Abstract
The discovery of symbiotic associations extends our understanding of the biological diversity in the aquatic environment and their impact on the host’s ecology. Of particular interest are nudibranchs that unprotected by a shell and feed mainly on sponges. The symbiotic association of the nudibranch Rostanga alisae with bacteria was supported by ample evidence, including an analysis of cloned bacterial 16S rRNA genes and a fluorescent in situ hybridization analysis, and microscopic observations. A total of 74 clones belonging to the phyla α-, β-, γ-Proteobacteria, Actinobacteria, and Cyanobacteria were identified. FISH confirmed that bacteriocytes were packed with Bradyrhizobium, Maritalea, Labrenzia, Bulkholderia, Achromobacter, and Stenotrophomonas mainly in the foot and notum epidermis, and also an abundance of Synechococcus cyanobacteria in the intestinal epithelium. An ultrastructural analysis showed several bacterial morphotypes of bacteria in epidermal cells, intestine epithelium, and in mucus layer covering the mollusk body. The high proportion of typical bacterial fatty acids in R. alisae indicated that symbiotic bacteria make a substantial contribution to its nutrition. Thus, the nudibranch harbors a high diversity of specific endo- and extracellular bacteria, which previously unknown as symbionts of marine invertebrates that provide the mollusk with essential nutrients. They can provide chemical defense against predators.
Similar content being viewed by others
Introduction
Symbiotic associations between marine invertebrates and microbes are ubiquitous and play an important role in the ecology and evolution of species1,2,3. In the marine environment, the symbiosis of invertebrates with photoautotrophic microalgae in the photic zone and the symbiosis with chemoautotrophic bacteria, found in extreme habitats such as deep-sea water and hydrothermal vents or cold seeps, are the most widely distributed and well-studied associations1,4. Associations with heterotrophic bacteria are also known, although they are described to a much lesser extent. These are bone-eating polychaete of the genus Osedax with bacteria from the order Oceanespirillales that are known for heterotrophic aerobic degradation of complex organic compounds5, wood-boring mollusks with cellulose-degrading bacteria6, and marine polychaete with N2-fixing Mesorhizobium sp. from an anoxic microbial mat community7.
The discovery of new symbiosis extends our understanding of the symbiotic diversity and their impact on the ecology and evolution of the marine invertebrates. Symbiotic microbes typically play a crucial role in supplying nutrients to their hosts8. In addition, some of symbionts provide the hosts with chemical defense against predators and environment2,9,10. Symbiosis is evidently expected in inhospitable environments for invertebrates with high concentrations of methane or hydrogen sulfide11,12, whereas marine organisms from shallow waters have rarely been investigated for symbiotic relationships13,14,15. Meanwhile, nudibranchs, which are not protected by a shell and feed mainly on sponges, may be of particular interest.
Nudibranchs are a common element of trophic webs in the benthic marine ecosystems. Found in almost all marine habitats, from the intertidal zone to the deep-sea, they have undergone a great evolutionary radiation, clearly influenced by dietary adaptation. A number of nudibranch species, belonging to two groups, Aeolidoidea and Dendronotoidea, have symbiotic relationships with photosynthetic dinoflagellates. They usually acquire them by sequestration from their prey and keep them inside cells of digestive gland16,17,18. For the first time, the presence of symbiotic bacteria in the mucus surrounding the egg capsules as well as between and partly aligned with the microvilli, in the vestibular gland of Dendrodoris nigra was detected by histological and ultrastructural analysis19.
The high abundance of odd and branched fatty acids specific to bacteria in their tissues led us to the hypothesis that nudibranchs may house symbiotic bacteria20,21. The nudibranch Dendrodoris nigra was found to harbor symbiotic bacteria enclosed in secondary vacuoles in the epithelial cells of the notum and the mantle edge20. Although these data support the hypothesis, they also raise many questions as to the prevalence of bacterial associations among nudibranch species, identification of specificity of bacteria, and determination of their responsibility in mollusks.
The present study focuses on the nudibranch mollusk Rostanga alisae Martynov, 2003, a member of the family Discodorididae that inhabits the intertidal zone to a depth of 15 m in the northwestern Sea of Japan and preys exclusively on the red sponge Ophlitaspongia pennata (Lambe, 1895) from the family Microcionidae (Fig. 1A).
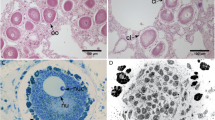
Morphology and localization of symbionts in Rostanga alisae. (A) Common view of the nudibranch in the natural habitat on the sponge. (B) Histological organization, longitudinal section. (C–E) are enlarged views of the areas indicated by the corresponding arrows: (C) Bacteria (b) in the mucus layer covering the body. Bacterial clusters (bc) in the epidermis of the notum (D) and foot (E). Nuclei (nu) of epithelial cells are visible. Combination of fluorescence microscopy and Phase microscopy, DNA stained with DAPI (blue fluorescence).
The hypothesis that R. alisae harbors symbiotic bacteria was tested by molecular phylogenetic analyses, epifluorescence microscopy, transmission electron microscopy, fluorescent in situ hybridization (FISH) analysis, and fatty acid analyses of tissues. This investigation was aimed to assess the diversity and phylogenetic composition of symbionts’ populations and to assign the nature of the nudibranch–bacteria symbiosis. The results provide evidence for an endo- and extracellular bacterial consortium, which is more complex than other marine symbioses described before. The results are discussed with a special emphasis on the bacterial diversity and on the potential role of the discovered symbionts. This study is the first multifaceted investigation of symbiotic association of bacteria with a nudibranch species.
Methods
Specimens
Specimens of the nudibranch R. alisae (Fig. 1A) and sponge O. pennata were collected in the Peter the Great Bay, Sea of Japan, by SCUBA diving, at the depth of 1–6 m year-round in 2017–2019. They were then kept in a tank or used immediately for analyses. The nudibranchs were dissected by razor and separate tissues were excised for the analyses under stereomicroscope Leica ES2 (Germany).
DNA amplification, cloning, and sequencing
Total genomic DNA was extracted from the foot, intestine, and notum tissues using the DNeasy Plant MiniKit protocol (Qiagen, Hilden, Germany). The procedures of DNA amplification, cloning, and sequencing were described previously13,14. A 1.5-kb fragment of the 16S rRNA bacterial genes was amplified with universal primers22: 5’-agagtttgatcatggctcag-3’ (27F, forward) and 5’-ggttaccttgttacgactt-3’ (1492R, reverse). The PCR reactions were carried out in final volumes of 25 µl using TaKaRa Ex Taq™ in accordance with the manufacturer’s description (Takara Biotechnology Co., Ltd.). The PCR reaction mixtures were placed in a DNA thermal cycler (Eppendorf, Mastercycler Gradient), incubated for 5 min at 94 °C and subjected to 32 cycles of denaturation, annealing, and extension: at 94 °C for 30 s, at 52 °C for 30 s, and at 72 °C for 1.5 min, with a final 7-min extension period at 72 °C. The PCR products for the 16S rRNA gene were cloned (TOPO TA cloning kit, Invitrogen, Calif.) and sequenced by the dideoxy chain-termination technique using a Dye Terminator chemistry and separated on an ABI PRISM 377 automated DNA sequencer (Perkin Elmer). The sequences of both strands were determined for each clone, using overlapping internal primers spaced, on average, 500 nucleotides. At least two independent PCR amplifications were sequenced in both directions to correct for possible cloning or sequencing errors. The 16S rRNA sequences have been deposited in GenBank under accession numbers MZ410589–MZ410616. The sequences were assembled using the program SeqMan (Lasergene, DNASTAR, Inc.). Multiple alignment was carried out manually and using the program CLUSTAL W23. Putative chimeras were identified using the Bellerophon software24. Two programs available online, RDP Classifier25 and Greengenes26, were used to uncover the bacterial affinities. The maximum-likelihood tree of the 16S rRNA sequences of Rostanga alisae symbionts was constructed with the program IQ-TREE27 using the TIM3 + F + I + G4 model of nucleotide substitution.
