Abstract
We have previously reported evidence that Nogo-A activation of Nogo-receptor 1 (NgR1) can drive axonal dystrophy during the neurological progression of experimental autoimmune encephalomyelitis (EAE). However, the B-cell activating factor (BAFF/BlyS) may also be an important ligand of NgR during neuroinflammation. In the current study we define that NgR1 and its homologs may contribute to immune cell signaling during EAE. Meningeal B-cells expressing NgR1 and NgR3 were identified within the lumbosacral spinal cords of ngr1+/+ EAE-induced mice at clinical score 1. Furthermore, increased secretion of immunoglobulins that bound to central nervous system myelin were shown to be generated from isolated NgR1- and NgR3-expressing B-cells of ngr1+/+ EAE-induced mice. In vitro BAFF stimulation of NgR1- and NgR3-expressing B cells, directed them into the cell cycle DNA synthesis phase. However, when we antagonized BAFF signaling by co-incubation with recombinant BAFF-R, NgR1-Fc, or NgR3 peptides, the B cells remained in the G0/G1 phase. The data suggest that B cells express NgR1 and NgR3 during EAE, being localized to infiltrates of the meninges and that their regulation is governed by BAFF signaling.
Similar content being viewed by others
Introduction
Multiple sclerosis (MS) is a neuroinflammatory disease of the spinal cord, brain and optic nerve manifesting as demyelination and progressive neurodegeneration1. In recent years, a number of vital insights into the immunopathogenesis of MS have provided considerable scope for the development of novel therapeutics to be formulated. Mechanisms involving immune cell adhesion and extravasation into the central nervous system (CNS) have, for instance, led to the development of a humanized monoclonal antibody against the cell adhesion molecule α4-integrin, known as natalizumab. Other immunomodulatory drugs have included interferon-β, glatiramer acetate, dimethyl fumarate, alemtuzumab and fingolimod2. These treatments are used to diminish the patient’s relapses both in frequency and severity2. Recently ocrelizumab has shown promise in the biological’s ability to reduce the relapse rate in MS patients by its ability to specifically target CD20+ maturing B-cells that may propagate the disease, with documented potential to limit progression3,4,5. However despite these benefits, chronic-active MS lesions do not respond effectively to the currently available immunomodulatory treatments6. This highlights the critical need to develop novel disease modifying agents that can address the gap in treating progressive MS with profound neurodegenerative changes.
Recently, B-cells have emerged as potential important pathogenic players in MS7,8,9. In individuals living with MS, B-cells have been identified to reside within the CNS for protracted periods and are present within the cerebral spinal fluid (CSF). These B-cells may also produce pathogenic autoantibodies intrathecally10. In addition, B-cells can be competent (auto) antigen-presenting (APC) cells to effector pathogenic T-cells. Indeed, animal models in which B-cells are specifically lacking MHC-II are resistant to experimental autoimmune encephalitis (EAE), suggesting that the antigen presenting cell (APC) function of B-cells is a key pathogenic mechanism governing disease11. B-cells also produce IL-6, a cytokine participating in the emergence of pathogenic Th17 T-cells in MS12. In fact, B-cells from MS patients have been demonstrated to produce elevated levels of IL-612. Furthermore, B-cells from MS patients but not from healthy controls produce tumor necrosis factor α (TNFα) along with neurotoxic factors that propagate oligodendrocytopathy13,14.
Accumulating evidence thus indicates that blunting the pathogenic effects of B cells could have therapeutic potential in relapsing–remitting multiple sclerosis (RRMS). Development of atacicept, a soluble fusion protein decoy receptor (incorporating the ligand-binding domain of the transmembrane activator and cyclophilin ligand activator (TACI, also known as TNFRSF13B)), designed to block two B-cell cytokines B-cell activating factor of the TNF family (BAFF, also known as BLyS, TNFSF13B) and a proliferation-induced ligand (APRIL, TNFSF13), unexpectedly led to severe adverse events and exacerbated symptoms and lesions when tested in clinical trials in MS patients15. The rationale for using this treatment was the observation of elevated BAFF levels in MS lesions and the general knowledge that elevated levels of BAFF can drive autoimmunity16,17. This surprising outcome highlighted the complexity of the role of B-cells in MS but also suggests that alternate receptor signaling mechanisms may exist to promote their survival and maturation.
Although the role of BAFF, a known stimulator of B-cell activity18,19,20, has been reported to promote NgR-dependent activation21, there seems to be further stratification in the activity related to neuroinflammatory lesions, with antagonists of APRIL/BAFF exacerbating the severity of EAE, an outcome recently reported to relate to APRIL’s anti-inflammatory effects22. This deleterious outcome observed during the therapeutic targeting of APRIL has been reported to be a consequence of the ligand’s pleotropic effects in astrocytes, derived from active macrophages during disease progression22.
Collectively these findings indicate that APRIL and BAFF are important in regulating autoimmunity23. Recent evidence suggests that serum levels of BAFF and APRIL are elevated in MS patients with grey matter neurodegenerative pathology and meningeal inflammation compared with healthy controls24. Notably it was reported that these cytokines are reduced in the CSF, during the provision of disease modifying therapies (DMTs)25, although surprisingly, they are elevated in the peripheral circulation26. Of importance to neurodegenerative changes during injury or disease was the finding that BAFF/BlyS can bind to Nogo Receptor (the high affinity receptor for the myelin associated inhibitory factor, Nogo-A)21,27,28, a common receptor expressed in neurons that can limit axonal regrowth and even signal degeneration, in a model of MS29,30,31.
A role for BAFF and BAFF-R in the regulation of primary neurodegeneration without overt inflammatory cell infiltration has been reported but clear mechanistic evidence is still lacking32. Both BAFF and BAFF-R are expressed in mouse spinal cord motor neurons and Transgenic SOD-1 mutant mice that were deficient for BAFF-R (Baffrm/m), exhibited increased neurological decline and reduced survival time, implicating neuronal BAFF-R expression as a survival signal for neurons. However, how this relates to neuronal survival signaling during primary neurodegeneration is unclear.
The myelin associated inhibitory factors (MAIFs), consist of Nogo-A, myelin-associated glycoprotein (MAG), and oligodendrocyte-myelin glycoprotein (OMgp) and are expressed on myelin and oligodendrocyte membranes. MAIFs have been demonstrated to be potent inhibitors of axonal regeneration during disease (for review see33). Indeed, we previously reported that Nogo-A and its cognate receptor NgR1, may promote axonal degeneration in the animal model of MS, EAE30,31. It has also been hypothesized that remyelination failure observed in progressive MS may be a result of co-activation of MAIFs within extracellular myelin debris, acting as potent endogenous inhibitory cues to block the recruitment and maturation of oligodendroglial precursor cells (OPCs) around demyelinated plaques34,35,36. MAIF-dependent axonal outgrowth inhibition is promulgated through activation of the small GTPase RhoA, inducing actin and tubulin disassembly via a signaling pathway which may be kinase-dependent37,38,39. Downstream signaling occurs following high-affinity ligand-mediated tripartite receptor complexing of the NgR1, low-affinity neurotrophin receptor (p75NTR), and LINGO-1; all expressed on axons40,41,42. However, how the proposed BAFF/NgR1 interaction can regulate active demyelination of lesions that are pathognomonic for MS, has not been elucidated.
