Cys (P = 0.002, 0.0008, 0.0011 and 1.97•10−7). Finally, we found that a model built with steroid hormone-related SNPs significantly improved the prediction of erosive disease in seropositive patients (PRF+ = 2.46•10−8) whereas no prediction was detected in seronegative patients (PRF− = 0.36). Although the predictive ability of the model was substantially lower in the replication population (PRF+ = 0.014), we could confirm that CYP1B1 and CYP2C9 SNPs help to predict erosive disease in seropositive patients. These results are the first to suggest a RF-specific association of steroid hormone-related polymorphisms with erosive disease.
Similar content being viewed by others
Introduction
Rheumatoid arthritis (RA) is a chronic and disabling chronic immune-mediated inflammatory disease that affects approximately 1% of the worldwide population1. Although the etiology of this autoimmune disease remains largely unknown, family- and population-based genome-wide association studies (GWAS) have consistently demonstrated that RA has a strong inherited component that influences not only the predisposition to develop the disease2,3,4,5. Even though there are no relevant predictors for treatment response in RA6, recent studies have suggested that inherited genetic factors might influence the response to both classical disease-modifying anti-rheumatic or biological drugs7,8,9,10,11 and even disease progression12,13,14. From epidemiological studies, it has also been proposed that in addition to inherited factors and environmental factors certain hormonal events might help to promote the onset of the disease15,16,17. It has been suggested that concentrations of steroid hormones and circulating immunocomplexes (CICs) in the synovial fluid and cartilage may contribute to promote gender-specific inflammatory responses, a differently controlled and sustained production of autoantibodies between men and women18,19,20 and a different release of a wide range of cytokines and pro-inflammatory mediators that trigger sustained and chronic inflammatory responses21,22,23. However, there is still controversy about the effect of steroid hormones on the risk of developing RA or disease progression since the administration of different hormone replacement therapies or the use of oral contraceptives has not been associated with the risk of RA and its progression in most of the epidemiological studies conducted to date24,25,26,27.
Some authors have hypothesized that these controversial results might be, at least in part, due to the presence of certain factors such as the HLA-DRB1 shared epitope or specific autoantibodies such as antibodies to cyclic citrullinated peptide (anti-CCP)28,29. Furthermore, there are contradictory results concerning to the role of steroid hormones in the modulation of immune responses. Some studies have reported, for instance, that vitamin D3 has immunomodulatory properties that may influence autoimmune disease risk and disease progression30 whereas some other have suggested that estrogens can induce both tolerogenic and pro-inflammatory responses at multiple levels and that this may result in remarkable sex differences on immune function. Under appropriate circumstances, estrogens may inhibit Th1- and Th17-mediated immune functions31,32 and stimulate Treg cell development33 and the activation of Th2-mediated immune responses. However, it has been also demonstrated that estrogens may induce pro-inflammatory responses. For instance, it has been suggested that estrogens influence FcγR3A mRNA gene expression and induce the FcγR3A-mediated release of tumor necrosis factor (TNF) and IL1β from monocytes34, thereby modulating degranulation, antibody-dependent cellular cytotoxicity (ADCC), transcription of cytokine genes, rapid release of inflammatory mediators and reactive oxygen species, and phagocytosis35,36,37. Considering the role of FcγR proteins in modulating autoimmune responses but also the plausibility of a gender-specific effect of estrogens to modulate immune responses, we aimed at analyzing whether the presence of single nucleotide polymorphisms (SNPs) within steroid hormone signaling (ESR1, ESR2, NR1I2, PGR, and SHBG), phase I- and II-metabolizing enzyme (CYP1A1, CYP1A2, CYP1B1, CYP17A1, CYP2C9, CYP2C19, CYP3A4, GSTP1, HSD17B1 and SULT1A1) and Fc gamma receptor (FcγR3A and FCGR2A) genes influence disease progression in RA. We assessed whether 41 potentially functional SNPs within these genes are associated with the risk of developing erosive disease and whether the effect of the SNPs on disease progression was modified by the presence of rheumatoid factor (RF) or anti-CCP. In order to confirm the consistency of our results, we performed fixed-effect meta-analyses with data from the DREAM registry. Finally, we also evaluated whether selected polymorphisms correlated with steroid hormone and cytokine levels and whether genotyping of selected SNPs might help us to improve the prediction of the appearance of bone erosions.
Patients and Methods
Study population
This retrospective cohort study included 816 RA patients ascertained through the REPAIR consortium (567 showing erosive disease and 249 without bone erosions). RA patients fulfilled the 1987 revised American College of Rheumatology (ACR)38 and the ACR/EULAR 2010 classification criteria39. A detailed description of the population has been reported elsewhere40,41,42. Briefly, 518 RA patients were recruited at the department of Rheumatology of the Virgen de las Nieves Hospital (Granada, Spain), the Reina Sofia Hospital (Córdoba, Spain), and the University Clinical Hospital of Santiago de Compostela (Santiago de Compostela, Spain). Two hundred and ninety-eight RA patients were additionally recruited from the Santa Maria Hospital–CHLN (Biobanco-IMM; Lisbon Academic Medical Centre, Lisbon, Portugal). The study was performed according to the Helsinki Declaration. All participants were of European ancestry and gave their written informed consent to participate in the study. The Ethics committee of each participant institution approved the study protocol: Virgen de las Nieves University Hospital (2012/89); Santa Maria Hospital-CHLN (CE 877/121.2012); University Clinical Hospital of Santiago de Compostela (2013/156). A detailed description of demographic and clinical variables of this population is included in Table 1.
Bone erosions
Bone erosions were visible in plain radiographs and defined as the interruption of the cortical bone surface within the joint region or underlying the cartilage43,44. Bone erosions were then coded as present or absent. All radiographs were assessed by, at least, an experienced radiologist or rheumatologist.
SNP selection and genotyping
We conducted an extensive literature search concerning the mechanism of action of estrogen and progesterone receptor, hormone transporter, and hormone-metabolizing enzyme genes was performed to select candidate genes that might affect the risk of developing bone erosions. SNPs were assessed on the basis of NCBI data and were selected according to their known or putative functional consequences, i.e. their modifying influence on the structure of proteins, transcription level, or alternative splicing mechanisms. SNPs within the same gene were also selected on the basis of linkage disequilibrium (LD) data. In total, 41 SNPs in 17 genes were selected for this study (Table 2).
Genotyping of selected steroid hormone-related SNPs was performed at GENYO (Granada, Spain) using KASPar® probes with the exception of the FcγR3Ars396991 and FCγR2Ars1801274 SNPs that were determined using TaqMan® SNP Genotyping Assays (Life Technologies, Carlsbad, CA, USA). Both KASPar® and Taqman® assays were assayed according to the manufacturer’s specifications for a 384-well plate format. Genomic DNA was extracted from peripheral blood mononuclear cells (PBMCs) using Qiagen Mini kit (Qiagen, Valencia, CA, USA) and PCR products were analyzed with ABI Prism 7900HT detection system using the SDS 2.4 software (Applied Biosystems). Five percent of samples were included in the PCR plates as duplicates and concordance between the analyzed original and duplicated samples was >99.0%.
Statistical analysis
Hardy-Weinberg Equilibrium (HWE) was assessed in the control group by a chi-square (χ2) test. Logistic regression analysis adjusted for age, sex and country of origin was used to assess the main effect of the selected SNPs on disease progression (defined as the presence of bone erosions). RF− and anti-CCP-stratified analyses were also carried out and we included RF as interaction term in the overall logistic regression analysis to evaluate whether there was any statistically significant effect modification by these factors. Haplotype analysis using the same variables for adjustment was conducted using the R package Haplo.stats45. In order to facilitate eventual meta-analyses, the major allele was set as reference allele. All tests were conducted using the statistical software STATA (v.12) and R (http://www.r-project.org). In order to account for multiple testing, we set a P value of 0.00074 as significance study-wide threshold. The P value was calculated considering the number of independent polymorphisms analyzed (n = 34, MeffLi method)46 but also the number of inheritance models tested (dominant and recessive).
