Abstract
Tritrophic interactions allow plants to recruit natural enemies for protection against herbivory. Here we investigated genetic variability in induced responses to stemborer egg-laying in maize Zea mays (L.) (Poaceae). We conducted a genome wide association study (GWAS) of 146 maize genotypes comprising of landraces, inbred lines and commercial hybrids. Plants were phenotyped in bioassays measuring parasitic wasp Cotesia sesamiae (Cameron) (Hymenoptera: Braconidae) attraction to volatiles collected from plants exposed to stemborer Chilo partellus (Swinhoe) (Lepidoptera: Crambidae) eggs. Genotyping-by-sequencing was used to generate maize germplasm SNP data for GWAS. The egg-induced parasitoid attraction trait was more common in landraces than in improved inbred lines and hybrids. GWAS identified 101 marker-trait associations (MTAs), some of which were adjacent to genes involved in the JA-defence pathway (opr7, aos1, 2, 3), terpene biosynthesis (fps3, tps2, 3, 4, 5, 7, 9, 10), benzoxazinone synthesis (bx7, 9) and known resistance genes (e.g. maize insect resistance 1, mir1). Intriguingly, there was also association with a transmembrane protein kinase that may function as a receptor for the egg elicitor and other genes implicated in early plant defence signalling. We report maize genomic regions associated with indirect defence and provide a valuable resource for future studies of tritrophic interactions in maize. The markers identified may facilitate selection of indirect defence by maize breeders.
Similar content being viewed by others
Introduction
Plants, being rooted to the ground and unable to flee from attack, have evolved highly sophisticated ways of defending themselves from insect herbivores, over a 400-million-year period of plant-insect interaction and coevolution1,2,3. These defences are wide ranging and can include physical barriers such as lignin, anti-nutritive substances like tannin, production of antibiotics and interactions with associated organisms, which function as natural enemies of the herbivores2,4. Some defences are constitutively expressed, whereas others are induced after exposure to herbivore attack, when elicitors from the insect saliva or insect eggs trigger defence responses5,6,7. When plants are exposed to herbivory, they emit herbivore induced plant volatiles (HIPVs)8,9,10. Plants tend to emit larger amounts of volatiles than the insect that is feeding on it and parasitoids and predators have evolved to use HIPVs as cues to locate the herbivores they use as hosts or prey10,11,12. Plant defence involving interaction with the third trophic level is referred to as “indirect defence”, in contrast to direct defences that make the plant less suitable for the herbivore10.
While wild plants in natural ecosystems are under natural selection for the ability to defend themselves, domesticated crops have been subjected to artificial selection and are often grown as monocrops13,14,15. These crop plants are vulnerable to attack by adapted insect herbivores, which become pests in this context16. Modern maize (corn), Zea mays (L.) (Poaceae), was domesticated approximately 9,000 years ago from wild teosinte species17. Maize is an essential staple and cash crop for millions of people, particularly in sub-Saharan Africa (SSA). Maize production remains severely constrained by Lepidopteran stemborers, devastating pests of staple cereals in SSA, which reduce yields by up to 80%, depending on the pest population density and the phenological stage of the crop at infestation18. Stemborers infest about 50% of the agricultural land in the SSA region, affect the lives of nearly 300 million people and cause yield losses of approximately US$ 1.5 billion per annum18.
Nearly all recent commercial maize breeding (artificial selection) has been undertaken in a pesticide treated background19. Conventional breeding for host plant resistance against attacking insect pests has largely been done by trial and error or by exposing different genetic lines of crops to the herbivore pests. Potential interactions between different crop genotypes and the natural enemies of the pests have received less attention. We hypothesised that locally adapted varieties (landraces) preferred by smallholder farmers who cannot afford pesticides might have better indirect defence than commercially bred genotypes. Evidence in support of this hypothesis was provided when we discovered that three farmer-selected landraces of maize emitted HIPVs in response to stemborer, Chilo partellus (Swinhoe) (Lepidoptera: Crambidae), egg laying whereas the two commercial varieties initially tested did not7. The HIPVs emitted by the landraces attracted Cotesia sesamiae (Cameron) (Hymenoptera: Braconidae) parasitoid wasps, which are a key natural enemy of the stemborer. These landraces were of South American origin and further studies provided evidence that the egg induced indirect defence trait also exists in some African maize landraces20 and in wild teosinte species21.
Given these promising initial findings, we embarked on a much larger genome wide association study (GWAS), involving 146 maize genotypes, reported here. The plant trait (phenotype) studied was the ability to “cry for help” by emitting HIPVs to attract C. sesamiae parasitic wasp bodyguards after egg deposition by C. partellus moths. Our objectives were 1. to determine how widespread this trait was in a wider germplasm collection comprising locally adapted landraces, improved breeding lines and higher yielding commercial varieties, and 2. to develop molecular markers for the indirect plant defence trait. We used GWAS to discover genomic regions and molecular markers associated with it. To the best of our knowledge, this is the first GWAS of parasitoid response to plants induced with insect eggs.