In this study, we show for the first time the effect of BAFF on NgR-expressing B-cells isolated at the peak stage of EAE in the MOG35-55-induced mouse model of MS. We found that both NgR1 and NgR3 may play a role in modulating the adaptive immune response during the progression of MOG35-55-induced EAE and demonstrate that mature B-cells express NgR3 within spinal cord infiltrates, localized to the meninges during EAE. Our systematic dissection of the immune function of NgR during CNS inflammation has identified that the NgR-expressing B-cells can be stimulated by BAFF to induce proliferation and differentiation that can be antagonized by the provision of Nogo receptor fusion proteins (NgR-Fc and NgR3-P). We further demonstrate that these leptomeningeal NgR-expressing B-cells can actively secrete anti-myelin glycoprotein antibodies that bind to myelin and oligodendrocytes in the mouse spinal cord white matter. Our data highlight the complexity of BAFF signaling during neuroinflammation and suggest that both NgR1 and NgR3 homologs can be important regulators of the neurodegenerative process governing EAE and by extension MS.
Results
MOG35-55 and rMOG-induction of EAE in ngr1 −/− exhibit reduced clinical severity, demyelination and axonal loss
We have previously reported that deletion of the ngr1 allele (exon 2) limits the progression of EAE and axonal degeneration30,31. In the current set of experiments we have interrogated further the immunological role of NgR1, by immunizing female wildtype (ngr1+/+) and NgR1 deficient (ngr1−/−) mice with either MOG35-55 peptide or recombinant MOG(1-125) [rMOG]. The effect of ngr1 gene deletion on the clinical course in MOG35-55 and rMOG-induction of EAE is shown in Table 1 (Table 1a, b), demonstrating a delay in the onset and severity of clinical disease signs. CFA inoculated animals did not develop EAE in any of two cohorts.
Consistent with our previous MOG35-55 data, a significant delay in the onset of clinical symptoms (dDO; 16.1 ± 0.6 in ngr1−/− vs 13.5 ± 0.3 in ngr1+/+, p < 0.05) and reduced disease burden (MMS; 2.3 ± 0.1 in ngr1−/− vs 3.2 ± 0.2 in ngr1+/+, p < 0.05 and AUC; 22.2 ± 2.3 in ngr1−/− vs 39.2 ± 3.1 in ngr1+/+, p < 0.001) has been observed in the ngr1−/− compared to the ngr1+/+ mice (Fig. 1a; Two-way ANOVA, p < 0.001). Histopathological evaluation performed with LFB/PAS and Bielschowsky staining, illustrated a significantly reduced demyelination in ngr1−/− mice at disease onset and reduced axonal loss at the peak stage of disease, in comparison to ngr1+/+ mice (Fig. 1b).
The onset of MOG35-55-induced EAE mice is accompanied by an increase in NgR1+ and NgR3+ immune cell populations. (a) There was a significant delay in the onset of EAE and reduction in severity for the ngr1−/− (n = 32; red line) compared to ngr1+/+ mice (n = 76; purple line) within the 2 main phases studied; onset (days 10–20 post-immunization; 10–20 dpi) and peak (days 20–30 post-immunization; 20–30 dpi). A control group (CFA-injected only; n = 44; black line) was also included. Daily clinical scores represent mean ± SEM, ***p < 0.001 (from day 15 until end-point), two-way ANOVA. (b) The severity of demyelination and axonal loss was also reduced in the spinal cord of ngr1−/− EAE-induced mice, for both onset and peak phases, determined by Luxol fast blue (LFB)/Periodic acid-Schiff (PAS) and Bielschowsky silver stain, respectively. (c,d) Flow cytometric analysis of double-labeled cell suspensions is from the spleen of both ngr1+/+ and ngr1−/− mice showed an elevated NgR3 expression in B-cells (B220+) at the onset of EAE (n = 5; t test ***p < 0.001 and *p < 0.05 compared to controls, respectively) along with a concomitant increase of NgR1 in ngr1+/+ mice (n = 5; t test **p < 0.01). B-cells co-expressing NgR3 were subsequently reduced in the peak stage of EAE (n = 5; t test *p < 0.05) in ngr1−/− mice that exhibited disease signs. The NgR2 homolog did not manifest any discernible significant increase in either ngr1+/+ or ngr1−/− mice. (e,f) The percentages of double labeled immune cells isolated from the spinal cord in EAE-induced mice were again significantly elevated for NgR3 and NgR1 during the onset of disease, compared to controls (n = 5; t test ***p < 0.001 for both). Interestingly, deletion of the ngr1 gene in ngr1−/− mice incurred an NgR3 and NgR2 upregulation in T-cell (CD3e+) populations (n = 5; t test ***p < 0.001 and **p < 0.01 compared to controls, respectively). B220: B-cell marker; CD3e: T-cell marker. Bars represent mean ± SEM, *p < 0.05, **p < 0.01, ***p < 0.001.
Similar results were demonstrated in the rMOG-inoculated group, a model of EAE-induction commonly utilized to induce overt B-cell activation, differentiation and anti-myelin autoantibodies43,44,45. We again observed a significant delay in EAE onset (dDO; 20.6 ± 0.9 in ngr1−/− vs 14.1 ± 0.4 in ngr1+/+, p < 0.01) and reduced severity (MMS; 2.1 ± 0.2 in ngr1−/− vs 2.9 ± 0.2 in ngr1+/+, p < 0.05 and AUC; 15.3 ± 1.7 in ngr1−/− vs 33.9 ± 2.5 in ngr1+/+, p < 0.001) in the ngr1−/− mice following rMOG-immunization (Additional file 1: Fig. S1a; Two-way ANOVA, p < 0.001). Demyelination and axonal loss exhibited a similar reduction in the pattern of degenerative change in the ngr1−/− mice following rMOG induction as that observed in the MOG35-55 EAE model (Additional file 1: Fig. S1b).
B-cells isolated from spleen and spinal cord of ngr1 +/+ and ngr1 −/− mice following either MOG35-55 or rMOG EAE induction express the NgR1 and NgR3 homolog
To interrogate the differing immunopathogenic mechanisms potentially governing the EAE clinical outcomes attributed to ngr1+/+ and ngr1−/− mice following MOG35-55 peptide and rMOG protein, we defined the co-expression profiles of selected immune cells from isolated populations (B220 for B-cells and CD3e for T-cells) with flow cytometry analysis, according to the three NgR homologs namely NgR1 (Rtn4r), NgR2 (Rtn4rl2) and NgR3 (Rtn4rl1) [Fig. 1 and Additional file 2: Fig. S2].
In the spleen of MOG35-55 EAE-induced ngr1+/+ mice, B-cells expressing NgR1 and NgR3 were significantly increased upon disease onset (p < 0.01 and p < 0.001, respectively), but as the disease progressed beyond the peak stage of EAE NgR1+ B-cells were markedly reduced while NgR3+ B-cells remained elevated at the peak stage of disease (Fig. 1c; and Additional file 2: Fig. S2). In the spleen of ngr1−/− mice there were elevated numbers of B-cells expressing NgR3 during disease onset (9.5 ± 1.7 vs controls 5.2 ± 1.8, p < 0.05), but these numbers were significantly reduced (5.1 ± 1.7, p < 0.05) upon progression, at the peak stage of EAE (Fig. 1d; and Additional file 2: Fig. S2).
Interestingly, in the spinal cord of the ngr1+/+ mice, both T- and B-cell populations expressing NgR3 were elevated at the onset of EAE (2.2 ± 0.9 vs controls 0.2 ± 0.1, p < 0.05 for CD3e+ T-cells and 3.2 ± 0.7 vs controls 0.05 ± 0.02, p < 0.05 for B220+ B-cells) and remained elevated at the peak stage of disease (Fig. 1e; and Additional file 2: Fig. S2). This elevated NgR3 expression profile in T- and B-cells (p < 0.001 vs controls for both populations) was also replicated in the ngr1−/− mice at the onset of EAE (Fig. 1f; and Additional file 2: Fig. S2). Intriguingly, an upregulation in NgR2+ T-cells (p < 0.01 vs controls) was also observed in both ngr1+/+ and ngr1−/− mice.