Transmission electron microscopy (TEM)
For TEM, the tissue samples were fixed with a 2.5% glutaraldehyde solution in a 0.1 M cacodylate buffer (pH 7.2) for 4 h at room temperature. After being rinsed in the buffer, the samples were post-fixed for 1 h with a 1% osmium tetroxide solution and dehydrated in a graduated series of ethanol and acetone, and then embedded in a mixture of Epon and Araldit epoxy resin (Sigma, USA). Ultra-thin sections (ca. 50 nm) were prepared on a Leica EM UC6 ultramicrotome, post-stained with a 0.5% uranyl acetate and lead citrate solution, and viewed using a Libra 120 electron microscope (Carl Zeiss, Germany). For TEM, 15 nudibranch individuals were used.
Fluorescent in situ hybridization (FISH)
For the FISH analysis, the foot, intestine, and mantle tissues were initially fixed at 4 °C in a 4% paraformaldehdye solution (pH 7.0) in a phosphate-buffered saline (PBS) for 8 h. The fixed samples were rinsed 3 × 1 h, impregnated with a 15% sucrose solution on PBS, placed in NEG 50 ™ (Thermo Scientific, USA), and frozen at –50 °C. The frozen tissues were sectioned (12–14 µm thick) on a Histostar 500 HM (Thermo Scientific) and placed onto glass slides treated with polylysine. A bacterial universal probe EUB338 was used along with a group of specific probes designed as symbiotic bacteria dominant in tissues of R. alisae (Table S1). Hybridization was performed at 46 °C for 3.5 h in a solution containing 900 mM NaCl, 20 mM Tris/HCl, 35% formamide, 0.01% SDS, and 3 ng/ml of the EUB338 probe along with one of the specific probes. After hybridization, the slides were washed twice at 48 °C for 15 min in a buffer containing 100 mM NaCl, 20 mM Tris/HCl (pH 8.0), 5 mM EDTA, and 0.0001% SDS. The sections were stained with a dilute 4′6'-diamidino-2-phenylindole (DAPI) solution (5 µg ml–1) for 1 min and examined by epifluorescence microscope using a LSM 780 laser scanning confocal microscope. For FISH microscopy, 5 nudibranch individuals were used.
Lipid analysis
Lipid extracts were prepared from homogenized tissues according to28. Fatty acid methyl esters (FAME) were prepared by the treatment of lipids with 2% H2SO4/MeOH in a screw-capped vial (2 h, 80 °C) under argon and purified by preparative TLC on silica gel in benzene. FAMEs were analyzed on a Shimadzu GC-2010 chromatograph equipped with a flame ionization detector and a capillary column (Supelcowax 10, 30 m × 0.25 mm i.d.) at 210 °C. The injector and detector temperatures were 250 °C. Fatty acids were identified by comparison with equivalent chain length values and confirmed by GC–MS of their FAMEs and DMOX derivatives on a GCMS QP5050A Ultra instrument (Shimadzu, Kyoto, Japan) (electron impact at 70 eV) fitted with a MDN 5S capillary column (30 m × 0.25 mm). The carrier gas was He at 30 cm s−1. The GC–MS analysis of FAMEs was performed at 160 °C increased to 260 °C with a 2 °C min−1 ramp. The GC–MS analysis of DMOX was performed at 210 °C with a 3 °C min−1 ramp to 270 °C held for 40 min. For fatty acid analysis, 7 nudibranch individuals were used.
Statistical analysis
Significance of differences in mean contents of fatty acids between the nudibranch tissues and the sponge was tested by one-way analysis of variance (ANOVA). Statistical analysis was performed using STATISTICA 5.1 (StatSoft, Inc., USA). A statistical probability of p < 0.05 was considered significant. Values are represented as mean ± standard deviation, n = 7.
Results
Histological analysis
Morphological analysis using a differential interference contrast (DIC) and epifluorescence microscopy of tissues (Fig. 1B–E) revealed bacterial clusters (10 − 15 µm overage size), and individual single bacteria in the epidermis covering the notum (Fig. 1D) and foot (Fig. 1E). Bacteria were also visualized in the mucus layer covering the body surface (Fig. 1C).
Genetic affiliation of the bacterial 16S rRNA clones
The analysis of the cloned bacterial 16S rRNA genes allowed to obtain the most conservative and reliable scan of bacterial diversity, which made it possible to reveal bacteria that form mutualistic relationships with the host and to formulate the most parsimonious hypothesis concerning the functional importance of the symbiosis. On the basis of non-chimeric sequences (a total of 74 clones), 24 bacterial phylotypes were detected (with 97% identity cutoff) in tissues of R. alisae (Table S2). Phylogenetic analysis of each clone showed close matches with a multitude of bacteria (Fig. 2) belonging to the classes α-Proteobacteria (15 clones), β-Proteobacteria (13 clones), γ-Proteobacteria (16 clones), Cyanobacteria (19 clones), and Actinobacteria (7 clones). Three rare phylotypes were associated with the classes δ-Proteobacteria (1 clone), Planctomycetes (1 clone), and Fusobacteria (2 clones). Three phylotypes (Bradyrhizobiaceae, Rhodobacteraceae, and Alcaligenaceae) were classified at the family level only. The rest of the phylotypes (except for those represented by single clones) belonged to 13 genera: Bradyrhizobium, Maritalea, Labrenzia, Burkholderia, Achromobacter, Stenotrophomonas, Synechococcus, Arthrobacter, Ilumatobacter, Aquicella, Lysobacter, Legionella, and Leptotrichia (Table S2).
Maximum-likelihood tree of the 16S rRNA sequences of Rostanga alisae symbionts. The tree is based on the TIM3 + F + I + G4 model of nucleotide substitution. The numbers at the nodes are bootstrap percent probability values based on 10,000 replications (values below 75% are omitted). The number following the name of specimens (RF, RN, and RI) denotes the clone number. RF foot, RN notum, RI intestine.
The phylogenetic analysis placed the bacterial phylotypes recovered from R. alisae within a clade of α-Proteobacteria that contains Labrenzia, Maritalea, and Bradyrhizobium. Phylotypes of symbiotic Labrenzia (transferred from Stappia) belong to a single cluster with the other species and strains of this genus. The closest relative of the nudibranch’s symbiont Labrenzia is L. alba isolated from oyster and described as straight rods, motile by a subpolar flagellum, and chemoorganotrophic aerobe29. One of the three α-Proteobacterial symbionts was clustered with Maritalea porphyrae (transferred from Zhangella), isolated from the red alga Porphyra yezoensis30. Another member of α-Proteobacteria symbionts was clustered with B. pachyrhizi and B. elkanii, which were isolated from nodules of legume plants. Among the dominant β-Proteobacterial symbionts, one fell into the genus Burkholderia and another into Achromobacter. The dominant γ-Proteobacteria symbiont was phylogenetically located within the genus Stenotrophomonas, whose species are found ubiquitously worldwide, particularly in close association with plants. The cyanobacterial 16S rRNA sequences from the nudibranch were grouped with a clade of sequences of marine Synechococcus spp. isolated from the water column. A sequencing analysis and BLAST searches retrieved close relation between nudibranch symbionts and actinobacteria Arthrobacter, Iamia, Ilumatobacter, and Kocuria.