Results
Parasitoid attraction to egg-induced volatiles, trait distribution across 146 maize genotypes
A diverse collection of 146 genotypes was tested (Table 1), comprising 9 landraces, 116 inbred lines and 21 hybrids. These were screened, to establish the presence/absence of the egg-induced parasitoid attraction trait. For each genotype, volatiles were sampled, from at least 4 plants with eggs and 4 plants without eggs. Responses of C. sesamiae parasitoid wasps to these volatiles were measured in an olfactometer bioassay, with at least 9 parasitoid wasp bioassays per genotype. DNA samples were collected from the same plants (see below). Olfactometer bioassay data are shown in Supplementary Table 1, which details the mean time spent by wasps in the different arms of the olfactometer arm (i.e. arms containing volatiles from plants with eggs; volatiles from plants without eggs, and the mean of the two solvent control arms). Egg-induced parasitoid attraction was observed when wasps spent significantly more time in the “with eggs” zone than in the “without eggs” zone. The trait was normally distributed in the whole population, as well as in the various classification groups (Supplementary Figure 1).
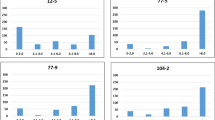
Analysis of variance (ANOVA) revealed significant differences in parasitoid wasp responses (\(\textit{P} < 0.05\)) for the time spent in the different olfactometer arms for 43 genotypes (Supplementary Table 1). We plotted the means of these 43 genotypes and observed a clear difference between the mean time spent in the olfactometer arm containing volatiles collected from plants with stemborer eggs, in comparison with the controls (Supplementary Figure 2). By comparing mean time spent in “with eggs” and “without eggs” arms, we found 42 genotypes in which the means of observations were significantly different (Table 2) i.e. there was attraction to egg-induced volatiles. Figure 1 shows differences between “with eggs” and “without eggs” observations per plant for these 42 genotypes, of which 6 were landraces, 33 were inbred lines and 3 were hybrids. Landraces, therefore, gave the highest proportion of number of plants having the trait (6 out of 15 screened = 40%) in comparison with inbred lines (33 out of 130 = 25.4%) and hybrids (3 out of 23 = 13%).
(A) Diagram of the 4-arm olfactometer. Insects were allowed to walk freely between 4 discrete odour zones (Zone 1: volatiles from plant without eggs; Zone 3: volatiles from plant with eggs; Zones 2 and 4: solvent blank). Time spent (min) in each zone was recorded. (B) Chilo partellus eggs. (C) Cotesia sesamiae parasitoid wasp. (D) Olfactometer bioassay response of parasitoid wasp, C. sesamiae, to volatiles from maize plants with and without stemborer, C. partellus, eggs. Parasitoids could choose between a zone containing volatiles from a plant with eggs (“WithEgg”) and a zone containing volatiles from a plant without eggs (“WithoutEgg”). Mean time spent (min ± SE) is shown for each genotype. Only genotypes that were significantly attractive (\(\textit{P} < 0.05\), ANOVA) are shown.
SNP discovery, distribution, heterozygosity and linkage disequilibrium
Genotyping-by-sequencing (GBS) data were generated from 1018 individual maize plants (4-6 replicates per genotype), which were representative of 146 diverse accessions (Table 1). In total, 2.1 billion reads were generated at an average of 2.06 million reads per maize genotype. We called 316,127 (0.32M) raw SNPs from all the plants genotyped, and later filtered the raw SNPs to 54,311 (54K) for subsequent analysis. The distribution of the 54K SNPs across the maize genome is shown in Supplementary Figure 3 and Supplementary Table 2. The number of SNPs per chromosome ranged from 3748 (Chr10) to 9275 (Chr1). The filtered SNPs resulted in an average marker density of 27 SNPs/Mbp of the maize genome. The average heterozygosity proportion for the whole maize population was 0.048 but was higher in hybrid (0.094) and landrace (0.080) subpopulations and lower in the inbred subpopulation (0.037). Linkage disequilibrium (LD) and LD decay distance in the 10 maize chromosomes are summarised in Supplementary Figures 4 and 5. The average whole genome LD decay is shown in Supplementary Figure 4. The genetic distance at which the estimated \(\hbox {R}^{2}\) fell below 0.2 ranged from 0.9 kb to 1 kb in all the 10 maize chromosomes except chromosomes 4 and 8 (Supplementary Figure 5). LD decay for chromosomes 4 and 8 ranged from 1 kb to 1.5 kb at \(\hbox {R}^{2} < 0.4\).
Phylogenetic tree showing genetic diversity of the maize genotypes using neighbor-joining method. Scale represents genetic distance: 0.1 is 10% genetic difference between genotypes. Blue, green and red lines represent landraces, hybrids and inbred lines respectively (also denoted by -L, -H and -I suffixes on genotype names). Genotypes in bold indicate where genetic separation was found within a genotype. Clusters A–F represent discrete genetic groups with similar pedigree and origin.