Thus, deletion of the ngr1 gene in mice does not result in an altered infiltration of B-cells during disease induction but there are altered numbers of NgR3-expressing cells during the peak stage of EAE that may relate to the reduced severity observed in this genotype following MOG35-55 immunization. Collectively, these data indicate that NgR1 and NgR3 are expressed on specific numbers of immune cells particularly of the B cell lineage, within the CNS compartment upon the induction of EAE, and thus both of these receptors may influence the behaviour of these cells once resident in the CNS. This does not seem to be the case with the NgR2 homolog.
Next, we examined the subset of immune cells present in the spleen and spinal cord harvested from ngr1+/+ and ngr1−/− mice compared to controls, following EAE induction by immunization with rMOG. In the spleen, the percentage of B-cells expressing NgR (NgR1 and 3 in ngr1+/+ and NgR3 in ngr1−/− specifically) increased significantly (13.8 ± 1.0 vs controls 3.3 ± 1.0, p < 0.001 for NgR1 in ngr1+/+and 7.7 ± 1.9 vs controls 5.1 ± 1.8, p < 0.01 for NgR3 in ngr1−/−) at the onset of the disease (Additional file 1: Fig. S1c, d; and Additional file 2: Fig. S2).
The B-cells expressing NgR1 in the spinal cord of ngr1+/+ mice were elevated (2.4 ± 0.6 vs controls 0.09 ± 0.03, p < 0.001) at the disease onset (Additional file 1: Fig. S1e; and Additional file 2: Fig. S2). Moreover, NgR3+ B-cells were significantly elevated in the spinal cord of the ngr1+/+ (3.5 ± 0.6 vs controls 0.05 ± 0.02, p < 0.001) and ngr1−/− mice (1.0 ± 0.2 vs controls 0.06 ± 0.02, p < 0.01) (Additional file 1: Fig. S1e, f; and Additional file 2: Fig. S2). Intriguingly, the numbers of NgR3-expressing B-cells resident in the CNS during EAE progression at the peak stage of disease in ngr1−/− mice were substantially reduced back to basal levels prior to disease onset.
Collectively these results demonstrate that NgR3 is expressed on increased numbers of B-cells within the CNS during the onset of EAE symptoms following either, MOG35-55- and rMOG-induction, demonstrated in both ngr1+/+ and ngr1−/− mice. However, these NgR3 expressing B-cells demonstrated reduced numbers toward basal levels in the CNS during the peak stage of EAE when profound neurodegeneration is evident. The data implicate a potential signaling role for NgR3 in B-cells while in the CNS compartment at the induction of inflammatory disease.
Due to the similarities between the two MOG-induced experimental models with regards to the clinical/immunopathological and the co-expression outcome of the Nogo receptor homologs, we proceeded with the MOG35-55 EAE material for the remaining analysis. At the same time, we excluded NgR2 as a valuable parameter to be further investigated, due to the lack of strong evidence of its implication to the disease induction and progression.
NgR1+ and NgR3+ B-cells are localized within infiltrates of the spinal cord during the onset of EAE symptoms
Following the identification that NgR1 and NgR3 are expressed on a subset of B-cells, we next posited whether receptor expression was dynamic or static in nature and where these B-cells are localized within inflammatory CNS infiltrates. Using western blot analyses of spleen and spinal cord extracts from MOG35-55–induced mice at onset of disease (clinical score 1), NgR1 and NgR3 in ngr1+/+, and NgR3 in ngr1−/− mice respectively, were elevated (Fig. 2a). Quantification of NgR1 and NgR3+ B-cells per unit area (mm2) of spleen and spinal cord inflammatory lesions in ngr1+/+ and ngr1−/− mice (n = 34 ngr1+/+ mice; 108 sections, n = 15 ngr1−/− mice; 40 sections) during the onset of EAE was performed to define the number of these cells localized within the respective regions (Fig. 2b).
NgR1 and NgR3 are localized to B-cells within lymphoid-like follicles of the spinal cord. (a) Western blot of spleen and spinal cord extracts from ngr1+/+ and ngr1−/− mice showcasing the expression of NgR1 and NgR3. β-actin was used as a loading control. (b) Semi-quantitative analysis of both NgR1- and NgR3-positive B-cells per unit area (mmy) of spleen and spinal cord infiltrates for the ngr1+/+ and ngr1−/− mice (n = 34 ngr1+/+ mice; 108 sections, n = 15 ngr1−/− mice; 40 sections) during the onset of EAE. These B-cells demonstrated a highly inducible receptor expression (student’s t test with Bonferroni correction ***p < 0.001 and **p < 0.01) compared to controls. (c) Illustration of inflammatory infiltrates analyzed via immunofluorescence immunolabeling of follicle-like structures (FLS) within the lumbosacral spinal cords of EAE-induced mice in (d–k). (d) Representative NgR1 and B220 double-labeled images showing the expression on clusters of B-cells localized to infiltrates around the leptomeninges during the onset of EAE in ngr1+/+ mice. (e) Representative NgR3 and B220 double-labeled images showing the expression on clusters of B-cells localized to infiltrates around the leptomeninges during the onset of EAE in ngr1+/+ mice (hatched boxes). (f) Representative images for the ngr1−/− mice, highlighting the absence of the NgR1 protein expression in clusters of B-cells in similar infiltrates of the leptomeninges during the onset of EAE (hatched boxes). (g) Representative images for the ngr1−/− mice, highlighting the strong expression of the NgR3 protein expression in clusters of B-cells in similar infiltrates of the leptomeninges during the onset of EAE (hatched boxes). (h) Representative NgR1 and CD19 double-labeled images showing the expression on clusters of B-cells localized to infiltrates around the leptomeninges during the onset of EAE in ngr1+/+ mice. (i) Representative NgR3 and CD19 double-labeled images showing the expression on clusters of B-cells localized to infiltrates around the leptomeninges during the onset of EAE in ngr1+/+ mice. (j) Representative images for the ngr1−/− mice, highlighting the absence of the NgR1 protein expression in clusters of CD19 + B-cells in similar infiltrates of the leptomeninges during the onset of EAE (hatched boxes). (k) Representative images for the ngr1−/− mice, highlighting the strong expression of the NgR3 protein expression in clusters of CD19 + B-cells in similar infiltrates of the leptomeninges during the onset of EAE. Scale = 50 μm.
We showed that the numbers of NgR1+ and NgR3+ B-cells were significantly increased in infiltrates around the leptomeninges (Fig. 2c-e B220 + and Fig. 2h-I CD19+) at the onset of EAE symptoms induced by the MOG35-55 peptide in the ngr1+/+ mice when compared to controls (279.4 ± 43.9 cells per mm2 for NgR1 and 336.4 ± 50.1 cells per mm2 for NgR3, p < 0.001 vs controls for both; Fig. 2b).
We also demonstrated that increased numbers of NgR3+ B-cells were also present in the leptomeningeal infiltrates of EAE-induced ngr1−/− mice while NgR1 was absent (Fig. 2f–g B220 + ; Fig. 2j-k CD19 +), as expected, at the onset of disease (240.2 ± 34.3 cells per mm2 for NgR3, p < 0.001 vs controls; Fig. 2b). These data demonstrate that NgR1 and NgR3 are expressed on significant numbers of B-cells present within leptomeningeal infiltrates at the clinical onset of EAE and that this expression is inducible.