Mosaic sets of common and tissue-specific bacterial phylotypes were detected in the foot, notum, and intestine. The clones were unequally distributed between the tissues studied. The deviation from equal proportions was highly significant (χ2 = 25.33, df = 1, P < 0.001). Synechococcus (Cyanobacteria) had the most abundant clones, especially in the intestine. Maritalea, Labrenzia, and a member of Rhodobacteriacea were located exclusively in the foot, which was especially rich in bacterial clusters. They, consequently, were specific symbionts of the foot tissue. Bradyrhizobium and Burkholderia were predominantly detected in the notum. Phylotypes of Stenotrophomonas were localized exclusively in the integumentary tissues, foot, and notum, and only Achromobacter clones were evenly distributed over the tissues. Phylotypes of Actinobacteria were found only in integumentary tissues; therefore, the species-specific tissue distribution was evident. Thus, Iamia, Ilumabacter, and Kocuria were found only in the foot; in contrast, another member of Actinobacteria, Arthrobacter, could be considered as a specific bacterium of the notum. Other bacterial clones had mosaic distribution with no clear preference for a tissue.
In addition to predominant bacterial phylotypes, members of γ-Proteobacteria such as Coxiella, Aquicella, Acinetobacter, and Aliivibrio, as well as Desulfovibrio, Planctomycetes, and Leptotrichia, were also present as single clones mainly in the intestine. These are known as intracellular parasites of protozoa and animal cells31, which suggest that these species are unlikely to be true symbiotic bacteria, and their presence most probably resulted from the contamination of this tissue as reported for Aquicella siphonis parasitizing an amoeba31.
FISH observations
A bacterial universal probe EUB338 revealed that the bacterial clusters were more abundant in cytoplasm of the foot epithelium in the skin fold between the foot and the notum (Fig. 3A). The presence and localization of bacterial symbionts in tissues was confirmed by FISH analysis using the genus-specific oligonucleotide probes (Table S1) for Maritalea, Labrenzia, Bradyrhizobium, Burkholderia, Achromobacter, Stenotrophomonas, and Cyanobacteria (Fig. 3B–I).
Fluorescent in situ hybridization microscopy of bacteria forming bacteriocytes (bc) in the epidermis and cyanobacteria in the intestinal epithelium of Rostanga alisae. (A) Accumulation of bacteriocytes (bc) in the skin fold between the foot and notum, transverse section of the body. Red fluorescence of bacteria hybridized with Cy3 universal UEB388 probe set. Green fluorescence of bacteria hybridized with bacteria-specific probes for (B) Achromobacter, (C) Stenotrophomonas, (D) Labrenzia, (E) Maritalea, (F) Bradyrhizobium, (G) Burkholderia, (H,I) Synechococcus (arrows). DNA stained with DAPI (blue fluorescence).
The group-specific probes for Achromobacter and Stenotrophomonas showed discrete patches of hybridization in epithelial cells of the foot and notum (Fig. 3B,C), whereas oligonucletide probes for Labrenzia and Maritalea hybridized strongly with bacteria that settle in the foot epithelium (Fig. 3D,E). Bacterial signals of Bradyrhizobium and Burkholderia were found only in notum epithelial cells (Fig. 3F,G). Each of the symbionts was localized in clusters and occurred within a single cluster. FISH analysis also revealed the presence of cyanobacteria in the epithelial cells of all studied tissues, but they were more represented in the intestinal epithelium (Fig. 3H,I). Thus, the characteristic tissue specificity of certain bacteria was revealed, which is consistent with the results of the 16S rRNA analysis (Table S2). The most common bacteriocytes were formed by Labrenzia and Stenotrophomonas, while Maritalea and Achromobacter were detected less frequently.
The number of bacterial clusters detected by Eub388 was higher than that of clusters labeled with each of the group-specific probes. The total number of bacteriocytes detected by the group-specific oligonucleotide probes was generally comparable to the number of bacteriocytes detected by EUB338, which is consistent with the assessment of the phylogenetic composition of the symbionts’ population.
TEM observations
To examine the spatial organization of the bacterial symbionts, we performed TEM of the foot, notum, and intestine tissues. TEM revealed dense clusters of rod-shaped gram-negative bacteria enclosed in the epithelium covering the foot and notum of R. alisae (Fig. 4). Bacteria were separated by a membrane and located within specialized vacuoles, referred to as bacteriocytes, which could occupy most of cytoplasm of a host cell. The number of bacteria per bacteriocyte reached a few dozen (Fig. 4A–C). Two main bacterial morphotypes formed bacteriocytes; however, bacteria within each vacuole were monomorphic.
Endosymbiotic bacteria housed in bacteriocytes of Rostanga alisae. (A) Cells of integument epithelium of the foot containing bacteriocytes with several bacterial morphotypes of bacteria. Phagosome (ph) is visible. (B) Bacteriocyte containing curved-rod bacteria with lophotrichous flagella (fl). Vacuoles with electron-transparent contents are visible in the cytoplasm of bacteria. (C) Bacteriocytes containing rod-shaped bacteria with flagella (black asterisk) and without flagella (white asterisk). (D) Dividing flagellate bacterium in the bacteriocyte (arrow). (E) Bacteriocyte containing large rod-shaped bacteria, which forming a chain as a result of incomplete division. Arrows show the contacts of bacteria. Nucleus (nu) of the epithelial cell is visible. (F) Higher magnification of bacteria of this morphotypes indicated in (E). A tortuous outer membrane, extensive nucleoid zone, and vacuoles are visible. Arrows show the contact points of the outer membranes of adjacent individuals.
The most common bacteriocytes were those containing gram-negative curved-rod lophotrichous bacteria (0.3–0.4 μm) having up to six flagella (Fig. 4B,C). Less common were bacteriocytes with large gram-negative rod-shaped bacteria (0.7 × 1.5 μm), many of which appeared to be dividing (Fig. 4D–F). In some bacteriocytes, membranes of neighboring bacteria came into contact. As a result of incomplete division, the rod-shaped bacteria can form chains of individuals connected to each other, suggesting their clonal origin within a single specialized vacuole (Fig. 4E,F).
Epithelial cells also contained vacuoles with single gram-negative bacteria (Fig. 5A). Their morphotypes were diverse and similar to that described from bacteriocytes above. The similarity between the morphotypes of single bacteria and bacteria forming clusters, as well as the presence of dividing bacteria in the bacteriocytes, suggests that the observed single bacteria are the initial stage of formation of a dense bacterial cluster within bacteriocyte.
Endosymbiotic single bacteria of Rostanga alisae, TEM images. (A) Numerous Single gram-negative bacteria within secondary vacuole (light arrows) in the apical zone of the cytoplasm of the epithelium of the foot. As a result of division (dark arrow indicates a dividing bacterium), bacteriocytes contain two or more bacteria. Cilia (c), phagosome (ph), mitochondria (m), and nucleus (nu) of epithelial cells are visible. (B,C) Cyanobacteria (arrows) in the epithelium of the notum and (D) foot; a developed polysaccharide capsule and concentrically arranged membranes of the thylakoid type (th) are visible. (E) Different morphotypes of cyanobacteria containing inclusions in the intestinal epithelium cells: bacteria with a developed capsule (light arrow); bacteria with a narrow capsule (dark arrow). The apical surface of epithelial cells has microvilli (mv).