Genetic diversity and population structure in the maize population
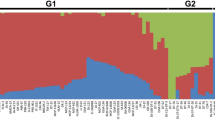
A similarity cladogram across the maize genotypes revealed 6 clusters (Fig. 2), generally grouped according to their pedigree and origin. Cluster A was composed mainly of landraces, inbred lines and hybrids developed at a local breeding program in Kenya (Maseno University). There was distinct clustering of inbred lines whose names started with the acronym CKSPL (Fig. 2, Cluster B). Landraces from Cuba, Brazil, Haiti and Venezuela clustered together (Cluster F). The rest of the genotypes clustered according to their pedigree and breeding history. We observed significant genotypic differences among plants of two landraces (“Nyamula” and “Jowi-red”) and four inbred lines (Ext-STR-150, MSMP-ZEBRA-2 and X87/02/312-F4-5, CML-395). Each genetically diverse plant was further treated as an independent genotype, bringing the total number of distinct genotypes used for genetic analysis to 167 lines. Furthermore, principal component analysis (PCA) confirmed a similar pattern of genetic diversity among the maize genotypes (Supplementary Figure 6) with the first three principal components (PC1, PC2 and PC3) explaining 14.5%, 5% and 3.2% of total genetic variance respectively. The ADMIXTURE model (K = 6) also predicted an optimum population number of 6 (Supplementary Figure 7). An admixed population is one that has multiple ancestral genetic proportions i.e. there is evidence of outbreeding. Populations A and D consisted mainly of inbred lines and advanced crosses being used in the Maseno University breeding program. Other than populations A, B and D, in which the accessions did not have significant admixtures, the rest of the clusters comprised mainly of admixed populations (Supplementary Figure 7). The genotypes that were considered to have the indirect defence trait did not cluster in any preferential manner but were distributed across the various populations.
Marker-trait associations
A total of 101 significant SNP-trait associations were identified (Supplementary Table 3) using both GLM + PCA and MLM + PCA + K analysis approaches, after FDR correction (q-value of 0.05)(GLM = General Linear Model, PCA = Principal Component Analysis, MLM = Mixed Linear Model, K analysis = Cluster analysis, FDR = False Discovery Rate). The P-value threshold was 9.23 × 10-5. The Manhattan plot of associated SNPs (for parasitoid wasp response to stemboer egg-induced plant volatiles, analysed with the 54,311 SNPs) is shown in Fig. 3 for MLM + PCA + K analysis, and Supplementary Figure 8 for GLM + PCA analysis. All 101 identified SNPs were located across all 10 maize chromosomes. More than half of the significant markers were located on chromosomes 1 (21 SNPs), 5 (12 SNPs), 8 (10 SNPs) and 10 (15 SNPs) (Supplementary Table 3). The QQ plots (Fig. 3 and Supplementary Figure 8) revealed that both GL and ML models successfully controlled any false positive associations that may have resulted from underlying population structure. The phenotypic variation (\(\hbox {R}^{2}\)) explained by the associated SNPs in GLM and MLM approach ranged from 0.099 – 0.498 and 0.123 – 0.409, respectively. These high \(\hbox {R}^{2}\) values and their consistency in both GLM and MLM approaches provide more confidence to the identified SNPs and are an indication that the association is not merely by chance.
(A) Manhattan plot using MLM approach indicating SNPs significantly associated with the egg induced parasitoid attraction trait (shown in red). SNP density is indicated by the colour scale on the bar next to the X-axis (scale given in inset). The X-axis is the genomic position of the SNPs in the genome, and the Y- axis is -log10 of the P-values. Each chromosome is coloured differently. The grey horizontal line represents the minimal significant level at the cutoff of FDR 0.05 (MLM Mixed Linear Model, SNP Single Nucleotide Polymorphism, FDR False Discovery Rate) (B) Quantile-quantile plot.
We retrieved 33 candidate genes (Table 3) within 10 Mbp up- and downstream of 23 associated SNP QTL (quantitative trait locus) positions using the ZmB73 RefGen v2 database (https://www.maizegdb.org/gbrowse). These genes have previously been annotated with a plant defence function. The phenotypic variation (\(\hbox {R}^{2}\)) of the 23 associated SNPs ranged from 0.099 – 0.498 with the GLM + PCA approach and 0.123 – 0.409 with the MLM + PCA + K approach. The distance between the 33 candidate genes and SNP positions ranged from 0.0007 mb (cdpk13 gene) to 9.8 mb (bx7 gene). Detailed information about candidate genes and their roles in plant defence is given in Supplementary Table 4. We also provide, in Supplementary Table 5, a listing of 202 genes located within a 10 mb region of the top 16 SNPs (selected based on having an \(\hbox {R}^{2}\) value of above 25% (0.25) with the MLM approach).
Discussion
Multitrophic interactions with natural enemies of herbivores allow plants to increase herbivore mortality by recruitment of “bodyguards” after changing their volatile emission profile to become more attractive to the natural enemies12,22. This “call for help” signalling is known as indirect defence10. The genetic basis for variation in insect egg-induced indirect defence between crop genotypes is poorly understood and therefore the current study was designed to identify regions of the maize genome associated with it, using C. partellus as the herbivore and C. sesamiae as the natural enemy. Our current study bridges the gap between studies of the chemical ecology of multitrophic interactions and plant genomics. Our previous studies7,20 showed that certain maize landraces responded to egg laying, the earliest stage of attack by maize stemborer, C. partellus insects, by emitting volatiles attractive to parasitoid wasps that are key natural enemies of the herbivore. However, this indirect defence trait was absent in the limited number of improved hybrids we initially tested. Here we provide a much larger analysis of 146 maize genotypes, comprising landraces, inbred lines and commercial hybrids, in a genome wide association study (GWAS).