BAFF may play a role in stimulating B-cells that express NgR1 and NgR3 during the disease onset of EAE
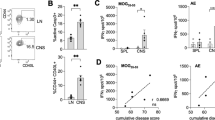
Since BAFF has been previously shown to act as a ligand for Nogo receptor21 and we identified the expression of NgR1 and 3 on B-cells within the CNS, an investigation towards a BAFF/NgR-dependent mechanism operative during the induction of MOG35-55 EAE, was conducted (Fig. 3a). Spinal cord sections from EAE-induced ngr1+/+ mice were stained for NgR1, NgR3, BAFF and BAFF-R. Substantial numbers of clustered immune cell infiltrates that co-labeled for either, BAFF and NgR1 or, BAFF and NgR3, were identified within the spinal cord of ngr1+/+ EAE-induced mice at disease onset (Fig. 3b,c). However, we could not detect double-positive immune cells that co-expressed BAFF-R with any of the Nogo receptors localized to the leptomeninges of the spinal cord (Fig. 3d,e).
BAFF play a role in stimulating B-cells that express NgR1 and NgR3 during the disease onset of EAE. (a) Schematic representation of the potential BAFF/NgR-dependent mechanism operative during the induction of EAE. (b,c) Substantial numbers of clustered immune cell infiltrates that co-labeled either BAFF and NgR1 or, BAFF and NgR3, were found within the spinal cord of ngr1+/+ EAE-induced mice at disease onset. (d,e) No double-positive immune cells co-expressing BAFF-R with NgR1 and NgR3 were identified in infiltrates of the spinal cord in ngr1+/+ mice. Hatched boxes are further apposed in separate channels; scale = 50 μm. (f) Cell cycle assessment (G1, gap 1 phase; S, synthesis; G2, gap 2 phase; M, mitosis) with flow cytometric analysis showing the gating for the cell cycle phases; G0/G1 (purple color), S phase (Blue), G2/M phase (yellow); BrdU, 5-Bromo-2′-deoxyuridine; 7AAD, 7-aminoactinomycin D (7AAD). (g,h) NgR1+ and NgR3+ B-cell population was either stimulated with 50 ng/mL recombinant BAFF (with BAFF) or left untreated (without BAFF). BAFF stimulation led to a significant increase in DNA synthesis (S phase compared to G0/G1 phase; n = 12; t test ***p < 0.001) of isolated cells from ngr1+/+ mice while cells remained in the G0/G1 phase (n = 12; t test *p < 0.05) in the absence of BAFF. Isolated cells from ngr1−/ −mice also exhibited significant differences between the G0/G1 and S phases in the BAFF-treated cultures (n = 10; t test *p < 0.05) and the untreated ones (n = 10; t test **p < 0.01), highlighting the role of NgR3 in the absence of NgR1. (i) To identify NgR interactions with BAFF, rBAFF was blocked with an excess amount of either rBAFF-R, NgR1-Fc or NgR3-P. All three blocking peptides were efficacious at blocking the cell cycle-dependent BAFF activity when compared to the BAFF-treated cultures. Bars represent mean ± SEM; #p < 0.05, ##p < 0.01, ###p < 0.001 comparisons with the G0/G1 phase of the BAFF-treated cultures, t test (with Bonferroni correction); $p < 0.05, $$p < 0.01, $$$p < 0.001 comparisons with the S phase of the BAFF-treated cultures, t test (with Bonferroni correction).
Similarly, BAFF+ NgR1+ and BAFF+ NgR3+ immune cell populations were exhibited within the splenic follicles of EAE-induced ngr1+/+ mice at the onset of EAE, while BAFF-R could only be detected on occasional immune populations of the spleen (Additional file 3: Fig. S3a–d, arrows). In order to define how BAFF may modulate B-cells, which express NgR1 and NgR3 during the onset of EAE, cells from the spinal cord and spleen of the ngr1+/+ mice were isolated by FACS, and cultured (spinal cord; n = 12, spleen; n = 10) in the presence or absence of recombinant BAFF (rBAFF) [50 ng/mL]. Stimulation of B-cells positive for either NgR1 or NgR3 with rBAFF led to a significant increase in DNA synthesis (Fig. 3f) [S phase, 71.6 ± 8.2% vs G0/G1 phase 18.2 ± 5.8%, p < 0.001; Fig. 3g] compared to the cultures that were not stimulated with BAFF (G0/G1 phase 52.7 ± 5.9% vs S phase, 36.8 ± 6.3%, p < 0.05; Fig. 3h). Analogous results were retrieved from ngr1−/− mice (with BAFF: S phase, 48.1 ± 5.1% vs G0/G1 phase 32.1 ± 7.9%, p < 0.05; without BAFF: G0/G1 phase 60.8 ± 7.4% vs S phase, 34.3 ± 8.3%, p < 0.01). No significant differences in the percentage of apoptosis were observed with or without BAFF stimulation for both cohorts of mice. Likewise, there were significant differences between the G0/G1 and S phases in the cells isolated from the spleens of ngr1+/+ and ngr1−/− mice; isolated B-cells were mainly residing in S phase with BAFF stimulation and preferred G0/G1 over S phase when they were devoid of BAFF (Additional file 3: Fig. S3e,f).
To further illustrate this, we identified that isolated cells also remained in G0/G1 phase in the spinal cords of ngr1+/+ mice when BAFF activity was blocked by pre-incubating the culture medium with either (i) rBAFF-R (n = 8 independent experiments) or; (ii) NgR3-peptide (n = 6 independent experiments); or (iii) NgR-Fc fusion protein (n = 6 independent experiments) (Fig. 3i). More specifically, 27.7 ± 5.3% of the isolated cells from the rBAFF-R cohort, 27.5 ± 6.3% from the NgR3-P and only 20.6 ± 3.4% from the NgR-Fc group were identified in the S phase, and were significantly lower in comparison to the control group, treated with BAFF (p < 0.01 for all comparisons). Interestingly, NgR3-P and NgR-Fc fusion protein were efficacious at blocking the cell cycle-dependent BAFF activity (53.6 ± 6.5% and 55.8 ± 1.5% of the isolated cells stayed in the G0/G1 phase, respectively) in a similar manner to that achieved with rBAFF-R antagonism (Fig. 3i).
In parallel experiments using cells isolated from spleens, we demonstrated that the NgR3-P and NgR-Fc fusion proteins were extremely efficacious at blocking the cell cycle-dependent BAFF activity halting 86.8 ± 4.7% and 85.5 ± 6.3% of the isolated cells in the G0/G1 phase, while only 7.7 ± 3.6% and 8.0 ± 4.5% could enter the S phase, respectively (Additional file 3: Fig. S3g). These data suggest that both NgR1 and NgR3 are active contributors to B-cell maturation during EAE and in fact may contribute to the signal transduction elicited by BAFF during spinal cord inflammation.
Immunoglobulin-specific phenotyping of B-cell infiltrates expressing NgR1 and NgR3 during EAE at the onset of EAE
B-cells expressing either NgR1 and/or NgR3 were isolated from the spinal cords of EAE-induced ngr1+/+ and ngr1−/− mice and phenotyped according to their membrane-bound immunoglobulins, to investigate potential humoral/immunopathogenic roles of NgR.
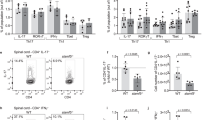
Flow cytometric analysis was performed on immune cells isolated from lumbar spinal cord at disease onset were stained with fluorescently tagged anti-NgR1 and 3, anti-B220, anti-IgM and IgD antibodies. Representative results showed that the majority of cells (> 80%) expressed high levels of cell surface IgG and low levels of IgM with a minor proportion (~ 12%) co-expressing high amounts of IgG and IgM (Fig. 4a,b). Furthermore, IgD was co-expressed on small proportion (~ 1%) of cells expressing high levels of IgM. Similar results were generated in ngr1−/− mice. The majority of double-positive IgG+ B-cells (96.6%) were not cycling based on Ki-67 labeling (Fig. 4c,d). These data suggest that the isolated spinal cord B-cells from EAE-induced mice that express NgR have matured and underwent immunoglobulin class switching.