Apart from bacteriocytes packed with bacteria, large bacteria (diameter 0.5–0.7 μm, length up to 2 μm) with a well-developed polysaccharide envelope were found mainly in the intestinal epithelium and, more rarely, in the integumentary epithelium (Fig. 5B–E). Their cytoplasm contained concentrically arranged membranes of the thylakoid type, vacuoles, and granules (Fig. 5C–D). Bacteria were single or formed chains of several individuals (Fig. 5C,D). Some individuals in the colony had a thick, hypertrophied peptidoglycan fibrillar envelope (Fig. 5D). The presence of thylakoid membranes, the ability to form chains, and the hypertrophied peptidoglycan envelope in some individuals, which is typical of heterocysts, allows classifying these microbes as cyanobacteria.
An ultrastructural analysis showed that the nudibranch bore various extracellular bacteria in the mucus layer of the foot and notum that were identified by a 16S rRNA gene sequence analysis as close relatives of the genera Arthrobacter, Iamia, Ilumatobacter, and Kocuria (Fig. 6). The most abundant were gram-negative curved rod-shaped bacteria (0.5 × 1.5–2 μm) (Fig. 6A) with a long (up to 1 μm) contact stalk-like extension, usually directed to the apical surface of epithelial cells and providing a primary contact of bacteria with the apical membrane of host’s epithelial cells before penetrating into cytoplasm (Fig. 6B). This confirms the presence of a similar bacterial morphotype in bacteriocytes in epithelial cells. Bacteria inhabiting the mucus layer often come into contact with the apical membrane of epithelial cells, which is probably the initial stage of their inoculation (Fig. 6C). Large gram-negative rods (0.7 × 3 μm) (Fig. 6D) with numerous electron-translucent vacuoles were much less common. Bacteria forming long branching filaments resembling fungal hyphae were observed on the surface of ciliated cells of the foot epithelium (Fig. 6E). Unusually thin, from 10 to 20 nm, non-septate filaments branched and formed round bubbles of 50–100 nm in diameter at the apex. Similar branching bacteria were also found in the apical cytoplasm of epithelial cells that indicates the presence of air and substrate mycelium of branching bacteria.
TEM images of epithelium cells and adjacent mucous layer of Rostanga alisae inhabited by exosymbiontic bacteria. (A) Bacteria having a long contact stalk-like extension directed towards the apical membrane of epithelial cells (white arrow). Dark arrows indicate similar bacteria, which are present both in the mucous layer and in vacuoles near the apical membrane of the integumentary epithelial cells. (B) The bacterium with a long process (dark arrows) in contact with the surface of the microvilli (cross section) of the integumentary epithelial cells. (C) Long rods localized between the microvilli (mv) of the integumentary epithelial cells. The arrow indicates the site of adhesion of the bacterial cell to the outer cell membrane. (D) Large bacteria with numerous electron-translucent vacuoles (dark arrow) (E) Processes of branching bacteria on the surface and in the cytoplasm of cilliated cells of the foot epidermis (dark arrows); branched processes are visible bearing spore-like vesicles on the surface (light arrow). Cilia (c) and microvilli (mv) are visible.
The presence of these symbionts was typical for all adult animals studied, although the pattern of distribution of bacteriocytes varied between individuals. The general occurrence of individuals with symbionts indicates an obligate rather than a facultative association between symbionts and the nudibranch.
Fatty acid analysis
Fatty acid analysis is a useful tool to ascertain original nutrient sources acquired by partners of the association5,32. The integumentary tissues of notum and foot that were rich in symbiotic bacteria and the intestine with an abundance of cyanobacteria were analyzed separately but their fatty acid compositions showed no significant differences (Table S3). Both tissues exhibited exceptionally high concentrations of odd- and branched fatty acids (OBFA) typical for bacteria (13.7 and 12.5%, respectively), and also an abundance of saturated (19.6 and 20.8%) and monounsaturated fatty acids (20.8 and 21.4%), mainly due to 16:0 and 18:1n-7, along with high levels of n-6 polyunsaturated fatty acids (PUFA) (20.2–20.5%) (Fig. 7A). Marked differences were found between the nudibranch and its prey, the sponge O. pennata. The sponge contained the highest concentration of very long chain fatty acids (VLCFA) (28.1%). The nudibranch was much more enriched in PUFA n-6, in particular in linoleic acid (18:2n-6), 20:4n-6, and 22:4n-6, while PUFA n-3 such as 22:6n-3 and 22:5n-3, were more abundant in the sponge. The level of 18:2n-6, typical for cyanobacteria33, and essential for the mollusks, was almost by an order of magnitude higher in R. alisae than in the sponge. The difference in the OBFA content between nudibranch and sponge was also pronounced (Fig. 7B). Moreover, the composition of these bacterial markers differed between the nudibranch and its prey, the sponge. The most noticeable difference was in the content of 17:0 (2.1% in R. alisae vs. 0.3% in the sponge) and anteiso-16:0 (2.0% vs. 0.2%, respectively). The acids, iso-15:0, 17:1∆5, iso-18:0, anteso-18:0, and cyclo-19:0, were found only in R. alisae. The predominance of OBFA, 16:0, and 18:1n-7, which are typical for bacteria, as well as differences in the composition of bacterial markers between the nudibranch and the sponge O. pennata indicates the important bacterial contribution to the diet of the nudibranch.
Distribution of the fatty acids among the notum and intestine of Rostanga alisae and its prey sponge Ophlithaspongia pennata (mean ± SD; n = 7). (A) Composition of principal fatty acids. (B) Composition of odd and branched fatty acids (OBFA) as markers of bacteria. Data on fatty acid composition and significant differences (one-way ANOVA) in their concentration between the nudibranch and its prey sponge are provided in Supplementary Table S3.
It seems unlikely that the sponge O. pennata is the only source of fatty acids in R. alisae, since their levels in the predator nudibranch were significantly higher than in its prey sponge. R. alisae tissues also contained twice as much bacterial marker cis-vaccenic acid (18:1n-7) as compared to that in the sponge. Moreover, some bacterial indicators (mainly anteiso-15:0, 11-Me 14:0, 7Me-16:1n-10, 17:0, 17:1n-12, iso-18:0, anteiso-18:0, 19:0, and cyclo-19:0) were identified only in the nudibranch, thus, confirming the different bacterial associations in the nudibranch and the sponge.
Discussion
Symbiont diversity and distribution
The present study provides the first evidence of symbiosis in R. alisae, a species of nudibranchs. This is the most multiple symbiosis that have ever been recorded for marine invertebrates. While many organisms establish an exclusively one-on-one relationship with a single microbial species or microbes belonging to the same functional group5,12, there are also organisms that harbor multiple microbial species, in which symbiont–symbiont and host–symbiont interactions occur. Six phylotypes of chemoautotrophic bacteria were reported for mussel Idas sp. from a cold seep area11 and five extracellular symbionts for the gutless oligochaete worm Olavius algarvensis34. However, in these cases, symbioses involving bacteria and marine invertebrates are either endosymbiotic microbes co-occurring inside the host bacteriocytes5,11 or ectosymbiotic microbes associated with the external surfaces of the animals3,4,9,15,34, with the exception of scaly-foot snail from hydrothermal vents having partnerships simultaneously with epi- and endosymbiontic microbes35.