Linkage disequilibrium (LD) and haplotype analysis
We performed haplotype frequency estimation and haplotype association analysis adjusted for age, sex and country of origin using the haplo.stats45. Haplotype frequencies were determined using the Expectation-maximization (EM) algorithm and haplotypes were reconstructed using SNPtools47 and Haploview48. Block structures were determined according to the method of Gabriel et al.49 (Supplementary Fig. 1).
Replication population and meta-analysis
With the aim of assessing the consistency of the overall and RF-specific associations between SNPs and the risk of developing bone erosions, we genotyped the most interesting markers in a replication population from the DREAM registry consisting of 436 RA patients (307 RA patients with bone erosions and 129 patients without erosive disease). Demographic and clinical parameters of this population are also included in Table 1. We performed a meta-analysis of the data obtained in the discovery population with those from the DREAM registry and we pooled the Odds Ratios (ORs) for the most interesting polymorphisms using a fixed-effect model. Coefficients with a P-value ≤ 0.05 were considered significant. I2 statistic was used to assess heterogeneity between studies.
Functional analysis of the estrogen-related variants
Cytokine stimulation experiments were conducted in the 500 Functional Genomics (500FG) cohort from the Human Functional Genomics Project (HFGP; http://www.humanfunctionalgenomics.org/), which was designed to determine the influence of genomic variation on the variability of immune responses. The HFGP study was approved by the Arnhem-Nijmegen Ethical Committee (no. 42561.091.12) and biological specimens were collected after informed consent was obtained. We investigate whether any of the 41 estrogen-related SNPs correlated with cytokine levels (IFNγ, IL1β, IL6, TNFα, IL17, and IL22) after the stimulation of peripheral blood mononuclear cells (PBMCs), macrophages or whole blood from 408 healthy subjects with LPS (1 or 100 ng/ml), PHA (10 μg/ml), and Pam3Cys (10 μg/ml). After log transformation, linear regression analyses adjusted for age and sex were used to determine the correlation of selected SNPs with cytokine expression quantitative trait loci (cQTLs). All analyses were performed using R software (http://www.r-project.org/). In order to account for multiple comparisons, we used a significant threshold of 0.00025 (0.05/34 independent SNPs × 6 cytokines).
Details on PBMCs isolation, macrophage differentiation and stimulation assays have been reported elsewhere50,51,52. Briefly, PBMCs were washed twice in saline and suspended in medium (RPMI 1640) supplemented with gentamicin (10 mg/mL), L-glutamine (10 mM) and pyruvate (10 mM). PBMC stimulations were performed with 5 × 105 cells/well in round-bottom 96-wells plates (Greiner) for 24 hours in the presence of 10% human pool serum at 37 °C and 5% CO2. Supernatants were collected and stored in −20 °C until used for ELISA. LPS (100 ng/ml), PHA (10 μg/ml) and Pam3Cys (10 μg/ml) were used as stimulators for 24 or 48 hours. Whole blood stimulation experiments were conducted using 100 μl of heparin blood that was added to a 48 well plate and subsequently stimulated with 400 μl of LPS and PHA (final volume 500 ul) for 48 hours at 37 °C and 5% CO2. Supernatants were collected and stored in −20 °C until used for ELISA. Concentrations of human IFNγ, IL1β, IL6, TNFα, IL17, and IL22 were determined using specific commercial ELISA kits (PeliKine Compact, Amsterdam, or R&D Systems), in accordance with the manufacturer’s instructions.
Once we assessed the correlation of estrogen-related SNPs with cytokine levels, we used the HaploReg SNP annotation tool (http://www.broadinstitute.org/mammals/haploreg/haploreg.php) to further investigate the functional consequences of each specific variant. We also assessed whether any of the potentially interesting markers correlated with mRNA expression levels of their respective genes using data from public eQTL browsers (GTex portal; www.gtexportal.org/home/ and Blood eQTL browser; https://genenetwork.nl/bloodeqtlbrowser/).
Correlation between steroid hormone levels and hormone-related SNPs
We also measured serum levels of seven steroid hormones (androstenedione, cortisol, 11-deoxy-cortisol, 17-hydroxy progesterone, progesterone, testosterone and 25 hydroxy vitamin D3) in the 500FG cohort, which includes 531 healthy subjects. Complete protocol details of steroid hormone measurements have been reported elsewhere52. Hormone levels and genotyping data were available for a total of 406 subjects.
After log-transform, correlation between steroid hormone levels and steroid hormone-related SNPs was evaluated by linear regression analysis adjusted for age and sex (or for age when men and women were analysed separately). In order to avoid a possible bias, we excluded from the analysis those subjects that were using oral contraceptives or those subjects in which this information was not available. A total of 279 healthy subjects (107 women and 272 men) were finally available for analysis. Significance threshold was set to 0.00021 considering the number of independent SNPs tested (n = 34) and the number of hormones determined (n = 7).
Predictive models and discriminative accuracy
The value of steroid hormone-related variants for prediction of prognosis and disease progression in seropositive and seronegative RA patients was assessed using stepwise logistic regression. Models were built including demographic variables (age and sex) and genetic polymorphisms that showed significant associations with erosive disease in the single-SNP analysis (P < 0.05). The genetic model was then compared with the reference model including demographic variables. The area under the curve (AUC) of a receiver operating characteristic (ROC) curve analysis and −2 log likelihood ratio (LR) tests were used to assess whether the genetic models fitted significantly better the data compared to their respective reference models. Finally, we run randomization tests to confirm whether the improved predictive ability of each genetic model was consistent after 50.000 iterations. All tests were conducted using R software (http://www.r-project.org/).
Ethics approval
The study was approved by the ethical review committee of each participant institution (Virgen de las Nieves University Hospital, Granada, Spain; Reina Sofia Hospital, Córdoba, Spain; University Clinical Hospital of Santiago de Compostela, Santiago de Compostela, Spain; Biobanco-IMM, Lisbon Academic Medical Centre, Portugal. Cytokine stimulation experiments and hormone analysis were approved by the Arnhem-Nijmegen Ethical Committee.
Results
Erosive RA patients had a similar age than those patients without bone erosions (59.66 ± 12.47 vs. 58.95 ± 14.30, P = 0.50) and had a significantly higher female to male ratio (462/105 = 4.4 vs. 182/67 = 2.71, P = 0.007; Table 1). Overall, the percentage of RA patients with positive RF and anti-CCP was 70.58% and 72.80% respectively, and these percentages were slightly higher among those patients with bone erosions (72.52% and 74.21%) than in those without erosive disease (66.12% and 69.39%). The mean of disease follow-up was 18.30 years whereas the mean of DAS28 was 5.63. Six hundred and three patients received methotrexate (79.24%) and 632 patients (77.45%) were treated with biologic therapies. With the exception of gender, none of the demographical or clinical variables significantly differ between patient with and without erosive disease (Table 1).