NgR1+ and NgR3+ B-cells exhibit increased secretion of IgG during the EAE onset. (a) Representative dot plots of NgR3+ B-cells isolated from both ngr1+/+ EAE-induced mice exhibited increased levels of IgG than IgM on their cell surface. (b) IgM+ B-cells found to be more abundant in comparison to IgD+ B-cell populations. (c) NgR1+ and NgR3+ B-cells were found to be mainly IgG+ with the representative estimation of NgR3+ isolated B-cells exhibiting ~ 96.6% IgG+, when gated against the cell proliferation marker Ki-67. (d) The data are represented in a bar graph of multiple experiments with mean ± SEM (n = 6; t test ***p < 0.001). (e–g) The titers of the secreted immunoglobulins from isolated and cultured NgR1+ and NgR3+ B-cells were also evaluated from isolated supernatant samples by ELISA for both ngr1+/+ and ngr1−/− mice. Lower titers yielded a higher concentration of the respective antibodies. ngr1−/− mice showed a reduced capacity for production of IgG, IgM and IgD but with no statistically significant differences. Results represent mean ± SEM.
Supernatants from the spinal cord and spleen of ngr1+/+ and ngr1−/− EAE-induced mice at disease onset were collected from sorted and cultured cells before BAFF stimulation, and the IgG, IgM and IgD secreted immunoglobulin profiles were examined using ELISA (Fig. 4e–g). The responses of IgA, IgE and IgG subclasses (IgG1, IgG2b, IgG2c and IgG3) were also evaluated (data not shown).
Upon interrogation of titre levels present within the culture medium supernatants for all the secretory immunoglobulins from the cells isolated from the spinal cord, the dilution that yielded the most abundant fraction was found to be 1:10. Spleen isolates showed comparable efficiency. At this titre, B-cells expressing NgR1 from ngr1+/+ genotype, were found to secrete 135.1 ± 47.1 μg/mL IgG, 129.0 ± 17.0 μg/mL IgM and 76.6 ± 13.0 μg/mL IgD, while B-cells expressing NgR3 from ngr1+/+, were found to secrete 88.9 ± 40.4 μg/mL IgG, 120.0 ± 14.0 μg/mL IgM and 53.9 ± 12.0 μg/mL IgD. In the ngr1−/− group, we failed to detect any immunoglobulin secreted from NgR1+ B-cells but NgR3+ B-cells could yield 64.9 ± 18.1 μg/mL IgG, 94.8 ± 2.1 μg/mL IgM and 43.2 ± 0.1 μg/mL IgD although it did not show any significant difference when compared with the titers of the ngr1+/+ group (Mann–Whitney test; p > 0.05). Based on these findings, CNS-infiltrating B-cells expressing NgR1 and NgR3 undergo class switching, with a profound IgG humoral response.
Secreted IgG from ngr1 + and NgR3+ isolated spinal cord B cells recognize epitopes on myelin and oligodendrocyte glycoproteins
In order to determine whether secreted IgG antibodies generated by NgR1+ or NgR3+ B-cells isolated from diseased spinal cords were reactive against myelin, immunostaining was performed on spinal cord tissue sections obtained from non-diseased ngr1+/+ mice (naïve controls).
We identified that clusters of spinal cord white matter MBP+ and Nogo-A+ cells were colocalized with secreted IgG present within the supernatant from the cultured NgR3+ B-cells of EAE-induced ngr1+/+ mice (Fig. 5a,b), suggesting that these antibodies are recognizing antigenic epitopes present on oligodendroglial lineage cells. Interestingly, IgG+ and Nogo-A+ co-immunoreactivity detected nodal myelin labeling, as demonstrated under high magnification insets (Fig. 5ai,bi).
Secreted immunoglobulin responses by BAFF-responsive NgR1+ and NgR3+ B-cells are directed against CNS myelin. (a,b) Double immunofluorescence labeling of NgR1+ and NgR3+ B-cell-secreted IgG (green) co-localized with MBP and Nogo-A, respectively (red), within adjacent spinal cord sections of EAE mice. DAPI (blue) stained cell nuclei. ai: inset of hatched box in a, bi: inset of hatched box in b. White arrow indicates co-localization. Scale = 5 μm. (c) Western Immunoblot analysis of secreted IgG in the supernatant from isolated cells. Purified myelin from lumbar spinal cord of ngr1+/+ and ngr1−/− mice were differentiated on a 4–12% Bis–Tris gel. IgG presented in the supernatant could directly bind to the major myelin proteins MBP, MOG, PLP and CNPase, annotated in the gel, respectively. The three lanes per box are replicates. No immunoreactive bands were detected when a commercial IgG antibody was utilized as a control.
The ability of these secreted IgG antibodies from isolated NgR1+ or NgR3+ B-cells (which bind to spinal cord white matter myelin), were further examined for their ability to bind to specific myelin epitopes (n = 3 independent experiments). Our results indicated that these IgG anti-myelin antibodies from isolated NgR1+ or NgR3+ B-cells (from either the spleen or spinal cord of ngr1+/+ mice at clinical score 1) could bind directly to all major myelin glycoproteins including MBP, MOG, PLP and CNPase (Fig. 5c). These results highlight that the secretory form of IgG derived from isolated peripheral and intrathecal NgR1+ and NgR3+ B-cells, directly interact with myelin, suggestive of a humoral response by BAFF-sensitive NgR1+ and NgR3+ B-cells.
Discussion
Current MS treatments predominantly target the inflammatory aspects of the disease, however, chronic-active MS lesions do not respond effectively to monotherapies6. This highlights the critical need to develop novel disease modifying agents that can address the gap in treating acute and/or progressive MS with profound neurodegenerative changes. Our previous work has shown that deletion of NgR1 can protect against axonal degeneration, and thus disease progression, in EAE-induced mice30. These data were further supported with signal transduction evidence to show that NgR1 may ultimately potentiate neurodegeneration in EAE via a direct mechanism that stalls axonal transport. We identified that despite inflammatory infiltrates present during the peak stage of EAE, conditional deletion of NgR1 in neurons limits axonal degeneration and demyelination31. In the current study, we investigated the role of NgR1 and its homologs, specifically during B-cell maturation and differentiation. We identified infiltrates localized to the CNS during the onset of EAE that exhibited NgR1 and NgR3 expression. These NgR1+ and NgR3+ B-cells were significantly elevated at the acute onset stage of EAE, following immunization of mice either with MOG35-55 or rMOG.
For the first time, we identified that both NgR1+ and NgR3+ are active contributors to B-cell maturation during EAE and may participate in the signal transduction events elicited through BAFF-specific stimulation to enhance the proliferation of B-cells within EAE lesions. Importantly we show that mature secretory NgR1+ and NgR3+ B-cells can generate immunoglobulins that bind to spinal cord white matter myelin and the integral myelin membrane glycoproteins. Our findings emphasize that NgR1+ and NgR3+ may contribute to signaling in B-cells during the onset of EAE and can generate autoantibodies against myelin. The relevance of these B-cells in the pathogenesis of inflammatory demyelination that govern MS progression requires investigation since if we could therapeutically target this B-cell population during the course of MS, it may serve as a more specific disease modifying regimen than depleting B-cells on a broader scale.