Our earlier studies7,20revealed the suite of plant volatiles induced by C. partellus eggs in maize. Thus, identification of the HIPVs was not the focus of the current study. Volatile samples in the current study were analysed by gas chromatography (data not shown) and similar key compounds, in particular (3E)-4,8-dimethyl-1,3,7-nonatriene (DMNT), were induced. Here we aimed to identify molecular markers for parasitoid attraction and obtain insight into adjacent potential candidate genes underpinning this indirect defence trait. Availability of molecular markers, provided in the current study, could facilitate accelerated breeding for improved maize cultivars with the indirect defence trait through marker assisted selection (MAS). We used the parasitoid bioassay response itself to directly measure parasitoid attraction, rather than use a proxy in terms of HIPVs. Our study used a biodiverse collection of maize genotypes, which were exposed to C. partellus eggs prior to volatile collection. Volatile samples were then used in large scale parasitoid bioassays, for all 146 genotypes, in a choice test, testing if volatiles from egg exposed plants were significantly preferred to volatiles from unexposed control plants. We found the indirect defence trait was more widespread in landrace germplasm (40% of genotypes) but, because these were not genetically uniform, considerable variation between individual plants was observed. The trait was found in 25% of inbred lines and in 13% of hybrids. These lines were more consistent because they were genetically uniform. Furthermore, discovering the indirect defence trait in improved lines opens up the prospect of introgressing the trait into other higher yielding maize cultivars with desirable agronomic characteristics.
Data were subjected to a GWAS analysis which revealed 101 SNPs strongly associated with the trait. Within a 10mb region of the genome next to these SNPs, there were 33 candidate genes that may code for the trait. Of these, 7 are terpene synthase genes (tps2, tps3, tps4, tps5, tps7, tps9 and tps10). This is not surprising because the indirect defence trait operates by emission of volatiles. Previous studies have linked terpene synthases to indirect defence23,24. Genes implicated in DMNT emission, induced by a synthetic jasmonic acid (JA) analogue, were investigated in an earlier GWAS by Richter et al.24 who found a strong association with tps2. Farnesyl diphosphate synthase3 (fps3) is another candidate gene and catalyses biosynthesis of precursor molecules for terpene biosynthesis. Several of our other candidate genes are implicated in plant secondary metabolism. The most notable of these are 12-oxo-phytodienoic acid reductase7 (opr7), allene oxide synthesis1 (aos1) allene oxide synthesis2 (aos2) and allene oxide synthesis3 (aos3) which encode key enzymes in the JA-defence pathway25,26. Another candidate gene that potentially plays a role is methionine S-methyltransferase1 (mmt1) as methyltransferases can be involved in plant volatile biosysnthesis27.
To trigger the plant defence cascade culminating in release of herbivore induced volatiles, the plant needs to detect the presence of the insect eggs through molecular recognition of the egg elicitor. A putative receptor gene, GRMZM2G438840, is strongly associated with the trait. It is annotated as a leucine-rich repeat transmembrane protein kinase family protein and was identified by28 as a putative immune receptor gene. A topic for future studies would be to investigate if silencing this gene prevents molecular recognition of C. partellus eggs. A clade I L-type lectin receptor kinase LecRK-I.8 has recently been shown to be involved in detection of Pieris brassicae insect eggs in Arabidopsis29. There is also a chitinase2 (chn2) which could play a role in interactions with eggs that contain chitin.
Two of our candidate genes are implicated in early plant defence signalling: calcium dependent protein kinase13 (cdpk13) has been shown to be a component of touch- and wound-induced pathways involved in early stages of local and systemic responses in maize30. Calcium-dependent protein kinases (CDPKs) play a vital role in stress signalling by detecting increases in \(\hbox {Ca}^{2+}\) and transducing them into phosphorylation events31. We also found a mitogen-activated protein kinase, MAP kinase15 (mpk15), associated with the indirect defence trait. Reducing the function of MAP kinases has been reported to impair the synthesis of secondary stress signals, including JA, and loss of MAPK function results in reduced resistance of plants to herbivorous insects32. It thus seems plausible that cdpk13 and/or mpk15 play a role in egg-induced signal transduction. We also found two pectin methylesterases (PMEs) - pectin methylesterase1 (pme1) and pectin methylesterase31 (pme31). These are noteworthy because PMEs are involved in cell wall modification and pectin catabolic processes33.
Interestingly, some of our candidate genes are associated with direct defence. These include maize insect resistance1 (mir1) and maize insect resistance2 (mir2) which encode cysteine proteinases (key defensive proteins against chewing insect pests in maize)34; benzoxazinone synthesis7 (bx7) and benzoxazinone synthesis9 (bx9), genes for benzoxazinoid biosynthesis35, and brown midrib 2 (bm2) which is associated with lignin synthesis36, a physical defence. Although we used an unbiased approach in selecting SNPs via the GWAS procedure, the selection of candidate genes was limited by searching for genes annotated with defence functions and it is likely that more genes are known for direct defence than are currently known for indirect defence. Another explanation is that plants that can recognise insect eggs have a suite of defences that are triggered upon detection of eggs which include direct as well as indirect defences. It is possible that genes encoding direct and indirect defences could cluster together in the genome but this would require further study.
Given the above candidate genes, we would like to suggest a hypothetical model by which the egg sensitive maize genotypes respond: Firstly, there is a molecular recognition process by which the C. partellus egg elicitor is detected; secondly, the JA-defence pathway is triggered, and, thirdly, JA-associated defences, including HIPV emission are triggered. Thus, the egg sensitive genotypes elicit a suite of defences following stemborer oviposition that comprise both direct and indirect defences which will protect the plant against caterpillars emerging from the eggs.