Association of steroid hormone-related polymorphisms with the risk of having bone erosions
Selected polymorphisms did not show deviation from HWE in the control population (patients without erosive disease; P > 0.001). Logistic regression analysis adjusted for age, gender and country of origin revealed that carriers of the ESR2rs1271572T/T genotype tended to have a decreased risk of developing erosive disease than those subjects carrying the G allele (OR = 0.55, P = 0.004: Table 3). Although the association of the ESR2rs1271572T/T genotype with a decreased risk of having bone erosions remained only marginally significant after correction for multiple testing, we found a significant RF-specific effect of this SNP to modulate the risk of having erosive disease. Seropositive patients carrying the ESR2rs1271572T/T genotype had a significantly reduced risk of developing erosive disease (OR = 0.38, P = 0.0002) whereas an opposite but not significant effect was found in seronegative patients (OR = 1.08, P = 0.83; PInt = 0.018; Table 3). Importantly, the association of the ESR2rs1271572 SNP with a reduced risk of developing bone erosions in seropositive patients remained significant after correction for multiple testing (P < 0.00074). Although the association was not replicated in the DREAM cohort, the meta-analysis of our data and those from the DREAM registry, including 1252 RA patients, confirmed that seropositive patients carrying the ESR2rs1271572T/T genotype had a decreased risk of developing erosive disease (ORRF+ = 0.52, P = 0.00074) whereas a totally opposite but not statistically significant effect was found in seronegative patients (ORRF− = 1.28, P = 0.42; PHet = 0.33; Table 4). No significant anti-CCP effect modification was found for this SNP to modulate the risk of developing bone erosions (PInt = 0.11; Supplementary Table 1), which suggest that ESR2 locus might play a relevant role in determining disease progression in a RF-dependent manner. In agreement with these results, we found a RF-specific effect of the ESR2CGTA haplotype to determine the risk of developing erosive disease. Seropositive RA patients carrying the ESR2CGTA haplotype (and, therefore, not harboring the ESR2rs1271572 protective allele) showed an increased risk of developing bone erosions (ORRF+ = 1.63, P = 0.0051) whereas an opposite but not significant effect was detected in seronegative patients (ORRF− = 0.93, P = 0.99; Supplementary Table 2). An overall haplotype analysis including 1252 RA patients from the discovery and replication populations confirmed the RF-specific association of the ESR2CGTA haplotype with an increased risk of developing bone erosions (ORRF+ = 1.44, 95%CI 1.13–1.84; P = 0.0036 and ORRF− = 0.89, 95%CI 0.62–1.26; P = 0.51). According to publicly available gene expression datasets (GTex portal and Haploreg), the ESR2rs1271572 variant strongly correlate with ESR2 mRNA expression levels in whole peripheral blood (P = 3.1•10−9) but also in primary B cells, lymphoblastoid cell lines (from P = 1.98•10−6 to P = 3.47•10−10) and several tissues (ranging from P = 2.60•10−5 to P = 8.33•10−23; Supplementary Table 3). Intriguingly, a similar level of correlation with gene expression was found for other variants belonging to the ESR2CGTA haplotype (Supplementary Table 3), which strongly suggested that the ESR2rs1271572 SNP or ESR2CGTA haplotype might represent an eQTL for ESR2.
Similarly, we also found a RF-specific effect of the CYP2C9rs1799853 and CYP1B1rs10012 SNPs to determine the risk of developing bone erosions. Seropositive patients carrying the CYP2C9rs1799853T/T or CYP1B1rs10012G/G genotypes had a significantly reduced chance of developing bone erosions (OR = 0.16, P = 0.0007 and OR = 0.42, P = 0.0040) whereas an opposite but not significant effect was observed in seronegative patients (OR = 2.71, P = 0.23; PInt = 0.003 and OR = 1.70, P = 0.35; PInt = 0.031; Table 3). The effect of the CYP2C9rs1799853 polymorphism on the risk of developing erosive disease in seropositive patients remained statistically significant after correction for multiple testing (P < 0.00074), which suggested a role of the CYP2C9 gene in modulating disease progression in RA. In accordance with these findings, we found that seropositive patients carrying the CYP2C9AT haplotype had a significantly decreased risk of developing erosive disease (ORRF+ = 0.61, P = 0.0075) whereas no effect was observed in seronegative patients (ORRF− = 0.87, P = 0.57; Supplementary Table 2). No significant anti-CCP effect modification was found for CYP2C9 and CYP1B1 variants to determine the appearance of bone erosions (PInt = 0.88 and PInt = 0.27) underlying the importance of considering RF when evaluating the impact of the CYP2C9 and CYP1B1 loci on the risk of developing erosive disease. Importantly, when we attempted to validate the RF-specific association of the CYP1B1rs10012G/G genotype with a decreased risk of having erosive disease in the replication population, we found that seropositive patients carrying the CYP1B1rs10012G/G genotype had a significantly decreased risk of developing bone erosions (ORRF+ = 0.30, P = 0.0051) whereas an opposite but not significant effect was found in seronegative patients (ORRF− = 5.97, P = 0.10; PInt = 0.012; Table 4). The meta-analysis of both populations confirmed the strong RF-specific effect of this SNP to determine the risk of developing bone erosions (ORRF+ = 0.38, P = 0.000081; PHet = 0.52 vs. ORRF− = 2.22, P = 0.11; PHet = 0.31). Although we attempted to validate the association of the CYP2C9rs1799853T/T genotype with a decreased risk of having erosive disease, the relatively small size of the replication population did not allow us to perform the association analysis according to a recessive model of inheritance. However, we found a RF-specific effect on the risk of having erosive disease for a neighboring SNP (rs1057910), which suggested a RF-dependent effect of the CYP2C9 locus to modulate the risk of erosive disease (ORRF+ = 2.75, P = 0.027 vs. ORRF− = 0.54, P = 0.47; Table 4). The meta-analysis of both cohorts confirmed the RF-specific effect of the CYP2C9rs1057910 SNP on the risk of developing bone erosions (ORRF+ = 2.68, P = 0.0022 vs. ORRF− = 1.08, P = 0.83; PHet = 0.34; Table 4).
In line with these findings, we also observed an additional RF effect modification of the FcγR3Ars396991 and SHBGrs6259 SNPs to determine the risk of having erosions. Seronegative patients carrying the FcγR3Ars396991C allele had a significantly reduced chance of developing bone erosions (OR = 0.45, P = 0.013) whereas an opposite but not significant effect was observed in seropositive patients (OR = 1.18, P = 0.46; PInt = 0.028; Table 3). Furthermore, seropositive subjects carrying the SHBGrs6259A allele showed an increased risk of developing bone erosions (ORRF+ = 1.87, P = 0.015) whereas an opposite but not significant effect was detected in seronegative patients (ORRF− = 0.66, P = 0.19). Interestingly, although the effect was stronger in seronegative patients, we could validate the RF-specific effect of the SHBGrs6259 SNP on the risk of developing erosions in the replication population (ORRF+ = 1.17, P = 0.63 vs. ORRF− = 0.22, P = 0.009; PInt = 0.013; Table 4) and the meta-analysis of both cohorts confirmed that the effect of this marker was dependent on the RF status (ORRF+ = 1.55, P = 0.033 vs. ORRF− = 0.48, P = 0.0087; PHet = 0.14; Table 4). Although we could not validate the RF-specific association of the FcγR3Ars396991 SNP with bone erosions in the DREAM registry, the meta-analysis of both cohorts confirmed the RF-specific effect of this SNP to modulate the risk of developing erosive disease (ORRF− = 0.47, P = 0.0067 vs. ORRF+ = 1.02, P = 0.93). None of these two SNPs showed a significant effect modification by anti-CCP (PInt = 0.85) suggesting again that RF, rather than anti-CCP, is a driver factor influencing the impact of the steroid hormone-related loci on disease progression in RA.