Bacterial symbionts in R. alisae have appeared to be more diverse than was previously known for marine invertebrates. It is evident that the detected symbiont phylotypes differ greatly from all other known symbionts found in marine invertebrates. Labrenzia (Rodobacteriales) and Maritalea (Rhizobiales) have not been recorded as forming symbiotic associations with invertebrates or plants so far, although other members of the families Rodobacteriales and Rhizobiales are well known symbionts14. Strains of Bradyrhizobium, Burkholderia, Achromobacter, and Stenotrophomonas are reported as symbionts of plants, interacting with a vast majority of nodulating legume species and efficient in biological nitrogen fixation36. This may be important when considering the nature of these symbionts in the nudibranch. Symbioses between cyanobacteria and marine organisms are commonly found among marine plants, fungi, sponges, ascidians, corals, and protists37,38. Synechococcus, identified as dominant symbiont clones of R. alisae (Table S2), is a unicellular cyanobacterium common in the marine environment, providing a range of beneficial functions including photosynthesis, nitrogen fixation, UV protection, and production of defensive toxins8,9,37. Symbiotic interactions between actinobacteria and their host have been observed in insects, human, animals, and plants, where the bacteria provide the host with protection against pathogens and produce essential nutrients39. However, none of the members of the clade Actinobacteria recorded in R. alisae are known to live symbiotically.
Arrangement of symbiotic association
Despite the high diversity of bacteria, they are well organized in the host. Dense clusters of rod-shaped bacteria, Labrenzia, Maritalea, Bradyrhizobium, Burcholderia, Achromobacter, and Stenotrophomonas, were found within host-derived vacuoles, referred to as bacteriocytes, inside epithelial cells of R. alisae (Fig. 3). Although such arrangement differs from that typical of bacteriocytes, which are usually considered as specialized cells of the hosts for harboring bacteria, it resembles that reported for scaly-food snail from hydrothermal vents, which harbor symbionts in the esophageal gland35. Bacteriocytes in the gastropod Lurifax vitreus found near hydrothermal vents also constitute a portion of the mantle epithelium; they have large vacuoles containing many live and dividing bacteria40. Each bacteriocyte was densely packed with certain symbionts, and the bacteriocytes were randomly distributed within the epithelium cells. A distinctly regular distribution pattern was observed in the gill epithelium of the mussel Bathymodiolus sp.: the thiotrophic symbionts occupy the apical region, and the methanotrophic symbionts are more abundant in the basal region of bacteriocytes4. In the mussel Idas sp., however, there is no spatial pattern of the six distinct bacterial phylotypes, and the symbionts are mixed within bacteriocytes11.
Synechococcus dominated the cytoplasm of intestinal epithelium and, more rarely, epidermis cells, mainly as specialized cell type referred to as nitrogen-fixing heterocysts. They are visually similar to cyanobacteria from corals and sponges8,37.
The phylogenetic diversity and the spatial organization of the symbiotic community in R. alisae were determined by the 16S rRNA analysis, which was consistent with the results of FISH and TEM. Unlike most symbioses of marine invertebrates when bacteria house specialized host cells5,11 or cover epidermis7,15, symbiotic association of R. alisae exhibited spatial partitioning between symbionts, which were unevenly distributed between the tissues (Table S2). It has been established that different members of the microbial community can complement each other in acquisition of various restrictive nutrients, confirming the importance of the functional diversity of symbionts41. Thus, Stenotrophomonas rhizophila and Bradyrhizobium build a beneficial association in the rhizosphere and can act synergistically on promoting growth and nutrient uptake of soybean36. Cyanobacteria can interact synergistically with beneficial members from the endophytic microbiome of rice seedlings42. The location of bacterium in the organism of R. alisae may, in fact, depend on the specific metabolic and ecological roles that the symbionts play, and also on the interaction with bacterium belonging to different physiological groups.
Nature of symbiosis
Symbiotic associations between microbes and invertebrates are acquired mainly in a nutrient-depleted environment where symbionts usually provide their hosts with essential nutrients and high-energy compounds1. In contrast to known symbioses between microbes and gutless invertebrates, which obtain nutrients exclusively from the bacteria, R. alisae, like most nudibranch species, is a sponge-eating predator. However, due to the lack of adipose tissue, sponges are distinguished by a low lipid content (0.4 to 1.5% of wet weight)43 and also by specific proteinaceous spongin fibers and chitin, a polysaccharide similar to cellulose that can be indigestible for some predators, which together indicate their low nutritional value. Furthermore, R. alisae feeds exclusively on the sponge O. pennata; therefore, in habitats with low prey availability, this nudibranch has to survive starvation while searching for sponge assemblages. We suppose that symbiotic bacteria of R. alisae contribute to the utilization of low-quality food, similarly to symbiotic bacteria from the genera Rhodobacter, Burkholderia, and Aeromonas associated with the detritivorous isopod Asellus aquaticus44.
A fatty acid analysis, as a useful approach to clarifying the nature of symbiosis5,20,32, has confirmed the trophic interaction between symbionts and the nudibranch host (Table S2). Among the fatty acids of symbiotic bacteria in R. alisae, OBFA are a major acyl constituent of membranes in Stenotrophomonas45 and also in Actinobacteria, Arthrobacter, Iamia, Ilumatobacter, and Kocuria46. Cis-vaccenic acid is a major component of Maritalea30. Omega-cyclohexyl tridecanoic acid (cyclo19:0) is specific to Bradyrhizobium47, Burkholderia, and Achromobacter48. Linoleic acid is produced by cyanobacteria including marine species of Synecoccocus33; in nudibranch, it obviously serves as a precursor in the synthesis of arachidonic acid (20:4n-6), thus, providing additional evidence for the transfer of fatty acids from symbionts to the host. Mollusks are capable of converting linoleic acid to arachidonic acid, since they have enzymes required for its synthesis21. The presence of these bacteria-specific markers and the abundance of arachidonic acid confirm the metabolic role of symbionts in the nudibranch host.
Among nutrients, biologically available nitrogen can be considered a restrictive nutrient for the sponge-eating R. alisae, which can be acquired with the help of nitrogen-fixing symbionts, also referred to as diazotrophs. R. alisae harbors Bradyrhizobium, Burkholderia, Achromobacter, and Stenotrophomonas that are efficient in biological nitrogen fixation previously found to be associated with nodulating legume species36. Symbiotic nitrogen fixers are known to be associated with a variety of marine invertebrates such as wood-boring bivalves, corals, sponges, sea urchins, tunicates, and polychaetes7,8,37. Moreover, the protection of the enzyme nitrogenase that catalyzes N2 fixation against oxygen is an important physiological requirement in bacteria such as symbiotic Bradyrhizobium, Burkholderia, Achromobacter, and Stenotrophomonas that are located in bacteriocytes and provide this protection. Synechococcus is known as a nitrogen-fixer37,49. It performs N2 fixation in heterocysts where nitrogenase is restricted under oxic conditions. Indeed, heterocysts of Synechococcus are abundant in the intestine cells of R. alisae (Fig. 5B–D).
Nitrate assimilation is one of the major processes of nitrogen acquisition by many heterotrophic bacteria and cyanobacteria50,51. Symbionts of R. alisae can play an important role in the process of nitrate utilization through denitrification, dissimilatory nitrate reduction, and assimilatory nitrate reduction as a nitrogen source and synthesize it into organic nitrogen. The nitrate reducers, Labrenzia52, Stenotrophomonas53, Maritalea30, and Rhodobacteraceae29 are widely represented in R. alisae. Synechococcus also utilizes nitrate, nitrite, or ammonium for growth50. Thus, symbiotic bacteria may play a significant role in the N-budget of the nudibranch mollusk.