The SNP molecular markers we have identified provide a resource for future studies of the underpinning genetics involved in indirect defence. We have highlighted regions of the genome associated with parasitoid attraction and have identified candidate genes already annotated with plant defence functions. However, it is likely that there are further genes, not yet annotated, that play a role. A particularly interesting opportunity is to discover a plant receptor used for recognition of the egg elicitor. Novel genes could be discovered that play a role in plant signal recognition, particularly of small lipophilic molecules reviewed in37, and biosynthetically related to the egg elicitor (currently under structural elucidation by some of the authors here). Thus, we hope our dataset will allow identification of novel genes involved in indirect defence signalling between maize plants provoked by herbivore (C. partellus) eggs and parasitoid wasp “bodyguards” that have not previously been annotated as having roles in plant defence. We provide information (in Supplementary Table 5) about genes in areas of the maize genome in the vicinity of the top 16 SNPs most closely associated with the indirect defence trait.
There are global pressures to reduce pesticide use in agriculture and in any case few African smallholder farmers in the study region have access to pesticides. The current findings will help develop improved maize varieties with indirect defence against stemborers because we have already identified improved lines and hybrids possessing the trait. Preliminary field trials indicate an increase in parasitism of maize stemborers in genotypes with the indirect defence trait. The indirect defence trait was rarer in improved lines than in landraces, perhaps because selection for yield and quality in commercial crop breeding environments could have compromised defence traits because the value of any defence traits would not be realised when plants were treated with insecticide38,39. However, it was less rare than expected. Our current findings open up the prospect of breeding crops that enhance biological control of insect pests by natural enemies, such as C. sesamiae, through marker assisted selection (MAS). For example, the CIMMYT ESA hybrid maize breeding program is mainly based on four parental lines (CML444, CML395, CML312 and CML442)40. We found that CML312 and CML442 possess the egg-induced parasitoid attraction trait, whereas CML395 and CML444 do not. Therefore our study identifies germplasm that could be used to introgress the trait into improved crops. Such crops would be more resilient to insect attack, difficult for insect to develop resistance and less dependent on pesticide application. They would, however, require natural enemies of pests to be present in the agricultural ecosystem as an ecosystem service. A recent meta-analysis41 found that “top-down” control of herbivorous insect populations by natural enemies is at least as important as “bottom-up” control by the plant and, thus, breeding crops for increased tritophic interaction with natural enemies42 could be a promising approach. Future work should investigate if the genetics identified in the current study with C. partellus stemborers is also involved in indirect defence against a new threat to maize in Africa—the invasive fall armyworm, Spodoptera frugiperda.
Methods
Plant material
A diverse collection of 146 maize genotypes comprising 9 landraces, 116 inbred lines and 21 commercial hybrid varieties were obtained from local farmers (farmer preferred landraces), Maseno University (Kenya), the International Maize and Wheat Improvement Center (CIMMYT, Nairobi, Kenya) and commercial seed suppliers (Table 1). Plants were grown individually in pots filled with fertilised soil in an insect-proof screen house at icipe-Thomas Odhiambo campus (ITOC), Mbita Point (0°25’S, 34°12’E; c. 1200 m above sea level), western Kenya. All plants were grown under natural conditions (c. 25 °C, 65% RH, 12L:12D).
Insects
Field-collected C. partellus were reared on a semi-synthetic diet containing sorghum (Sorghum bicolor) leaf powder43. The larval parasitoid C. sesamiae was reared on stemborer larvae using methodologies described previously44. Experimental insects were maintained at the insect mass rearing unit of icipe-Thomas Odhiambo campus (\(24 \pm 3\) °C, \(70 \pm 5\)% RH, 12L: 12D). The insect culture was infused with field-collected insect population every 3 months to avoid genetic decay and maintain the original behavioural characteristics of the species. Naïve, 1-day old mated female parasitoids obtained from the fourth to fifth generation were used in experiments.
Volatile collection
Volatile compounds from whole maize plants, with and without stemborer eggs, were collected by headspace sampling7. Volatiles were collected from at least 4 plants with and 4 plants without eggs per genotype. Prior to volatile collection, 4-week old maize seedlings were placed inside oviposition cages (\(80 \times 40 \times 40\) cm) into which six gravid female stemborer moths were introduced and kept overnight for oviposition. Concurrently, control plants were kept inside similar cages, but without stemborer moths. Volatiles were collected the following day, starting from the last 2 h of photophase, for 24 h. Leaves of plants with or without eggs were enclosed in polyethyleneterephthalate (PET) bags (volume 3.2 L, \(\simeq \) 12.5 mm thickness) heated to 150 °C before use and fitted with Swagelock inlet and outlet ports. Charcoal-filtered air was pumped (500 mL min−1) through the inlet port. Volatiles were collected on Porapak Q (0.05 g, 60/80 mesh; Supelco, Bellefonte, PA, USA) filters inserted in the outlet port through which air was drawn at 300 mL min−1. After entrainment, volatiles were eluted with 0.5 mL dichloromethane (Sigma Aldrich) for use in subsequent bioassays. Volatiles were collected from 1,168 plants representing 146 genotypes.