Finally, an overall association analysis revealed that carriers of the ESR1rs1801132G allele showed a decreased risk of developing bone erosions (OR = 0.71, P = 0.034). Although we could not validate this association in the replication population, we found that this SNP showed a significant RF-specific effect to modulate the risk of developing bone erosions but according to a recessive model of inheritance. Thus, seropositive carriers of the ESR1rs1801132G/G genotype showed a decreased risk of developing bone erosions (ORRF+ = 0.39, P = 0.004) whereas an opposite but not statistically significant effect was observed in seronegative subjects (ORRF− = 1.43, P = 0.57; Table 4). Furthermore, we found a similar RF-specific effect for the ESR1rs9340799 SNP that was not detected in the discovery population (ORRF+ = 0.42, P = 0.009 vs ORRF− = 8.33, P = 0.011). Considering that none of these associations survived after correction for multiple testing and that the effect of ESR1 SNPs on the risk of developing erosive disease seemed to depend on the inheritance model applied, these results suggested a complex relationship between the ESR1 locus and bone erosion probably mediated by more than one SNP. In support of this notion, we found that 3 large ESR1 haplotypes (ESR1CTATTTTCTA, ESR1CTATTCTTCA, and ESR1GTATTCTCTA) were significantly associated with a decreased risk of having erosive disease (P = 0.0094, P = 0.0021 and P = 0.0023, respectively; Supplementary Table 2).
Correlation of selected SNPs with steroid hormone levels
Besides the strong genetic association with erosive disease identified for the CYP2C9rs1799853 SNP and its known role in controlling the metabolism of a wide range of drugs (with the T allele acting as poor metabolizer), we found that this coding variant strongly correlated with serum vitamin D3 levels in women (P = 0.00085 and P = 0.0019; Fig. 1) whereas no effect was detected in men. Although the association of the CYP2C9rs1799853 SNP with reduced levels of vitamin D3 in women remained borderline significant, this finding suggested that this marker might have a role in the modulation of bone homeostasis and vitamin D3-mediated immune responses.
Correlation of the CYP2C9rs1799853 and ESR1rs2881766 SNPs with vitamin D3 and progesterone levels in women (n = 107) and men (n = 172). Patients using oral contraceptives were excluded from the analysis. After log transformation, linear regression analyses were adjusted for age. NS; non-significant.
On the other hand, we found that the ESR1rs2881766G/G genotype or G allele weakly correlated with progesterone levels (P = 0.0076 and P = 0.0071) and that the ESR1rs851984, ESR1rs2077647, ESR1rs2071454, ESR1rs3798577 and ESR1rs910416 variants mapped among histone marks in several cell types including osteoblasts and a wide variety of immune cells.
Correlation of steroid hormone SNPs with cytokine levels
Interestingly, we also found that carriers of the ESR2rs4986938T allele had reduced levels of TNFα after the stimulation of PBMCs with Pam3Cys for 24 h (P = 0.0022; Fig. 2A). These results along with those reporting that the ESR2rs4986938 and ESR2rs1271572 SNPs map among histone marks in multiple cell types including osteoblasts and different subsets of immune cells, suggest a possible functional role of the ESR2 variants in modulating the risk of developing bone erosions likely through the modulation of ESR2 expression. In addition, we found that the presence of the CYP2C9rs1799853T allele correlated with an increased production of IL1β after the stimulation of PBMCs with LPS or PHA for 24 h or 48 h (P = 0.0057 and P = 0.0058; Fig. 2B,C), which also pointed to a functional role of this marker in determining the presence of bone erosions.
In addition, we found that the ESR1rs3798577 variant correlated with TNFα and IL6 levels after the stimulation of human macrophages with LPS for 24 h (P = 0.00083 and 0.0011; Fig. 2D,E). Finally, we found that carriers of the FcγR3Ars396991C allele showed a significantly increased production of TNFα after stimulation of macrophages with LPS for 24 h (P = 1.97•10−7; Fig. 2F). Of note, the association of the FcγR3Ars396991C allele with increased levels of TNFα in macrophages survived after correction for multiple testing, which strongly suggested a functional effect of this variant to modulate macrophage-mediated immune responses, a key factor influencing the risk of developing erosive disease. On the contrary, although it was tempting to speculate that ESR1, ESR2, CYP2C9 SNPs might also exert their effect on the risk of developing erosive disease through the modulation of steroid hormones or steroid hormone-mediated immune responses, it is important to mention that none of the associations between ESR1, ESR2 or CYP2C9 SNPs and cytokine levels survived after correction for multiple testing, which suggested a modest functional impact of these polymorphisms on the risk of developing bone erosions.
Usefulness of steroid hormone-related SNPs to predict erosive disease
As a whole, our data suggest that the attributable effect of the CYP1B1, CYP2C9, ESR1, ESR2, SHBG, and FcγR3A loci to modulate the risk of developing bone erosions in RA patients might be dependent on the presence of either missense or intronic polymorphisms that affect the immune responses to a greater or lesser extent. Considering the strength of the RF-specific associations found for SNPs within CYP1B1, CYP2C9, SHBG, ESR1, ESR2, FcγR3A and CYP3A4 loci in the discovery and/or replication populations, we decided to assess whether SNPs within these loci could be useful to differentially predict disease progression in seropositive and seronegative patients. Our results showed that the addition of 5 steroid hormone-related SNPs within the CYP1B1, CYP2C9, CYP3A4, ESR2 and SHBG loci to a model including demographic variables significantly improved the ability to predict the appearance of bone erosions in seropositive patients (AUCGenetic = 0.73 vs. AUCDemographic = 0.63; P = 2.46•10−8; Table 5) whereas no significant predictive value was found for these SNPs in seronegative patients (AUCGenetic = 0.61 vs. AUCDemographic = 0.59; P = 0.36; Fig. 3). The consistency of this predictive analysis was confirmed through a permutation test that showed that none of the 50.000 permuted models for each group showed a better prediction capacity than the genetic model (Average sorted AUC = 0.644, Z-score = 6.79 and PZ_score (50.000perm) = 5.67•10−12). Even though the lack of patients carrying the CYP2C9rs1799853T/T genotype and the relatively small size of the replication population hampered the validation of this 5-SNP model to predict erosive disease, we attempted to confirm the utility of this model in the DREAM registry. We found that a similar model slightly improved the ability to predict erosive disease in both seropositive and seronegative patients (AUCGenetic-RF+ = 0.63 vs. AUCDemographic-RF+ = 0.53; P = 0.014 and AUCGenetic-RF− = 0.78 vs. AUCDemographic-RF− = 0.54; P = 0.015; Fig. 3). Despite these interesting results, only the CYP1B1 and CYP2C9 SNPs seemed to have a consistent predictive value for the development of bone erosions in seropositive patients.
Receiver operating characteristics (ROC) curve analysis in the discovery and replication populations. ROC curves summarize the accuracy of prediction for genetic and demographic models in seropositive and seronegative patients. The genetic models (marked in blue) included SNPs that were significantly associated with erosive disease in seropositive patients (either in the single-SNP or haplotype analyses) whereas the demographic models included demographic variables (age and gender as covariates; marked in green) for seropositive and seronegative patients.