The symbiotic bacteria of R. alisae, including Bradyrhizobium, Maritalea, Labrenzia, Burkholderia, Achromobacter, Stenotrophomonas, Arthrobacter, Iamia, Ilumatobacter, and Kocuria, are known as carboxydotrophic or carbon monoxide (CO) oxidizers54,55. Despite the toxicity of CO for multicellular organisms, numerous aerobic and anaerobic microorganisms can use CO as a source of energy and/or carbon for cell growth56. The marine worm Olavius algarvensis establishes symbiosis with chemosynthetic bacteria using CO, a substrate previously not known to play a role in symbiotic associations with marine invertebrates, as an energy source57. We do not rule out that the R. alisae symbionts also might exploit CO as carbon and energy source. Despite this, assumption may seem impossible taking in account the CO toxicity, but, since many invertebrates (mollusks, tube worm, etc.) use toxic sulfate, thiosulfate, and methane as an energy source1,15, this hypothesis is worth to be addressed.
An important component of skeleton in marine sponges of the family Microcionidae, including O. pennata, is the structural polysaccharide chitin58. Some bacteria are capable of hydrolyzing chitin via the activity of chitinolytic enzymes and can utilize chitin as a source of carbon, nitrogen, and/or energy59. Chitinase activity was documented for strains of Labrenzia60, Burkholderia61, Arthrobacter62, Achromobacter63, Stenotrophomonas64, Alcaligenes65, and actinobacteria59 associated with R. alisae. Thus, these bacteria can work synergistically to digest chitin and spongin, contributing to feeding success of the host nudibranch which depends solely on low-quality, nitrogen- and carbon-deficient food available.
Furthermore, direct evidence has confirmed that many bioactive compounds from invertebrates are produced by symbiotic microorganisms66,67. Many biologically active compounds including toxic and deterrent secretions have been identified in nudibranchs of the family Discodorididae68. Symbiotic bacteria may exhibit toxic activity to provide the host nudibranch with chemical defense against predators and environment. Bacteria, especially actinobacteria, living in a symbiotic relationship with R. alisae may help the host in defense, since nudibranch lack a shell, and secondary metabolites of bacteria can provide them with chemical defense against predators and environment, as has been reported for some marine invertebrates2,9,10.
In complex associations, the integration and coexistence of symbionts depend on supplementary partnerships and mutual contribution to the host’s metabolism41. The most intensively studied cases are highly specialized associations, where both partners can only exist in close relationship with one another. The relatively high diversity of microbes in R. alisae complicates understanding the complex pattern of molecular and cellular interactions between the host and its symbionts.
Conclusion
The current study demonstrates that the nudibranch R. alisae is a unique species among the marine animals, as it harbors a wide range of phylogenetically distinct but physiologically similar bacteria. These bacteria, known as aerobic chemoheterotrophs or facultative anaerobes, are capable of nitrogen fixation and/or denitrification under anaerobic conditions, oxidizing of carbon monoxide, hydrolyzing of chitin, and can use chitin as a source of carbon, nitrogen, and/or energy. They may also exhibit toxic activity and may provide chemical defense for the host nudibranch. A combination of such characteristic features as the dense population of diverse endo- and extracellular bacteria, the specific diet that is extremely poor in organic nutrients and energy, and the abundance of diverse fatty acids specific for bacteria and cyanobacteria in its composition suggest the existence of nutritional symbiosis in this nudibranch species. Overall, the data obtained expand our understanding of the biological role of the symbiosis of Metazoa with bacteria.
Data availability
The datasets generated or analyzed during the current study are included in this published article and its Supplementary Information files. All sequences are available in GenBank under accession numbers MZ410589–MZ410616.
References
Dubilier, N., Berqin, C. & Lott, C. Symbiotic diversity in marine animals: The art of harnessing chemosynthesis. Nat. Rev. Microbiol. 6, 725–740. https://doi.org/10.1038/nrmicro1992 (2008).
Mascuch, S. & Kubanek, J. A marine chemical defense partnership. Science 364, 1034–1035. https://doi.org/10.1126/science.aax8964 (2019).
Uchiumi, Y. & Sasaki, A. Evolution of division of labour in mutualistic symbiosis. Proc. R. Soc. B 287, 1930. https://doi.org/10.1098/rspb.2020.0669 (2020).
Duperron, S., Quiles, A., Szafranski, K. M., Léger, N. & Shillito, B. Estimating symbiont abundances and gill surface areas in specimens of the hydrothermal vent mussel Bathymodiolus puteoserpentis maintained in pressure vessels. Front. Mar. Sci. 3, 16. https://doi.org/10.3389/fmars.2016.00016 (2016).
Goffredi, S. K. et al. Evolutionary innovation: a bone-eating marine symbiosis. Environ. Microbiol. 7, 1369–1378. https://doi.org/10.1111/j.1462-2920.2005.00824.x (2005).
Betcher, M. A. et al. Microbial distribution and abundance in the digestive system of five shipworm species (Bivalvia: Teredinidae). PLoS ONE 7, e45309. https://doi.org/10.1371/journal.pone.0045309 (2012).
Summers, M. M., Katz, S., Allen, E. E. & Rouse, G. W. Association of rhizobia with a marine polychaete. Environ. Microbiol. Rep. 5, 492–498. https://doi.org/10.1111/1758-2229.12043 (2013).
Fiore, C. L., Jarett, J. K., Olson, N. D. & Lesser, M. P. Nitrogen fixation and nitrogen transformations in marine symbioses. Trends Microbiol. 18, 455–463. https://doi.org/10.1016/j.tim.2010.07.001 (2010).
Lindquist, N., Barber, P. H. & Weisz, J. B. Episymbiotic microbes as food and defence for marine isopods: Unique symbioses in a hostile environment. Proc. R. Soc. B. 272, 1209–1216. https://doi.org/10.1098/rspb.2005.3082 (2005).
Lopanik, N. B. Chemical defensive symbioses in the marine environment. Funct. Ecol. 28, 328–340. https://doi.org/10.1111/1365-2435.12160 (2014).
Duperron, S., Halary, S., Lorion, J., Sibuet, M. & Gaill, F. Unexpected co-occurrence of six bacterial symbionts in the gills of the cold seep mussel Idas sp. (Bivalvia: Mytilidae). Environ. Microbiol. 10, 433–445. https://doi.org/10.1111/j.1462-2920.2007.01465.x (2008).
Goffredi, S. K. et al. Methanotrophic bacterial symbionts fuel dense populations of deep-sea feather duster worms (Sabellida, Annelida) and extend the spatial influence of methane seepage. Sci. Adv. 6, eaay8562. https://doi.org/10.1126/sciadv.aay8562 (2020).
Balakirev, E. S., Pavlyuchkov, V. A. & Ayala, F. J. DNA variation and endosymbiotic associations in phenotypically-diverse sea urchin Strongylocentrotus intermedius. Proc. Natl. Acad. Sci. USA 105, 16218–16223 (2008).
Balakirev, E. S., Krupnova, T. N. & Ayala, F. J. Symbiotic associations in the phenotypically-diverse brown alga Saccharina japonica. PLoS ONE; 7, e39587 (2012).