Olfactometer bioassay
To phenotype the egg-induced indirect defence trait, behavioural responses of parasitoids to volatiles from different maize genotypes were tested in a Perspex four-arm olfactometer (Fig. 1) described in7. Air was drawn through the four arms towards the centre at 260 mL min−1. Headspace samples (10 μL aliquots) were applied, using a micropipette (Drummond “microcap”, Drummond Scientific Co., Broomall, PA, USA), to a piece of filter paper (4 × 25 mm) subsequently placed in an inlet port at the end of each Olfactometer arm. Mated female parasitoids, without previous exposure to plants or hosts, were transferred individually into the central chamber of the Olfactometer using a custom-made piece of glass tubing. Time spent in each olfactometer arm was recorded with “Olfa” software (F. Nazzi, Udine, Italy) for 12 min.
The experiments were replicated 9 - 15 times per plant. A choice test was carried out to compare insect responses to headspace samples from oviposition-induced and control plants for all 146 maize genotypes. The two opposite arms held the test stimuli (10 μL aliquots of headspace sample) that had been collected from plants that had stemborer eggs and those without the eggs (see Fig. 1). This dose was approximately equal to the amount emitted by 12 plants over 10 min7. The remaining two arms were solvent controls. For each plant, we calculated the average proportion of time spent by the parasitoid in each olfactometer arm across all replications and compared the means using analysis of variance (ANOVA). The means from the two arms representing the solvent controls were analysed together. Comparisons were made: 1. between time spent in arms containing volatiles from solvent control and from a plant with eggs, and 2. between time spent in arms containing volatiles from “with eggs” and “without eggs” plants. Significant observations were determined using P \(\le \) 0.05. Means of significant observations were separated using Fisher’s LSD test with \(\alpha \) set at 0.05 (Genstat version 10, VSN International, Hemel Hempstead, UK). An attraction index was calculated by dividing proportion of time spent in the treated olfactometer area by time spent in the solvent blank control area and log10 transforming the data. These attraction index values were used to draw normal distribution curves using the ggplot2 package in R studio (Version 1.1.383) (Supplementary Figure 1). The calculated attraction index value was used for GWAS.
DNA extraction and genotyping
Fresh leaf samples were collected from assayed plants, immersed in liquid nitrogen, and crushed into fine powder using mortar and pestle. DNA was extracted from 1018 plants (146 maize genotypes, 4-6 plants per genotype) (Table 1) using the DNeasy mini kit (Qiagen, Hilden, Germany), according to manufacturer’s instructions, from at least four individuals per genotype. Purity and quantity of the extracted DNA was determined using gel electrophoresis and a Qubit 2.0 Fluorometer (Life Technologies, Carlsbad, CA, USA) respectively with final dilution to 30 ng/μL. The DNA was sent to Cornell University for library construction (ApeKI restriction enzyme) and genotyping-by-sequencing (GBS). The resulting raw reads were processed using the GBS pipeline of the Trait Analysis by aSSociation, Evolution and Linkage (TASSEL) 5.0 program45. Raw SNPs were further filtered using a minor allele frequency of \(\ge \) 0.05, minimum depth coverage of 5, maximum mismatch of 3 for alignment, and maximum missing data of 30%. Chromosomal assignment and position of SNPs on the physical map was deduced from the draft whole B73 genome sequence of ZmB73 RefGen v246. SNPs were designated based on chromosome number and position (e.g. Chr1_187669221 meaning SNP located at 187669221th position on chromosome 1).
Genetic fidelity, diversity, population structure and Genome Wide Association Study
A filtered SNP dataset was used for all molecular analysis in this study. Genetic fidelity was confirmed with identity-by-state distance matrix in Tassel 5.0. We used the filtered SNP data set to generate a Neighbor-Joining cladogram and estimated principal component analysis (PCA) with covariance and five components. The population structure of the genotyped plants was determined using the admixture model with correlated allele frequencies. The estimated proportions of each individual’s genome originating from each of the K ancestral populations (q) was calculated for K ranging from 1 to 10 ancestral populations (or clusters), with 10 runs for each K value. The structure harvester program was used to estimate optimum K value from admixture analysis results47. Linkage disequilibrium (\(\hbox {R}^2\)) was calculated from TASSEL 5.0 and LD decay plot generated using the R-program (http://www.R-project.org/)(version 3.6.2). Association mapping based on General Linear Model (GLM) with PCA as the fixed effect (GLM+PCA); and Mixed Linear Model (MLM) with PCA results and Kinship value (MLM+PCA+K) were conducted in TASSEL 5.0 software. The p values for each marker were adjusted for false discovery rate (FDR) or transformed to q-values using the R package (q-value)48. The q-value package has been widely adopted to control for multiple testing49,50. We used the positions of significant markers that had a positive effect on the trait as reference points and identified candidate genes falling within 10 Kbp up- and downstream from them on the database (https://www.maizegdb.org/gene_center/gene) of the maize reference genome, ZmB73 RefGen v2 (https://www.maizegdb.org/gbrowse). The selection of these candidate genes was limited by searching for genes annotated with defence functions. In addition, we selected, regardless of any existing annotation, the top 16 SNPs that had an \(\hbox {R}^2\) value of \(\ge \) 25% (using the MLM approach) out of 101 trait associated SNPs. A total of 202 candidate genes were identified within a 10 mb region of these top 16 SNPs that are closely linked with the indirect defense trait across 10 chromosomes of the maize genome (B73 RefGen v2 maize database).
References
Labandeira, C. C. A paleobiologic perspective on plant-insect interactions. Curr. Opin. Plant Biol. 16, 414–421 (2013).