Discussion
The present study reports, for the first time, both overall and RF-specific associations of steroid hormone-related polymorphisms with the risk of developing erosive RA. The most relevant effect was found for SNPs within CYP1B1, CYP2C9, and ESR2 genes. We observed that seropositive RA patients carrying the CYP1B1rs10012G/G, CYP2C9rs1799853T/T, and ESR2rs1271572T/T, genotypes had a significantly reduced risk of developing bone erosions during the course of the disease whereas an opposite but not significant effect was found in seronegative patients. Although the relatively small size of the replication population hampered the validation of these associations according to a recessive model of inheritance, we could validate the RF-specific association of the CYP1B1rs10012 SNP with the risk of developing erosive disease in the replication population and the meta-analysis of the discovery and replication cohorts confirmed the strong RF effect modification of this SNP to determine the risk of bone erosions. In addition, although we could not validate the RF-specific association of the CYP2C9rs1799853 variant in the replication population due to the lack of patients carrying the CYP2C9rs1799853T/T genotype, we found a RF-specific effect on the risk of having erosive disease for a neighbouring SNP within the CYP2C9 locus (rs1057910) that was further confirmed through meta-analysis. Although this SNP was not in linkage disequilibrium (LD) with the rs1799853 and, therefore, does not represent the same association signal, these results support the idea that the CYP2C9 locus might influence the risk of developing bone erosions in a RF dependent manner and likely through the modulation of the hormone metabolism and hormone-dependent immune responses.
Whilst the CYP1B1 locus is located on chromosome 2p21-22, CYP2C9 belongs to the CYP2C family, a gene cluster (CYP2C19-CYP2C9-CYP2C8) located on chromosome 10q23.3. Together with CYP1A2 and CYP3A4, CYP1B1 and CYP2C9 catalyze the conversion of estrogens to genotoxic catechol estrogens (estradiol 4- and 2-hydroxylation, respectively)53, which are key processes that allow the binding of catechol estrogens to ESR1 and ESR2. At low concentrations, CYP2C9 is also implicated in the 17beta-hydroxy dehydrogenation of estradiol creating estrone, which is one of the 3 natural estrogens with multiple immunomodulatory actions. Given that both the CYP1B1rs10012 and CYP2C9rs1799853 SNPs are coding variants that alters their respective protein amino acid sequences (R48G and Arg144Cys) and appear to decrease the activity of the enzyme but also proper folding and stability, it seems to be plausible to hypothesize that the presence of these SNPs could determinate estrogen-dependent immune responses and thereby modulate the risk of developing bone erosions. Our functional studies also demonstrated that the CYP2C9rs1799853 SNP correlated with serum vitamin D3 levels, which suggest that the CYP2C9rs1799853 SNP might also affect disease progression through the regulation of vitamin D3-mediated immune responses. However, we need to interpret these results with caution as we only found a significant correlation with vitamin D3 in women whereas no effect was seen in men. These results, together with those reporting that carriers of the CYP2C9rs1799853T allele have an increased production of IL1β after the stimulation of PBMCs or whole blood with LPS or PHA, suggest that the protective effect attributed to this coding variant might not only depend on vitamin D3 but other factors such as RF or even other stimuli or substrates. In this regard, our team has reported that the CYP2C9rs1799853 polymorphism is strongly associated with poor response to anti-TNF drugs in RA54, suggesting that this missense variant might modulate the strength of immune responses through the regulation of the metabolism of endogenous compounds but also compounds administered exogenously.
Although we also attempted to validate the RF-specific association of the ESR2rs1271572T/T genotype with a decreased risk of having erosion in the replication population, we only found a modest and not significant effect of this variant to determine erosive disease. However, the direction of the effect in seropositive and seronegative patients was similar to the one observed in the discovery population and the meta-analysis of both cohorts confirmed that the effect of this SNP on the risk of developing bone erosions was modified by RF. In support of a RF-specific effect of this variant to influence the risk of erosive RA, we found that seropositive carriers of the ESR2CGTA haplotype had a decreased risk to develop erosive RA whereas no effect was detected in seronegative patients. Interestingly, an overall haplotype analysis also revealed a significant association of 3 common haplotypes within the ESR1 locus (ESR1CTATTTTCTA, ESR1CTATTCTTCA, and ESR1GTATTCTCTA) with a decreased risk of developing bone erosions, which also pointed to a role of ESR1 SNPs in modulating the risk of erosions.
The ESR2 and ESR1 genes (14q23.2 and 6q25.1 respectively) encode the estrogen receptor beta (ESRβ) and alpha (ESRα) that are highly expressed in synovial cells55 and bone56 but also in most of the immune cells57. Although a number of experimental studies have shown that female RA patients have worse prognosis and higher disease activity and health assessment questionnaire scores in comparison with male patients, it is also well established that steroid hormones have both pro- and anti-inflammatory roles in RA. Although it has been reported, for instance, that the activation of ESRs by estradiol (E2) often leads to joint protection and the maintenance of bone density (by inhibiting bone resorption)58 and that the withdrawal of estrogens drastically increases the severity of the disease (by promoting joint destruction, bone erosions and physical disability)59, it has been also reported that RA patients have high levels of estrone in the synovial fluid compared to healthy individuals and that estrogens can also induce pro-inflammatory responses through the activation of different mechanisms involving humoral immunity60, multiple transcription factors (such as c/EBPβ, STAT-1, NFκB) and oxidative stress pathways (especially those involving iNOs)61,62. Furthermore, it has been reported that estrogens are able to promote pro-inflammatory pathways including B- and T-cell proliferation63, thymocyte maturation64, cell trafficking65 and the expression of specific adhesion molecules63. Although the existing paradox with respect to the immunomodulating role of steroid hormones in RA remain unresolved, it seems to be reasonable to hypothesize that the presence of ESR2 polymorphisms that correlate either with ESR2 mRNA expression levels may influence on the risk of developing bone erosions in RA likely through the modulation of ESR2-dependent tolerogenic immune responses. In addition, although the association of the ESR2 and ESR1 polymorphisms with serum hormone levels or TNF and IL6 levels in stimulated macrophages did not remain significant after correction for multiple testing, our functional findings were in agreement with the genetic results suggesting a protective effect of ESR2 polymorphisms and specific ESR1 haplotypes on the risk of developing erosive RA. In addition, our genetic and functional results were also concordant with data of previous studies reporting that the presence of certain SNPs, microsatellites or even specific haplotypes within estrogen receptor genes is associated with bone mineral density and influences the risk of developing bone erosions66,67 affecting RA patients68 but also subjects diagnosed with other chronic inflammatory diseases69 and bone degenerative diseases70,71.
Finally, this study also showed a weak but still interesting RF-specific effect of the SHBGrs6259 and FcγR3Ars396991 SNPs to determine the risk of having erosions. Importantly, we could validate the RF-specific effect of the SHBGrs6259 SNP on the risk of developing erosions in the replication population and the meta-analysis of both cohorts confirmed that the effect of this marker was strongly dependent on the RF status. On the other hand, although the RF-specific association of the FcγR3Ars396991 SNP with bone erosions was not statistically significant in the DREAM registry, the meta-analysis of both cohorts confirmed that the effect of this SNP on the risk of bone erosions was modified by the RF.
Whereas little is known about the role of the SHBG locus (17p13) in determining RA progression, a number of experimental studies have shown that the FcγR3A locus (1q23) is involved in the recognition of IgG1 and IgG3 by NK cells and macrophages and that the activation of this receptor by IgG and IgG-RF immunocomplexes might lead to the initiation of a range of sustained and harmful inflammation events that, if chronified, may cause joint and bone destruction and promote the onset of RA72,73,74. In this context and considering the number of studies reporting association of the FcγR3Ars396991 SNP with autoimmune diseases75,76,77,78,79,80,81,82, the response to a wide range of biologic drugs83,84,85,86,87,88,89 and an exacerbated production of TNFα after stimulation of macrophages with LPS for 24 h but also the reported association of the SHBGrs6259 with serum SHBG levels90, we hypothesize that these SNPs might also play a relevant role in determining bone erosions and disease progression.