Bellec, L. et al. Chemosynthetic ectosymbionts associated with a shallow-water marine nematode. Sci. Rep. 9, 7019. https://doi.org/10.1038/s41598-019-43517-8 (2019).
Kempf, S. C. A ‘primitive’ symbiosis between the aeolid nudibranch Berghia verrucicornis (A. Costa, 1867) and a zooxanthella. J. Molluscan Stud. 57, 75–85. https://doi.org/10.1093/mollus/57.Supplement_Part_4.75 (1991).
Burghardt, I., Stemmer, K. & Wägele, H. Symbiosis between Symbiodinium (Dinophyceae) and various taxa of Nudibranchia (Mollusca: Gastropoda), with analyses of long-term retention. Org. Divers. Evol. 8, 66–76. https://doi.org/10.1016/j.ode.2007.01.001 (2008).
Monteiro, E. A. et al. Evidence against mutualism in an aeolid nudibranch associated with Symbiodiniaceae dinoflagellates. Symbiosis 79, 183–189. https://doi.org/10.1007/s13199-019-00632-4 (2019).
Klussmann-Kolb, A. & Brodie, G. Internal storage and production of symbiotic bacteria in the reproductive system of a tropical marine gastropod. Mar. Biol. 133, 443–447. https://doi.org/10.1007/s002270050483 (1999).
Zhukova, N. V. & Eliseikina, M. G. Symbiotic bacteria in the nudibranch mollusk Dendrodoris nigra: Fatty acid composition and ultrastructure analysis. Mar. Biol. 159, 1783–1794. https://doi.org/10.1007/S00227-012-1969-7 (2012).
Zhukova, N. V. Fatty acids of marine mollusks: Impact of diet, bacterial symbiosis and biosynthetic potential. Biomolecules 9, 857. https://doi.org/10.3390/biom9120857 (2019).
Lane, D. J. 16S/23S rRNA sequencing. in Nucleic Acid Techniques in Bacterial Systematics. (eds. Stackebrandt, E. & Goodfellow, M.). 115–175. (Wiley, 1991).
Thompson, J. D., Higgins, D. G. & Gibson, T. J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 (1994).
Huber, T., Faulkner, G. & Hugenholtz, P. Bellerophon: A program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20, 2317–2319 (2004).
Cole, J. R. et al. The ribosomal database project (RDP-II): Introducing myRDP space and quality controlled public data. Nucleic Acids Res. 35, D169–D172 (2007).
DeSantis, T. Z. et al. NAST: A multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res. 34, W394–W399 (2006).
Nguyen, L. T., Schmidt, H. A., von Haeseler, A. & Minh, B. Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015).
Bligh, E. G. & Dyer, W. J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Phys. 37, 911–917 (1959).
Pujalte, M. J., Lucena, T., Ruvira, M. A., Arahal, D. R. & Macian, M. C. The family Rhodobacteriacea. in The Prokaryotes—Alphaproteobacteria and Betaproteobacteria (eds. Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E. & Thompson, F.). 440–512. https://doi.org/10.1007/978-3-642-30197-1_377 (Springer, 2014).
Fukui, Y. et al. Maritalea porphyrae sp. nov., isolated from a red alga (Porphyra yezoensis), and transfer of Zhangella mobilis to Maritalea mobilis comb. nov. Int. J. Syst. Evol. Microbiol. 62, 43–48. https://doi.org/10.1099/ijs.0.028704-0 (2012).
Santos, P. et al. Gamma-Proteobacteria Aquicella lusitana gen. nov., sp. nov., and Aquicella siphonis sp. nov. infect protozoa and require activated charcoal for growth in laboratory media. Appl. Environ. Microb. 69, 6533–6540. https://doi.org/10.1128/AEM.69.11.6533-6540.2003 (2003).
Zhukova, N. V., Kharlamenko, V. I., Svetashev, V. I. & Rodionov, I. A. Fatty-acids as markers of bacterial symbionts of marine bivalve mollusks. J. Exp. Mar. Biol. Ecol. 162, 253–263. https://doi.org/10.1016/0022-0981(92)90205-O (1992).
Los, D. A. & Mironov, K. S. Modes of fatty acid desaturation in cyanobacteria: An update. Life (Basel) 5, 554–567. https://doi.org/10.3390/life5010554 (2015).
Ruehland, C. et al. Multiple bacterial symbionts in two species of co-occurring gutless oligochaete worms from Mediterranean sea grass sediments. Environ. Microbiol. 10, 3404–3416. https://doi.org/10.1111/j.1462-2920.2008.01728.x (2008).
Goffredi, S. K., Warén, A., Orphan, V. J., Van Dover, C. L. & Vrijenhoek, R. C. Novel forms of structural integration between microbes and a hydrothermal vent gastropod from the Indian Ocean. Appl. Environ. Microbiol. 70, 3082–3090. https://doi.org/10.1128/AEM.70.5.3082-3090 (2004).
Egamberdieva, D., Jabborova, D. & Berg, G. Synergistic interactions between Bradyrhizobium japonicum and the endophyte Stenotrophomonas rhizophila and their effects on growth, and nodulation of soybean under salt stress. Plant Soil 405, 35–45. https://doi.org/10.1007/s11104-015-2661-8 (2016).
Adams, D. G., Duggan, P. S. & Jackson, O. Cyanobacterial symbioses. in Ecology of Cyanobacteria II: Their Diversity in Space and Time (ed. Whitton, B.). 593–647. (Springer, 2012).
Zehr, J. P. & Capone, D. G. Changing perspectives in marine nitrogen fixation. Science 368, eaay9514. https://doi.org/10.1126/science.aay9514 (2020).
Anteneh, Y. S. & Franco, C. M. M. Symbiosis and pathogenicity of Actinobacteria. in Biology and Biotechnology of Actinobacteria (eds. Wink J., Mohammadipanah F. & Hamedi J.). 233–268. https://doi.org/10.1007/978-3-319-60339-1_9 (Springer, 2017).
Hawe, A., Gensler, Y. & Haszprunar, G. Bacteriocytes in the mantle cavity of Lurifax vitreus Wareґn & Bouchet, (Orbitestellidae): The first case among heterobranch gastropoda. J. Mollusc Study 80, 337–340. https://doi.org/10.1093/mollus/eyu018 (2014).
van der Heijden, M., Bruin, S., Luckerhoff, L., van Logtestijn, R. S. & Schlaeppi, K. A widespread plant-fungal-bacterial symbiosis promotes plant biodiversity, plant nutrition and seedling recruitment. ISME J. 10, 389–399. https://doi.org/10.1038/ismej.2015.120 (2016).
Priya, H. et al. Influence of cyanobacterial inoculation on the culturable microbiome and growth of rice. Microbiol. Res. 171, 78–89. https://doi.org/10.1016/j.micres.2014.12.011 (2015).
Chanas, B. & Pawlik, J. R. Defenses of Caribbean sponges against predatory reef fish. II. Spicules, tissue toughness, and nutritional quality. Mar. Ecol. Prog. Ser. 127, 195–211. https://doi.org/10.3354/meps127195 (1995).
Zimmer, M. & Bartholmѐ, S. Bacterial endosymbionts in Asellus aquaticus (Isopoda) and Gammarus pulex (Amphipoda), and their contribution to digestion. Limnol. Oceanogr. 48, 2208–2213. https://doi.org/10.4319/lo.2003.48.6.2208 (2003).