Moles, A. T. et al. Correlations between physical and chemical defences in plants: tradeoffs, syndromes, or just many different ways to skin a herbivorous cat?. New Phytol. 198, 252–263 (2013).
Bruce, T. J. Interplay between insects and plants: dynamic and complex interactions that have coevolved over millions of years but act in milliseconds. J. Exp. Bot. 66, 455–465 (2014).
Howe, G. A. & Jander, G. Plant immunity to insect herbivores. Annu. Rev. Plant Biol. 59, 41–66 (2008).
Alborn, H. et al. An elicitor of plant volatiles from beet armyworm oral secretion. Science 276, 945–949 (1997).
Hilker, M. & Meiners, T. Early herbivore alert: insect eggs induce plant defense. J. Chem. Ecol. 32, 1379–1397 (2006).
Tamiru, A. et al. Maize landraces recruit egg and larval parasitoids in response to egg deposition by a herbivore. Ecol. Lett. 14, 1075–1083 (2011).
Vet, L. E. & Dicke, M. Ecology of infochemical use by natural enemies in a tritrophic context. Annu. Rev. Entomol. 37, 141–172 (1992).
Dicke, M. & Baldwin, I. T. The evolutionary context for herbivore-induced plant volatiles: beyond the ‘cry for help’. Trends Plant Sci. 15, 167–175 (2010).
Heil, M. Indirect defence via tritrophic interactions. New Phytol. 178, 41–61 (2008).
Dicke, M. & Sabelis, M. W. Infochemical terminology: based on cost-benefit analysis rather than origin of compounds?. Funct. Ecol. 2, 131–139 (1988).
Turlings, T. C., Tumlinson, J. H. & Lewis, W. J. Exploitation of herbivore-induced plant odors by host-seeking parasitic wasps. Science 250, 1251–1253 (1990).
Conway, G. R. Agroecosystem analysis. Agric. Admin. 20, 31–55 (1985).
Gepts, P. Plant genetic resources conservation and utilization. Crop Sci. 46, 2278–2292 (2006).
Ratnadass, A., Fernandes, P., Avelino, J. & Habib, R. Plant species diversity for sustainable management of crop pests and diseases in agroecosystems: a review. Agron. Sustain. Dev. 32, 273–303 (2012).
Bruce, T. J. Gm as a route for delivery of sustainable crop protection. J. Exp. Bot. 63, 537–541 (2012).
de Lange, E. S., Balmer, D., Mauch-Mani, B. & Turlings, T. C. Insect and pathogen attack and resistance in maize and its wild ancestors, the teosintes. New Phytol. 204, 329–341 (2014).
Kfir, R., Overholt, W., Khan, Z. & Polaszek, A. Biology and management of economically important lepidopteran cereal stem borers in Africa. Annu. Rev. Entomol. 47, 701–731 (2002).
Fess, T. L., Kotcon, J. B. & Benedito, V. A. Crop breeding for low input agriculture: a sustainable response to feed a growing world population. Sustainability 3, 1742–1772 (2011).
Tamiru, A. et al. Oviposition induced volatile emissions from African smallholder farmers’ maize varieties. J. Chem. Ecol. 38, 231–234 (2012).
Mutyambai, D. M. et al. Responses of parasitoids to volatiles induced by Chilo partellus oviposition on teosinte, a wild ancestor of maize. J. Chem. Ecol. 41, 323–329 (2015).
Dicke, M. et al. Isolation and identification of volatile kairomone that affects acarine predator prey interactions: involvement of host plant in its production. J. Chem. Ecol. 16, 381–396 (1990).
Fontana, A. et al. Attractiveness of constitutive and herbivore-induced sesquiterpene blends of maize to the parasitic wasp Cotesia marginiventris (cresson). J. Chem. Ecol. 37, 582 (2011).
Richter, A. et al. Characterization of biosynthetic pathways for the production of the volatile homoterpenes dmnt and tmtt in Zea mays. Plant Cell 28, 2651–2665 (2016).
Vick, B. A. & Zimmerman, D. C. Characterization of 12-oxo-phytodienoic acid reductase in corn: the jasmonic acid pathway. Plant Physiol. 80, 202–205 (1986).
Schaller, A. & Stintzi, A. Enzymes in jasmonate biosynthesis—structure, function, regulation. Phytochemistry 70, 1532–1538 (2009).
Köllner, T. G. et al. Herbivore-induced sabath methyltransferases of maize that methylate anthranilic acid using S-adenosyl-l-methionine. Plant Physiol. 153, 1795–1807 (2010).
Song, W. et al. Identification of immune related LRR-containing genes in maize (Zea mays L.) by genome-wide sequence analysis. Int. J. Genomics https://doi.org/10.1155/2015/231358 (2015).
Gouhier-Darimont, C., Stahl, E., Glauser, G. & Reymond, P. The arabidopsis lectin receptor kinase LecRK-i.8 is involved in insect egg perception. Front. Plant Sci. 10, 623 (2019).
Szczegielniak, J. et al. Maize calcium-dependent protein kinase (ZmCPK11): local and systemic response to wounding, regulation by touch and components of jasmonate signaling. Physiol. Plant. 146, 1–14 (2012).