B-cells play a significant role in MS pathogenesis, in addition to their ability for processing and presenting antigen46. Recent clinical trials and archival post-mortem analyses have established the benefit of targeting leptomeningeal-localized B-cell populations to limit MS relapse rates and progression14,47,48. However, a recent clinical phase II randomized, double-blind, placebo controlled trial with tabalumab (NCT0088299999), a IgG4 antagonizing monoclonal humanized antibody directed against membrane-bound and soluble BAFF, did not meet its primary endpoint of reduced gadolinium-enhancing MRI lesions in 197 RRMS patients that completed the 73 week study49,50. Moreover, the secondary endpoints of T2-weighted lesions and annualized relapse rates were not met. Therefore, the failure of this well powered and designed trial would suggest that antagonizing peripheral BAFF-dependent B-cell activity may not be relevant in limiting immune-mediated demyelination in humans and would advocate for greater mechanistic studies to be performed to understand the signaling dynamics of leptomeningeal follicular B-cells that may propagate inflammatory demyelinating lesions. Since NgR1 and its downstream signaling cascade has been defined as a critical mechanism governing axonal and myelin degeneration during the onset and progression of EAE, we set out to address the main question of how the receptor may influence immunopathogenic mechanisms (if any) that govern this model of inflammatory demyelination.
We have shown that specific B-cell populations express NgR1 and NgR3 clustered within leptomeningeal infiltrates of the lumbosacral spinal cords during the onset of EAE-induced inflammation. This finding was supported by data showing that antagonizing BAFF-dependent B-cell stimulation can be achieved by the provision of the NgR-Fc and NgR3-P fusion proteins, blocking B-cell proliferation. A previous study has demonstrated that NgR1 and NgR3, co-expressed on 293 T cells, can collaborate in the same receptor complex51. In CNS tissue, neurons have been shown to upregulate the mRNA expression of NgR3 during kainic acid treatment52, with chondroitin sulphate proteoglycans (CSPGs) able to ligate to NgR3 to regulate neurite outgrowth inhibition51. Importantly, it has been demonstrated that ablation of the putative CSPG receptor type protein tyrosine phosphatase (Rptpσ) can enhance axonal regeneration following optic nerve crush injury51. Although the role of NgR3 has not been defined in B-cells, Rptp (CD148) signaling has been demonstrated to regulate B-cell and macrophage lineage cell differentiation53. Moreover, CSPGs derived from either T-cells or reactive astrocytes are capable of stimulating B-cell differentiation54 and may play a role for antigen presentation during autoimmunity55 or even capable of binding APRIL to regulate B-cell maturation22. These data raise the tantalizing hypothesis that NgR1 and NgR3 may collaborate with various receptors in multimeric complexes regulating signal transduction cascades for B-cell activation. This is of particular importance in MS since the histopathological observations have demonstrated that B-cells are indeed presented in follicles at the meninges of MS patients47,56 and may regulate progressive neurodegeneration57.
The question whether or not NgR expression in B-cells and downstream signaling govern the formation of follicular-like structures in the CNS during progressive MS requires elucidation since our current findings implicate NgR1 and NgR3 as collaborative players in the induction of EAE. Although the exact mechanism by which B-cells contribute to the cellular and humoral responses that govern MS pathogenesis is still not fully understood, B-cell-targeted therapies have shown promising outcomes in clinical trials3,58 and so if the expression of NgR1 and NgR3 can be therapeutically targeted, then novel pathways for modifying disease specific B-cells may have been identified considering our current data. However, recent novel data generated in the EAE model of MS show that IgA generated from plasma cells localized to the CNS are derived from the gut58. This study has intriguingly demonstrated that removal of this population of IgA + plasma cells exacerbated EAE implicating their neuroprotective nature driven by IL10. Specifically, this study demonstrated that BAFF transgenic mice that overexpress BAFF and exhibit increased levels of circulating IgA, were resistant to the induction of MOG35-55 and recombinant human EAE59. These authors concluded that the Transgenic overexpressing BAFF studies along with the observations from TACI knock out mice (Tnfrsf13b-/-), may suggest that BAFF signaling through TACI provides a protective effect to limit immune-mediated demyelination through the maintenance of IgA + plasma cells in the gut and the CNS. Our data would support the notion that at peak stage of EAE we did not demonstrate high levels of IgA but only IgM and IgG, that we demonstrated could be generated from NgR1 + and NgR3 + B cells grown in vivo. Whether these Nogo Receptor expressing B cells can eventually derive BAFF-responsive IgA + Plasma cells remains to be determined.
With the emerging view that BAFF is a known stimulator of B-cell activation18,19,20, our investigations identifying the BAFF responsiveness of NgR1+ and NgR3+ B-cells during EAE induction, raises key questions surrounding the pathophysiological role of these receptors. Of particular importance is that BAFF has been highlighted as a potential target in MS because it is produced endogenously in the CNS by astrocytes during the pathogenesis and it is also associated with BAFF-R expressing cells, which are up-regulated within the meninges in ectopic lymphoid follicles16,60. Whether NgR1 and NgR3 can amplify the signaling of BAFF in B-cells expressing these receptors is yet to be established. However, our data has identified that we can block the B-cell proliferative response by antagonizing BAFF with recombinant NgR peptides implicating a signaling role.
It has been identified that BAFF can act as an alternate ligand for NgR1 to inhibit DRG neurite outgrowth in vitro21. However, our study is the first to demonstrate that there exists a BAFF-dependent B-cell pathophysiological response that is driven through NgR1 and NgR3. Our finding that BAFF can stimulate spinal cord-specific NgR1+ and NgR3+ B-cells to proliferate, confirm a signaling role for these receptors during neuroinflammation. Importantly, without BAFF stimulation these NgR1+ and NgR3+ B-cells were also able to generate IgG antibodies either expressed on the surface (defining maturation through class switching), or secreted as IgG myelin-specific antibodies. Whether these autoantibodies are capable of promulgating neurodegeneration or targeted deletion of NgR1+ and NgR3+ B-cells during neuroinflammation can ameliorate disease progression requires further investigation.
The current study has identified for the first time that both NgR1 and NgR3 are active contributors to B-cell maturation during EAE and may participate in the signal transduction events elicited through BAFF ligation that may enhance proliferation of these cells intrathecally. Although the direct signal transduction events within these B-cells are yet to be elucidated, the responsiveness of the NgR1+ and NgR3+ B-cells may be a major pathogenic event governing the cycle of neuroinflammation. The pathobiological consequences of myelin-specific immunoglobulins generated by this population of B-cells may be of great significance during the epitope spreading effects of EAE-induced neurodegeneration and by extension MS, raising the possibility of novel B-cell therapeutics targeting NgR1 and NgR3.
Materials and methods
Mouse breeding
All experiments were implemented in accordance with the guidelines of the Australian code of practice for the use and care of animals for scientific purposes and approved by the Monash University Animal Ethic Committee (Alfred Medical Research and Education Precinct; AMREP) and office of the Gene Technology Regulator of Australia (AEC AMREP Approval number E/1532/2015/M). Female ngr1−/− mice (aged 8–12 weeks) and their ngr1+/+ littermates were bred and maintained at the AMREP animal services. Exon 2 of the ngr1 gene was a double-targeted allele on a C57Bl/6 background and backcrossed more than 10 generations as mentioned previously61.