Ramos, P. L. et al. Screening for endophytic nitrogen-fixing bacteria in Brazilian sugar cane varieties used in organic farming and description of Stenotrophomonas pavanii sp. nov.. Int. J. Syst. Evol. Microbiol. 61, 926–931. https://doi.org/10.1099/ijs.0.019372-0 (2011).
Busse, H. J. Review of the taxonomy of the genus Arthrobacter, emendation of the genus Arthrobacter sensu lato, proposal to reclassify selected species of the genus Arthrobacter in the novel genera Glutamicibacter gen. nov., Paeniglutamicibacter gen. nov., Pseudoglutamicibacter gen. nov., Paenarthrobacter gen. nov. and Pseudarthrobacter gen. nov., and emended description of Arthrobacter roseus. Int. J. Syst. Evol. Microbiol. 66, 9–37. https://doi.org/10.1099/ijsem.0.000702 (2016).
Wang, J. Y. et al. Bradyrhizobium daqingense sp. nov., isolated from soybean nodules. Int. J. Syst. Evol. Microbiol. 63, 616–624. https://doi.org/10.1099/ijs.0.034280-0 (2013).
Coenye, T., Vancanneyt, M., Falsen, E., Swings, J. & Vandamme, P. Achromobacter insolitus sp. nov. and Achromobacter spanius sp. nov., from human clinical samples. Int. J. Syst. Evol. Microbiol. 53, 1819–1824. https://doi.org/10.1099/ijs.0.02698-0 (2003).
Mutalipassi, M. et al. Symbioses of cyanobacteria in marine environments: Ecological insights and biotechnological perspectives. Mar. Drugs 19, 227. https://doi.org/10.3390/md19040227 (2021).
Flores, E., Frías, J. E., Rubio, L. M. & Herrero, A. Photosynthetic nitrate assimilation in cyanobacteria. Photosynth. Res. 83, 117–133. https://doi.org/10.1007/s11120-004-5830-9 (2005).
Pajares, S. & Ramos, R. Processes and microorganisms involved in the marine nitrogen cycle: Knowledge and gaps. Front. Mar. Sci. 6, 739. https://doi.org/10.3389/fmars.2019.00739 (2019).
Bibi, F., Jeong, J. H., Chung, E. J., Jeon, C. O. & Chung, Y. R. Labrenzia suaedae sp. nov., a marine bacterium isolated from a halophyte, and emended description of the genus Labrenzia. Int. J. Syst. Evol. Microbiol. 64, 1116–1122. https://doi.org/10.1099/ijs.0.052860-0 (2014).
Heylen, K., Vanparys, B., Peirsegaele, F., Lebbe, L. & De Vos, P. Stenotrophomonas terrae sp. nov. and Stenotrophomonas humi sp. nov., two nitrate-reducing bacteria isolated from soil. Int. J. Syst. Evol. Microbiol. 57, 2056–2061. https://doi.org/10.1099/ijs.0.65044-0 (2007).
Weber, C. F. & King, G. M. Physiological, ecological and phylogenetic characterization of Stappia a marine CO oxidizing bacterial genus. Appl. Environ. Microbiol. 73, 1266–1276. https://doi.org/10.1128/AEM.01724-06 (2007).
Weber, C. F. & King, G. M. The phylogenetical distribution and ecological role of carbon monoxide oxidation in the genus Burkholderia. FEMS Microbiol. Ecol. 79, 167–174. https://doi.org/10.1111/j.1574-6941.2011.01206.x (2012).
King, G. M. & Weber, C. F. Distribution, diversity and ecology of aerobic CO-oxidizing bacteria. Nat. Rev. Microbiol. 5, 107–118. https://doi.org/10.1038/nrmicro1595 (2007).
Kleiner, M. et al. Use of carbon monoxide and hydrogen by a bacteria-animal symbiosis from seagrass sediments. Environ. Microbiol. 17, 5023–5035. https://doi.org/10.1111/1462-2920.12912 (2015).
Ehrlich, H. et al. Discovery of chitin in skeletons of non-verongiid Red Sea demosponges. PLoS ONE 13, e0195803. https://doi.org/10.1371/journal.pone.0195803 (2018).
Beier, S. & Bertilsson, S. Bacterial chitin degradation – Mechanisms and ecophysiological strategies. Front. Microbiol. 4, 149. https://doi.org/10.3389/fmicb.2013.00149 (2013).
Raimundo, I. et al. Functional metagenomics reveals differential chitin degradation and utilization features across free-living and host-associated marine microbiomes. Microbiome 9, 43. https://doi.org/10.1186/s40168-020-00970-2) (2021).
Kong, H., Shimosaka, M., Ando, Y. & Nishiyama, K. Species-specific distribution of a modular family 19 chitinase gene in Burkholderia gladioli. FEMS Microbiol. Ecol. 37, 135–141. https://doi.org/10.1111/j.1574-6941.2001.tb00861.x (2001).
Souza, P. C., Almeida, C. B., Colwell, R. R. & Rivera, G. N. The importance of chitin in the marine environment. Mar. Biotechnol. 13, 823–830. https://doi.org/10.1007/s10126-011-9388-1 (2011).
Subramanian, K. et al. Bioconversion of chitin and concomitant production of chitinase and N-acetylglucosamine by novel Achromobacter xylosoxidans isolated from shrimp waste disposal area. Sci. Rep. 10, 11898. https://doi.org/10.1038/s41598-020-68772-y (2020).
Jankiewicz, U., Brzezinska, M. S. & Saks, E. Identification and characterization of a chitinase of Stenotrophomonas maltophilia, a bacterium that is antagonistic towards fungal phytopathogens. J. Biosci. Bioeng. 113, 30–35. https://doi.org/10.1016/j.jbiosc.2011.08.023 (2012).
Annamalai, N., Veeramuthu Rajeswari, M., Vijayalakshmi, S. & Balasubramanian, T. Purification and characterization of chitinase from Alcaligenes faecalis AU02 by utilizing marine wastes and its antioxidant activity. Ann. Microbiol. 61, 801–807. https://doi.org/10.1007/s13213-011-0198-5 (2011).
Winters, A. E. et al. Distribution of defensive metabolites in nudibranch molluscs. J. Chem. Ecol. 44, 384–396. https://doi.org/10.1007/s10886-018-0941-5 (2018).
Flórez, L. V., Biedermann, P. H. W., Engl, T. & Kaltenpoth, M. Defensive symbioses of animals with prokaryotic and eukaryotic microorganisms. Nat. Prod. Rep. 32, 904–936. https://doi.org/10.1039/c5np00010f (2015).
Chen, L., Hu, J.-S., Xu, J.-L., Shao, C.-L. & Wang, G.-Y. Biological and chemical diversity of ascidian-associated microorganisms. Mar. Drugs 16, 362. https://doi.org/10.3390/md16100362 (2018).
Funding
DNA analysis was supported by the Bren Professor Funds at the University of California, Irvine to F.J.A.
Author information
Authors and Affiliations
Contributions
N.V.Z. conceived and designed the experiments. N.V.Z., M.G.E., E.S.B. performed the experiments and analyzed the data. N.V.Z, M.G.E., F.J.A. contributed reagents/materials/analysis tools. N.V.Z. wrote the manuscript with participation of all authors. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhukova, N.V., Eliseikina, M.G., Balakirev, E.S. et al. Multiple bacterial partners in symbiosis with the nudibranch mollusk Rostanga alisae. Sci Rep 12, 169 (2022). https://doi.org/10.1038/s41598-021-03973-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-03973-7
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.