Considering the noticeable RF-specific impact of the CYP1B1, CYP2C9, ESR2, FcγR3A and SHBG SNPs on the risk of developing erosive disease but also their functional implication in modulating hormone levels and/or immune responses, we decided to assess if the presence of steroid hormone-related SNPs could be useful to reliably predict the appearance of bone erosions in seropositive and seronegative patients separately. To do that, we built a genetic model including demographic variables and those SNPs that were consistently associated with the risk of developing bone erosions in seropositive patients. After removing the SNPs that were not significantly associated with erosive disease in the model, we obtained a model including 5 SNPs within the CYP1B1, CYP2C9, CYP3A4, ESR2, and SHBG loci that significantly improved the ability to predict the risk of developing erosive disease when compared with a reference model including demographic variables. The predictive capacity of these SNPs was restricted to seropositive patients since the addition of the same SNPs (or any other genetic marker) to a model built with demographic variables in seronegative patients did not show any predictive value. The predictive ability of the genetic model in seropositive patients was consistent as no similar models were found after performing a 50.000 permutation test. When we attempted to confirm the utility of this model in the DREAM registry, we found that the CYP1B1 and CYP2C9 SNPs in seropositive patients showed a consistent predictive value for the development of bone erosions. These results suggest that CYP1B1 and CYP2C9 SNPs alone or in combination with other clinical and genetic markers might help to improve the ability to predict the appearance of bone erosions in seropositive patients (~70% of RA patients). Additional studies including these and other genetic and clinical markers are urgently needed to improve our ability to predict disease progression in RA.
This study has strengths and weaknesses. The strengths of this study include a relatively large and well-characterized population and the meta-analyses conducted considering results from the DREAM registry. In the discovery population, we had 80% of power to detect an odds ratio of 1.68 (α = 0.00074) for a SNP with a frequency of 0.25, which underlined the feasibility of the study design. Another important strength of this study is the development of cytokine stimulation experiments and the measurement of seven serum steroid hormones in a large cohort of healthy subjects, which allowed us to investigate the functional role of the most interesting markers in modulating immune responses but also to test their impact on determining steroid hormone levels. A drawback is the multicenter nature of this study that placed inevitable limitations such as the impossibility of using available scores to better define bone erosions (Sharp van der Heijde, Genant, SENSE, and Ratingen scores). Given the cross-sectional approach of the study, we had also intrinsic limitations such as a possible bias due to variations in treatments and follow-up time among study participants. Finally, it is important to mention that the selection of SNPs for this study was influenced by the limited research funds and that the relatively small size of the replication population hampered the validation of the most interesting associations that were detected when a recessive model of inheritance was assumed.
Conclusion
In conclusion, this candidate gene association study suggests, for the first time, that the presence of CYP1B1, CYP2C9, ESR1, ESR2, SHBG and FcγR3A SNPs or haplotypes influence the risk of developing bone erosions in RA. This study also shows that the effect found for most of the SNPs or haplotypes was dependent on the RF status and that genotyping of hormone-related SNPs might help to reliably predict disease progression in seropositive patients.
References
Brooks, P. M. The burden of musculoskeletal disease–a global perspective. Clin Rheumatol 25, 778–781 (2006).
Gregersen, P. K., Silver, J. & Winchester, R. J. The shared epitope hypothesis. An approach to understanding the molecular genetics of susceptibility to rheumatoid arthritis. Arthritis Rheum 30, 1205–1213 (1987).
Wellcome Trust Case Control, C. et al. Association scan of 14,500 nonsynonymous SNPs in four diseases identifies autoimmunity variants. Nature genetics 39, 1329–1337 (2007).
Lettre, G. & Rioux, J. D. Autoimmune diseases: insights from genome-wide association studies. Hum Mol Genet 17, R116–121 (2008).
Okada, Y. et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature 506, 376–381 (2014).
Cuppen, B. V. et al. Personalized biological treatment for rheumatoid arthritis: a systematic review with a focus on clinical applicability. Rheumatology (Oxford) 55, 826–839 (2016).
Umicevic Mirkov, M. et al. Genome-wide association analysis of anti-TNF drug response in patients with rheumatoid arthritis. Ann Rheum Dis 72, 1375–1381 (2013).
Acosta-Colman, I. et al. GWAS replication study confirms the association of PDE3A-SLCO1C1 with anti-TNF therapy response in rheumatoid arthritis. Pharmacogenomics 14, 727–734 (2013).
Cui, J. et al. Genome-wide association study and gene expression analysis identifies CD84 as a predictor of response to etanercept therapy in rheumatoid arthritis. PLoS Genet 9, e1003394 (2013).
Plant, D. et al. Genome-wide association study of genetic predictors of anti-tumor necrosis factor treatment efficacy in rheumatoid arthritis identifies associations with polymorphisms at seven loci. Arthritis Rheum 63, 645–653 (2011).
Liu, C. et al. Genome-wide association scan identifies candidate polymorphisms associated with differential response to anti-TNF treatment in rheumatoid arthritis. Mol Med 14, 575–581 (2008).
Marinou, I., Maxwell, J. R. & Wilson, A. G. Genetic influences modulating the radiological severity of rheumatoid arthritis. Ann Rheum Dis 69, 476–482 (2010).
Maxwell, J. R. et al. Rheumatoid arthritis-associated polymorphisms at 6q23 are associated with radiological damage in autoantibody-positive RA. J Rheumatol 39, 1781–1785 (2012).
Scott, I. C. et al. Do Genetic Susceptibility Variants Associate with Disease Severity in Early Active Rheumatoid Arthritis? J Rheumatol 42, 1131–1140 (2015).
Klareskog, L. et al. A new model for an etiology of rheumatoid arthritis: smoking may trigger HLA-DR (shared epitope)-restricted immune reactions to autoantigens modified by citrullination. Arthritis Rheum 54, 38–46 (2006).
Symmons, D. P. et al. Blood transfusion, smoking, and obesity as risk factors for the development of rheumatoid arthritis: results from a primary care-based incident case-control study in Norfolk, England. Arthritis Rheum 40, 1955–1961 (1997).
Mahdi, H. et al. Specific interaction between genotype, smoking and autoimmunity to citrullinated alpha-enolase in the etiology of rheumatoid arthritis. Nature genetics 41, 1319–1324 (2009).
Mathsson, L., Lampa, J., Mullazehi, M. & Ronnelid, J. Immune complexes from rheumatoid arthritis synovial fluid induce FcgammaRIIa dependent and rheumatoid factor correlated production of tumour necrosis factor-alpha by peripheral blood mononuclear cells. Arthritis Res Ther 8, R64 (2006).
Newkirk, M. M. Rheumatoid factors: host resistance or autoimmunity? Clin Immunol 104, 1–13 (2002).
Dorner, T., Egerer, K., Feist, E. & Burmester, G. R. Rheumatoid factor revisited. Curr Opin Rheumatol 16, 246–253 (2004).
Blom, A. B. et al. Increased expression of Fcgamma receptors II and III on macrophages of rheumatoid arthritis patients results in higher production of tumor necrosis factor alpha and matrix metalloproteinase. Arthritis Rheum 48, 1002–1014 (2003).
Blom, A. B. et al. Fc gamma R expression on macrophages is related to severity and chronicity of synovial inflammation and cartilage destruction during experimental immune-complex-mediated arthritis (ICA). Arthritis Res 2, 489–503 (2000).
van Lent, P. L. et al. Role of Fc receptor gamma chain in inflammation and cartilage damage during experimental antigen-induced arthritis. Arthritis Rheum 43, 740–752 (2000).