EAE induction and clinical assessment
In accordance with the ARRIVE 2.0 guidelines, this original study compared EAE clinical scores, immune cell profiles and histopathological outcomes in 8–12 weeks old C57Bl/6 (ngr1+/+ or ngr1−/−) female mice with or without MOG35-55-induction using a blinded and randomised study design. To induce EAE, 8–12 weeks old C57Bl/6 (ngr1+/+ or ngr1−/−) female mice were immunized with either 200 μg MOG35-55 (Purar Chemical, China) or full-length recombinant mouse MOG (AnaSpec, USA) as previously described30,31,62. Mice with complete Freund’s adjuvant but without MOG peptide (CFA only), served as non-specific immunization controls. All EAE animals were examined for 28 consecutive days using a six-grade score scale consistent with well documented disease stages as previously described63,64: 0, asymptomatic; 1, partial loss of tail tonicity; 2, flaccid tail paralysis; 3, difficulty to roll over from a supine position; 4, hindlimb paralysis; 5, forelimb paresis. Animals without a score were excluded from further neuropathological analysis. The disease stages classically included: pre-onset (0–10 days post immunization (dpi); score 0), onset (10–20 dpi; mean score 1), peak (20–30 dpi; mean score 3). The following indexes were also calculated: Mean day of disease onset (dDO), mean maximal score (MMS) and area under the curve (AUC), which cumulatively measure the extent and burden of the disease. All groups of ngr1+/+ and ngr1−/− mice experimental groups were coded to perform a blinded study, to limit observer bias when assessing EAE clinical score. The codes and data were stored electronically and as hard copies by the senior investigator (SP) and then decoded upon collection of all data synthesized for analysis.
Tissue processing
Mice were euthanized at various stages of EAE via CO2 inhalation according to ethical guidelines. Spleens and spinal cords were dissected to isolate immune cells for immune-phenotyping analysis. Otherwise, an equal amount of 4% paraformaldehyde (PFA) fixative was injected into the left ventricle and tissues were then processed for immunohistochemistry. Longitudinal spleen and spinal cord sections were cut into 10 μm serial sections and processed for immunohistochemistry.
Isolation of immune cells and myelin from mouse tissues
Spleen isolation
B- and T-cells were isolated from the spleens of EAE-induced and non-immunized ngr1+/+ and ngr1−/− mice. Tissues were mashed by passing through a 40 μm nylon filter, red blood cells were lysed with red blood cell lysis buffer (Sigma-Aldrich) and the remaining cells were washed in D-PBS and subjected to flow cytometry analysis.
Spinal cord isolation
Spinal cord tissue was chopped into fine pieces and then enzymatically digested at 37 °C for 30 min with collagenase D (Roche)/Dnase I (Roche) in PBS. These were then subjected to Ficoll density gradient (Stem Cell Technologies). The gradient interface was collected upon centrifugation at 800 g max for 30 min at 20 °C. The remaining cells were analysed by flow cytometry analysis.
Myelin isolation
Purification of central nervous system myelin was performed as described previously65. Lumbosacral spinal cord was obtained from both ngr1+/+ and ngr1−/−, immunized with MOG35-55 and non-immunized mice. Fresh frozen spinal cord (immersed in liquid N2) was then grounded to a fine powder in liquid N2 and homogenized in 0.29 mol/L of sucrose solution (5% w/v). After preparing a discontinuous sucrose gradient by layering aliquots of the homogenate over of 0.85 mol/L sucrose, this was then spun for 45 min at 90,000 g max. The interface, including crude whole myelin, was collected and diluted 1:1 with 0.24 mol/L sucrose. Then this was spun for 30 min at 17,000 g max to collect the pellet, which was resuspended in dH2O (osmotic shock), and re-spun again for 30 min at 17,000 g max. A second discontinuous sucrose gradient was performed for further myelin purification, and to remove any immunoglobulin contamination that may provide a false positive signal. The purified myelin was treated with diethylamine-HCl (50 mmol/L, pH = 11.5), then pelleted in a microfuge, and thoroughly washed with dH2O to become lyophilized.
Flow cytometry analysis
Isolated cells were analyzed using flow cytometry to determine whether there existed disparate populations within disease-originating organ sites. Briefly, cells were washed with 1 mL FACS wash buffer [0.02% sodium azide, 1% bovine serum albumin (BSA) in PBS], centrifuged (3x) at 1500 rpm for 5 min. Mouse immune cell antigens were detected using the following fluorescently labeled antibodies (for a full list of antibodies used in all experiments see Additional File 4: Table S1: CD45/B220-PE (BD Biosciences) and its isotype control (BD Biosciences; PE labeled rat IgG 2b kappa); CD3e-Pe-Cy7 (eBioscience); CD11c-APC (BD Bioscience) and its isotype control (eBioscience; PE-Cy7 labeled hamster IgG); NgR1 (R&D systems) and its isotype control (BD Pharmingen; rat IgG 2b kappa); NgR2 (Santa Cruz Biotechnology); NgR3 (Santa Cruz Biotechnology); IgM-PerCP/Cy5.5 (BioLegend); IgD-APC (BioLegend), IgG-Biotin (eBioscience); Streptavidin APC (eBioscience): and Ki-67-eFluor 450 (eBioscience).
In addition to the single staining, each immune cell antigen was stained with the NgR homologs, in order to determine its co-expression on these isolated immune cells. Briefly, normal rat serum (Life Technologies) was added into all tubes to block non-specific binding and incubated at 4 °C for 30 min. The cells were then washed (3x) and centrifuged at 1500 rpm for 5 min at 4 °C, followed by the secondary antibody staining (Alexa fluor 488 goat anti-rat IgG (H + L); Life Technologies) at 4 °C for 30 min. Stained cells were washed and DAPI was added into each sample to label dead cells for 10 min before analysis. Finally, all tubes were analyzed by using the BD FACS Canto II (BD Biosciences).
Immunohistochemistry
Paraffin sections
Fixed (4% paraformaldehyde – PFA), paraffin-embedded tissues were cut into 10 μm serial sections on a conventional microtome and processed for immunohistochemistry. The sections were de-waxed and incubated with proteinase K (20 μg/mL) (Qiagen) for 1 h at 37 °C, then washed (3 × 10 min) with PBS. All sections were post-fixed with 4% PFA for 30 min at room temperature, followed by three washes with PBS. Tissues were then incubated with blocking buffer (5% goat serum, 0.1% triton X-100 in PBS) at 4 °C overnight. All sections were incubated with primary antibodies: anti-BAFFR – Biotinylated (R&D Systems); anti-CD19 (Alexa Fluor 594) (BioLegend); anti-CD45/B220 (BIOSS); anti-BLyS/BAFF (Novus Biologicals); anti-CD268/BAFF Receptor (eBioscience); anti-NgR1 (R&D systems and/or Millipore); anti-NgR2 (Santa Cruz Biotechnology); and anti-NgR3 (Santa Cruz Biotechnology) for overnight at 4 °C, followed by washing (3x) in PBS for 10 min. Appropriate secondary antibodies (Alexa Fluor 488 goat anti-rabbit IgG (H + L) antibody; Life technologies, Alexa Fluor 488 goat anti-rat IgG (H + L) antibody; Life technologies, Alexa Fluor 555 goat anti-rabbit IgG (H + L) antibody; Life technologies, Alexa Fluor 555 goat anti-rat IgG (H + L) antibody; Life technologies, Donkey anti-Goat 488 antibody; Life technologies, Donkey anti-Goat 555 antibody; Life technologies) were applied, and incubated for 2 h at room temperature. Streptavidin (BD Pharmingen) was added to the appropriate slides for 1 h at RT. Finally, DAPI was added after washing in PBS for 15 min and slides were cover slipped using fluorescent mounting medium (Dako). Subsequently, all tissue specimens were captured by the four-channel fluorescence imaging, using a confocal Nikon C1 upright microscope with 40 × and 60 × UPlanApo objective lens.
Frozen sections
OCT-embedded tissue blocks were cut into 10 μm serial sections on a cryostat (Leica Microsystems). The sections were post-fixed with 4% PFA for 30 min at room temperature, followed by three washes with PBS. Tissues were then incubated in the blocking buffer for 1 h at room temperature. All sections were incubated with primary and secondary antibodies, followed by DAPI, as indicated above. These were then scanned under an inverted confocal microscope (described above).