Singh, A., Sagar, S. & Biswas, D. K. Calcium dependent protein kinase, a versatile player in plant stress management and development. Crit. Rev. Plant Sci. 36, 336–352 (2017).
Stratmann, J. Map kinases in plant responses to herbivory. In Induced Plant Resistance to Herbivory, 329–347 (Springer, 2008).
Zhang, P. et al. Genome-wide identification, phylogeny and expression analysis of the pme and pmei gene families in maize. Sci. Rep. 9, 1–12 (2019).
Louis, J. et al. Ethylene contributes to maize insect resistance1-mediated maize defense against the phloem sap-sucking corn leaf aphid. Plant Physiol. 169, 313–324 (2015).
Gierl, A. & Frey, M. Evolution of benzoxazinone biosynthesis and indole production in maize. Planta 213, 493–498 (2001).
Tang, H. M. et al. The maize brown midrib2 (bm2) gene encodes a methylenetetrahydrofolate reductase that contributes to lignin accumulation. Plant J. 77, 380–392 (2014).
Pickett, J. A. & Khan, Z. R. Plant volatile-mediated signalling and its application in agriculture: successes and challenges. New Phytol. 212, 856–870 (2016).
Chen, Y. H., Gols, R. & Benrey, B. Crop domestication and its impact on naturally selected trophic interactions. Annu. Rev. Entomol. 60, 35–58 (2015).
Mitchell, C., Brennan, R. M., Graham, J. & Karley, A. J. Plant defense against herbivorous pests: exploiting resistance and tolerance traits for sustainable crop protection. Front. Plant Sci. 7, 1132 (2016).
Masuka, B. P. et al. Genetic diversity among selected elite cimmyt maize hybrids in east and southern Africa. Crop Sci. 57, 2395–2404 (2017).
Vidal, M. C. & Murphy, S. M. Bottom-up vs. top-down effects on terrestrial insect herbivores: a meta-analysis. Ecol. Lett. 21, 138–150 (2018).
Stenberg, J. A., Heil, M., Åhman, I. & Björkman, C. Optimizing crops for biocontrol of pests and disease. Trends Plant Sci. 20, 698–712 (2015).
Ochieng, R., Onyango, F. & Bungu, M. Improvement of techniques for mass-culture of Chilo partellus (swinhoe). Int. J. Trop. Insect Sci. 6, 425–428 (1985).
Overholt, W., Ochieng, J., Lammers, P. & Ogedah, K. Rearing and field release methods for Cotesia flavipes cameron (Hymenoptera: Braconidae), a parasitoid of tropical gramineous stem borers. Int. J. Trop. Insect Sci. 15, 253–259 (1994).
Bradbury, P. J. et al. Tassel: software for association mapping of complex traits in diverse samples. Bioinformatics 23, 2633–2635 (2007).
Schnable, P. S. et al. The b73 maize genome: complexity, diversity, and dynamics. Science 326, 1112–1115 (2009).
Evanno, G., Regnaut, S. & Goudet, J. Detecting the number of clusters of individuals using the software structure: a simulation study. Mol. Ecol. 14, 2611–2620 (2005).
Dabney, A., Storey, J. D. & Warnes, G. qvalue: Q-value estimation for false discovery rate control. R package version 1 (2010).
Storey, J. D. & Tibshirani, R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. 100, 9440–9445 (2003).
Storey, J. D., Taylor, J. E. & Siegmund, D. Strong control, conservative point estimation and simultaneous conservative consistency of false discovery rates: a unified approach. J. R. Stat. Soc. Ser. B 66, 187–205 (2004).
Acknowledgements
This Research was supported by BBSRC grant BB/J011371/1. We gratefully acknowledge the financial support for icipe research by European Union, UK’s Department for International Development (DFID); Swedish International Development Cooperation Agency (Sida); the Swiss Agency for Development and Cooperation (SDC); Federal Democratic Republic of Ethiopia and the Kenyan Government. We thank CIMMYT and Prof Mathews Dida of Maseno University for supplying seed. We thank Amos Gadi and Silas Ouko for assistance with insect rearing and screen house operations.
Author information
Authors and Affiliations
Contributions
A.T., R.P, D.O., Z.K., C.M., J.P. and T.B. conceived the experiments, A.T., R.P. and S.M. conducted the experiments, A.T., R.P, D.O. and T.B. analysed the results and wrote the paper. All authors discussed the results and reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Tamiru, A., Paliwal, R., Manthi, S.J. et al. Genome wide association analysis of a stemborer egg induced “call-for-help” defence trait in maize. Sci Rep 10, 11205 (2020). https://doi.org/10.1038/s41598-020-68075-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-68075-2
This article is cited by
-
Focused identification of germplasm strategy (FIGS): a strategic approach for trait-enhanced pre-breeding
Genetic Resources and Crop Evolution (2024)
-
Female accessory glands of Adoxophyes honmai contain elicitor inducing tea leaves to arrest the egg-larval parasitoid, Ascogaster reticulata
Arthropod-Plant Interactions (2024)
-
Genetic variation among elite inbred lines suggests potential to breed for BNI-capacity in maize
Scientific Reports (2023)
-
Genetic analysis reveals three novel QTLs underpinning a butterfly egg-induced hypersensitive response-like cell death in Brassica rapa
BMC Plant Biology (2022)
-
Molecular mechanisms, genetic mapping, and genome editing for insect pest resistance in field crops
Theoretical and Applied Genetics (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.