Karlson, E. W., Mandl, L. A., Hankinson, S. E. & Grodstein, F. Do breast-feeding and other reproductive factors influence future risk of rheumatoid arthritis? Results from the Nurses’ Health Study. Arthritis Rheum 50, 3458–3467 (2004).
Doran, M. F., Crowson, C. S., O’Fallon, W. M. & Gabriel, S. E. The effect of oral contraceptives and estrogen replacement therapy on the risk of rheumatoid arthritis: a population based study. J Rheumatol 31, 207–213 (2004).
Walitt, B. et al. Effects of postmenopausal hormone therapy on rheumatoid arthritis: the women’s health initiative randomized controlled trials. Arthritis Rheum 59, 302–310 (2008).
Hernandez-Avila, M. et al. Exogenous sex hormones and the risk of rheumatoid arthritis. Arthritis Rheum 33, 947–953 (1990).
Orellana, C. et al. Postmenopausal hormone therapy and the risk of rheumatoid arthritis: results from the Swedish EIRA population-based case-control study. Eur J Epidemiol 30, 449–457 (2015).
Salliot, C., Bombardier, C., Saraux, A., Combe, B. & Dougados, M. Hormonal replacement therapy may reduce the risk for RA in women with early arthritis who carry HLA-DRB1 *01 and/or *04 alleles by protecting against the production of anti-CCP: results from the ESPOIR cohort. Ann Rheum Dis 69, 1683–1686 (2010).
Adorini, L. Intervention in autoimmunity: the potential of vitamin D receptor agonists. Cell Immunol 233, 115–124 (2005).
Luo, C. Y., Wang, L., Sun, C. & Li, D. J. Estrogen enhances the functions of CD4(+)CD25(+)Foxp3(+) regulatory T cells that suppress osteoclast differentiation and bone resorption in vitro. Cell Mol Immunol 8, 50–58 (2011).
Lelu, K. et al. Estrogen receptor alpha signaling in T lymphocytes is required for estradiol-mediated inhibition of Th1 and Th17 cell differentiation and protection against experimental autoimmune encephalomyelitis. J Immunol 187, 2386–2393 (2011).
Tai, P. et al. Induction of regulatory T cells by physiological level estrogen. J Cell Physiol 214, 456–464 (2008).
Kramer, P. R., Winger, V. & Kramer, S. F. 17beta-Estradiol utilizes the estrogen receptor to regulate CD16 expression in monocytes. Mol Cell Endocrinol 279, 16–25 (2007).
Fossati, G., Bucknall, R. C. & Edwards, S. W. Insoluble and soluble immune complexes activate neutrophils by distinct activation mechanisms: changes in functional responses induced by priming with cytokines. Ann Rheum Dis 61, 13–19 (2002).
Robinson, J., Watson, F., Bucknall, R. C. & Edwards, S. W. Activation of neutrophil reactive-oxidant production by synovial fluid from patients with inflammatory joint disease. Soluble and insoluble immunoglobulin aggregates activate different pathways in primed and unprimed cells. Biochem J 286(Pt 2), 345–351 (1992).
Robinson, J. J., Watson, F., Bucknall, R. C. & Edwards, S. W. Stimulation of neutrophils by insoluble immunoglobulin aggregates from synovial fluid of patients with rheumatoid arthritis. Eur J Clin Invest 22, 314–318 (1992).
Arnett, F. C. et al. The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum 31, 315–324 (1988).
van Gestel, A. M. et al. Development and validation of the European League Against Rheumatism response criteria for rheumatoid arthritis. Comparison with the preliminary American College of Rheumatology and the World Health Organization/International League Against Rheumatism Criteria. Arthritis Rheum 39, 34–40 (1996).
Caliz, R. et al. Gender-specific effects of genetic variants within Th1 and Th17 cell-mediated immune response genes on the risk of developing rheumatoid arthritis. PloS one 8, e72732 (2013).
Canet, L. M. et al. Genetic variants within immune-modulating genes influence the risk of developing rheumatoid arthritis and anti-TNF drug response: a two-stage case-control study. Pharmacogenet Genomics 25, 432–443 (2015).
Canet, L. M. et al. Genetic variants within the TNFRSF1B gene and susceptibility to rheumatoid arthritis and response to anti-TNF drugs: a multicenter study. Pharmacogenet Genomics 25, 323–333 (2015).
Schett, G. & Gravallese, E. Bone erosion in rheumatoid arthritis: mechanisms, diagnosis and treatment. Nat Rev Rheumatol 8, 656–664 (2012).
Aletaha, D. et al. Rheumatoid arthritis classification criteria: an American College of Rheumatology/European League Against Rheumatism collaborative initiative. Arthritis Rheum 62, 2569–2581 (2010).
Schaid, D. J., Rowland, C. M., Tines, D. E., Jacobson, R. M. & Poland, G. A. Score tests for association between traits and haplotypes when linkage phase is ambiguous. American journal of human genetics 70, 425–434 (2002).
Nyholt, D. R. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet 74, 765–769 (2004).
Chen, B., Wilkening, S., Drechsel, M. & Hemminki, K. SNP_tools: A compact tool package for analysis and conversion of genotype data for MS-Excel. BMC research notes 2, 214 (2009).
Barrett, J. C., Fry, B., Maller, J. & Daly, M. J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21, 263–265 (2005).
Gabriel, S. B. et al. The structure of haplotype blocks in the human genome. Science 296, 2225–2229 (2002).
Li, Y. et al. A Functional Genomics Approach to Understand Variation in Cytokine Production in Humans. Cell 167, 1099–1110 e1014 (2016).
Schirmer, M. et al. Linking the Human Gut Microbiome to Inflammatory Cytokine Production Capacity. Cell 167, 1897 (2016).
Ter Horst, R. et al. Host and Environmental Factors Influencing Individual Human Cytokine Responses. Cell 167, 1111–1124 e1113 (2016).
Van Booven, D. et al. Cytochrome P450 2C9-CYP2C9. Pharmacogenet Genomics 20, 277–281 (2010).
Canet, L. M. et al. Polymorphisms at phase I-metabolizing enzyme and hormone receptor loci influence the response to anti-TNF therapy in rheumatoid arthritis patients. Pharmacogenomics J (2018).
Cutolo, M. et al. Androgen and estrogen receptors are present in primary cultures of human synovial macrophages. J Clin Endocrinol Metab 81, 820–827 (1996).
Bord, S., Horner, A., Beavan, S. & Compston, J. Estrogen receptors alpha and beta are differentially expressed in developing human bone. J Clin Endocrinol Metab 86, 2309–2314 (2001).
Cutolo, M. et al. Presence of estrogen-binding sites on macrophage-like synoviocytes and CD8+, CD29+, CD45RO+ T lymphocytes in normal and rheumatoid synovium. Arthritis Rheum 36, 1087–1097 (1993).
Recker, R. R., Davies, K. M., Dowd, R. M. & Heaney, R. P. The effect of low-dose continuous estrogen and progesterone therapy with calcium and vitamin D on bone in elderly women. A randomized, controlled trial. Ann Intern Med 130, 897–904 (1999).
Kuiper, S. et al. Influence of sex, age, and menopausal state on the course of early rheumatoid arthritis. J Rheumatol 28, 1809–1816 (2001).
Cutolo, M. et al. New roles for estrogens in rheumatoid arthritis. Clin Exp Rheumatol 21, 687–690 (2003).
Dai, R., Phillips, R. A., Karpuzoglu, E., Khan, D. & Ahmed, S. A. Estrogen regulates transcription factors STAT-1 and NF-kappaB to promote inducible nitric oxide synthase and inflammatory responses. J Immunol 183, 6998–7005 (2009).
Wihlen, B., Ahmed, S., Inzunza, J. & Matthews, J. Estrogen receptor subtype- and promoter-specific modulation of aryl hydrocarbon receptor-dependent transcription. Mol Cancer Res 7, 977–986 (2009).
Da Silva, J. A. Sex hormones and glucocorticoids: interactions with the immune system. Ann N Y Acad Sci 876, 102–117; discussion 117–108 (1999).
Erlandsson, M. C., Ohlsson, C., Gustafsson, J. A. & Carlsten, H. Role of oestrogen receptors alpha and beta in immune organ development and in oestrogen-mediated effects on thymus. Immunology 103, 17–25 (2001).
Mo, R. et al. Estrogen regulates CCR gene expression and function in T lymphocytes. J Immunol 174, 6023–6029 (2005).
Kobayashi, S. et al. Association of bone mineral density with polymorphism of the estrogen receptor gene. J Bone Miner Res 11, 306–311 (1996).
Willing, M. et al. Bone mineral density and its change in white women: estrogen and vitamin D receptor genotypes and their interaction. J Bone Miner Res 13, 695–705 (1998).
Masi, L. et al. The role of osteoprotegerin (OPG) and estrogen receptor (ER-alpha) gene polymorphisms in rheumatoid arthritis. Clin Cases Miner Bone Metab 4, 156–160 (2007).
Lian, K. et al. Estrogen receptor alpha genotype is associated with a reduced prevalence of radiographic hip osteoarthritis in elderly Caucasian women. Osteoarthritis Cartilage 15, 972–978 (2007).
Albagha, O. M., McGuigan, F. E., Reid, D. M. & Ralston, S. H. Estrogen receptor alpha gene polymorphisms and bone mineral density: haplotype analysis in women from the United Kingdom. J Bone Miner Res 16, 128–134 (2001).
Sano, M. et al. Association of estrogen receptor dinucleotide repeat polymorphism with osteoporosis. Biochem Biophys Res Commun 217, 378–383 (1995).
Karsten, C. M. & Kohl, J. The immunoglobulin, IgG Fc receptor and complement triangle in autoimmune diseases. Immunobiology 217, 1067–1079 (2012).
Wu, J. et al. A novel polymorphism of FcgammaRIIIa (CD16) alters receptor function and predisposes to autoimmune disease. The Journal of clinical investigation 100, 1059–1070 (1997).
Guilliams, M., Bruhns, P., Saeys, Y., Hammad, H. & Lambrecht, B. N. The function of Fcgamma receptors in dendritic cells and macrophages. Nature reviews. Immunology 14, 94–108 (2014).
Bournazos, S., Woof, J. M., Hart, S. P. & Dransfield, I. Functional and clinical consequences of Fc receptor polymorphic and copy number variants. Clinical and experimental immunology 157, 244–254 (2009).
Nimmerjahn, F. Activating and inhibitory FcgammaRs in autoimmune disorders. Springer seminars in immunopathology 28, 305–319 (2006).
Koene, H. R. et al. The Fc gammaRIIIA-158F allele is a risk factor for systemic lupus erythematosus. Arthritis and rheumatism 41, 1813–1818 (1998).
Zuniga, R. et al. Low-binding alleles of Fcgamma receptor types IIA and IIIA are inherited independently and are associated with systemic lupus erythematosus in Hispanic patients. Arthritis and rheumatism 44, 361–367 (2001).
Nieto, A. et al. Involvement of Fcgamma receptor IIIA genotypes in susceptibility to rheumatoid arthritis. Arthritis Rheum 43, 735–739 (2000).
Manger, K. et al. Fcgamma receptor IIa, IIIa, and IIIb polymorphisms in German patients with systemic lupus erythematosus: association with clinical symptoms. Annals of the rheumatic diseases 61, 786–792 (2002).
Latiano, A. et al. Evaluating the role of the genetic variations of PTPN22, NFKB1, and FcGRIIIA genes in inflammatory bowel disease: a meta-analysis. Inflamm Bowel Dis 13, 1212–1219 (2007).
Lee, Y. H., Ji, J. D. & Song, G. G. Associations between FCGR3A polymorphisms and susceptibility to rheumatoid arthritis: a metaanalysis. J Rheumatol 35, 2129–2135 (2008).
Montes, A. et al. FCGR polymorphisms in the treatment of rheumatoid arthritis with Fc-containing TNF inhibitors. Pharmacogenomics 16, 333–345 (2015).
Lee, Y. H., Bae, S. C. & Song, G. G. Functional FCGR3A 158 V/F and IL-6 -174 C/G polymorphisms predict response to biologic therapy in patients with rheumatoid arthritis: a meta-analysis. Rheumatol Int 34, 1409–1415 (2014).
Kim, D. H. et al. FCGR3A gene polymorphisms may correlate with response to frontline R-CHOP therapy for diffuse large B-cell lymphoma. Blood 108, 2720–2725 (2006).
Cartron, G. et al. Therapeutic activity of humanized anti-CD20 monoclonal antibody and polymorphism in IgG Fc receptor FcgammaRIIIa gene. Blood 99, 754–758 (2002).
Weng, W. K. & Levy, R. Two immunoglobulin G fragment C receptor polymorphisms independently predict response to rituximab in patients with follicular lymphoma. J Clin Oncol 21, 3940–3947 (2003).
Musolino, A. et al. Immunoglobulin G fragment C receptor polymorphisms and clinical efficacy of trastuzumab-based therapy in patients with HER-2/neu-positive metastatic breast cancer. J Clin Oncol 26, 1789–1796 (2008).
Robledo, G. et al. Association of the FCGR3A-158F/V gene polymorphism with the response to rituximab treatment in Spanish systemic autoimmune disease patients. DNA Cell Biol 31, 1671–1677 (2012).
Bhatnager, R., Senwal, A., Nanda, S. & Dang, A. S. Association of rs6259 polymorphism with SHBG levels and Poly Cystic Ovary Syndrome in Indian population: a case control study. Mol Biol Rep 46, 2131–2138 (2019).
Acknowledgements
The authors thank all participants who agreed to participate in this study. They thank María Dolores Casares, Ángeles Molina, and Carmen Olóriz for sample collection and technical support. They also thank Biobanco-IMM (Lisbon Academic Medical Center, Lisbon, Portugal) for providing blood samples from Portuguese RA patients and controls. This study was supported by a research grant from FIBAO foundation.
Author information
Authors and Affiliations
Contributions
R.C. and J.S. designed the study and drafted the manuscript. J.M.S.-M., L.C., C.B.L. and A.R.-R. were responsible for genotyping of the different populations. R.C., A.A.B., H.C., A.G., E.P.-P., A.G.-U., A.E., M.A.F., E.C.-S., M.A.L.-N., M.J., J.E.F., M.J.H.C. and J.S. coordinated the sample collection and H.C., J.S.-C. and M.J.S.-P. were involved in the records review and data acquisition. R.T.H., O.B.B., R.N.-M., Y.L., M.G.N. were implicated in providing functional data. M.M.-B. and J.S. did the statistical data analysis. All authors contributed to, seen, and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sánchez-Maldonado, J.M., Cáliz, R., Canet, L. et al. Steroid hormone-related polymorphisms associate with the development of bone erosions in rheumatoid arthritis and help to predict disease progression: Results from the REPAIR consortium. Sci Rep 9, 14812 (2019). https://doi.org/10.1038/s41598-019-51255-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-51255-0
This article is cited by
-
Sex steroids and autoimmune rheumatic diseases: state of the art
Nature Reviews Rheumatology (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.