Quantification of NgRs and B-cell populations in the white matter tracts of the lumbar and sacral spinal cord
Analysis of cell numbers localized within specific tissue areas were performed on all spinal cord sections of both ngr1+/+and ngr1−/− mice following immunohistochemistry. The number of double stained cells (NgR that is expressed on B-cells) was counted per unit area (per μm2) of spinal cord longitudinal-sections in all genotypes (3 different sections per mouse > 50 μm apart). The mean and standard error of the mean (SEM) was calculated for each group of mice at different clinical stages of the disease.
Demyelination and axonal degeneration
Post-tissue processing, Luxol fast blue/Periodic acid-Schiff (LFB/PAS) and Bielschowsky staining were used on spinal cord sections of ngr1+/+ and ngr1−/− mice at different clinical scores (Histology platform, Monash University) in order to demonstrate areas of demyelination and axonal injury in EAE-induced mice, respectively.
LFB/PAS was performed by dehydrating the tissue sections using 95% ethanol, then placed in Low Viscosity Nitrocellulose (LVN) for 1 min. The sections were then placed into 0.1% LFB solution (w/v) overnight at 40 °C incubator (cover staining dish in cling wrap to avoid excessive evaporation of the solution). Excess stain was rinsed off in 70% ethanol then washed in dH2O. The sections were briefly dipped in dilute lithium carbonate (0.5 mg/mL) then differentiated in 50–70% ethanol. Immediately, all sections were washed in dH2O (to stop the differentiation but if it over differentiated, were placed back into LFB solution for 5–6 h or overnight). The sections were counterstained with PAS for 5 s, then dehydrated rapidly through ethanol into xylene, then mounted in DPX.
Bielschowsky staining were performed by adding sections into 20% silver nitrate (w/v), incubated for 20 min within a 40 °C incubator. All sections were then washed with dH2O and returned into silver nitrate again. Ammonium hydroxide was added drop-by-drop until the precipitate formed had completely dissolved. Then drops of developer stock solution (37–40% formaldehyde, 0.5 g of citric acid (5 mg/mL), 2 drops of 20% nitric acid, and 100 mL of dH2O) were added to 20% of ammoniacal silver, then reduced to a visible metallic silver. The sections were dehydrated rapidly through ethanol and then xylene.
Protein biochemistry
Preparation of cell and tissue lysate for western blotting
Spleen and spinal cord samples were ground and lysed from both naïve ngr1+/+ and ngr1−/− mice using radioimmunoprecipitation assay (RIPA) buffer (Cell Signaling Technology), including PhosSTOP phosphatase inhibitors cocktail (Roche Applied Science) and 1% protease inhibitor (v/v) (Sigma-Aldrich) in a dounce homogenizer. The protein supernatants were collected from homogenates, after centrifuging them at 13,000 g max for 15 min at 4 °C. Protein concentrations were determined by using the bicinchoninic acid (BCA) protein assay kit (Thermo Fisher Scientific).
Western immunoblotting
10 μg of protein isolated from spleen, spinal cord and myelin samples were loaded into 4–12% graded sodium dodecyl sulphate polyacrylamide gels (SDS-PAGE) (Invitrogen). The proteins were blocked with 5% skim milk powder (w/v) in Tris-buffered saline-Tween (TBST) for 30 min at room temperature after transferring onto polyvinylidene fluoride (PVDF) membranes. The membranes were then incubated with primary anti-NgR1 antibody (Millipore and/or R&D systems), anti-NgR3 antibody (Santa Cruz Biotechnology), anti-Nogo-A (Merck-Millipore) and anti-myelin basic protein (MBP) antibody (Sapphire Bioscience) in 5% skim milk (50 mg/mL) at 4 °C, overnight. The membranes were then washed (3x) with 0.1% TBST for 10 min, followed by adding the secondary anti-rabbit antibody (HRP-conjugated; Merck-Millipore), anti-rat antibody (HRP-conjugated; Merck-Millipore), and anti-mouse antibody (HRP-conjugated; Merck-Millipore) diluted in 5% skim milk or TBST and incubated for 2 h at room temperature; washed (3x) in 0.1% TBST (v/v). The membranes were then incubated with 2 mL of Luminata Forte Western Reagent (Millipore) or Luminata ECL chemiluminescence (Merck-Millipore) for 3–5 min at room temperature for development. Using the Alpha Imager (Alpha Innotech, San Leandro), the films were scanned, and the intensities of the bands were determined using ImageQuant TL v2003 software (Nonlinear Dynamics Ltd, All Saints, Newcastle, UK). Some of the membranes were stained with coomassie blue (8 mg/mL) dye for 30 min to 1 h, and they were scanned after de-staining.
Cell culture
Immune cells were isolated from spleen and spinal cords of EAE mice, as previously described30. The cells were suspended into 2 mL FACS buffer (2% heat inactivated fetal calf serum (FCS) and DPBS) in 5 mL polypropylene round bottom tubes, then stained using only anti-CD45/B220-PE antibodies and either anti-NgR1 or anti-NgR3 antibodies. The double-positive cells were stained with DAPI, then sorted using either BD FACS ARIA or BD Influx sorter. The sorted cells were collected and cultured for 2–3 days in RPMI medium supplemented with 10% heat inactivated FCS (Clontech), 50 μM β-mercaptoethanol, L-glutamine (2 mM), 100 U/ml penicillin, and 100 U/ml streptomycin (Gibco) at 37 °C with 5% CO2 in a humidified incubator.
Cell cycle analysis
In order to determine the ability of BAFF to stimulate B-cells expressing NgR1 or NgR3 in the spleen and spinal cord of ngr1+/+ mice (n = 12) at disease onset, these isolated cells were analyzed with or without BAFF (50 ng/mL) in vitro. Briefly, 50 ng/mL of soluble recombinant mouse BAFF (rBAFF; Enzo) was added into all B-cell cultures and incubated for 24 h at 37 °C in a 5% CO2 humidified incubator. To identify NgR interactions with BAFF, rBAFF was blocked with an excess amount of either rBAFF-R, NgR1-Fc or NgR3-P by incubating them for 30 min at 37 °C. The supernatant was collected after spinning for 5 min with 16,000 g max and added to cultured cells for 24 h. 100 μL of supernatant was collected for all samples before and after stimulation with rBAFF, or following inhibition of rBAFF activity with either NgR1-Fc or NgR3-P peptides. The following day, the cell cycle was tested using a BrdU APC flow kit (BD Bioscience). First, 1 mM solution of BrdU was added for 1 h to cultured and treated cells. Second, BrdU-pulsed cells were transferred into FACS tubes and their counts were measured. Antibodies (CD45/B220, NgR and NgR3) were used to stain cells, described above. Cells were fixed and permeabilized with a BD Cytofix/Cytoperm Buffer for 20 min on ice, then washed with 1X BD Perm/Wash Buffer and spun for 5 min at 300 g max. Then the supernatant was discarded. Cells were next incubated with BD Cytoperm permeabilization buffer Plus for 10 min on ice and washed with 1X BD Perm/Wash Buffer again. Cells were re-fixed with BD Cytofix/Cytoperm buffer for 5 min on ice, then washed. Dnase was added into treated cells and incubated for 1 h at 37 °C, then washed with 1X BD Perm/Wash buffer. Finally, cells were stained using an anti-BrdU antibody for 20 min. These were stained with total DNA for cell cycle analysis, 7-aminoactinomycin D (7AAD), after washing it with 1X BD Perm/Wash buffer. Stained cell counts were acquired from the BD LSR II flow cytometer (BD Biosciences).
Enzyme-linked immunosorbent assay (ELISA)
Supernatant was collected from FAC sorted and cultured cells of ngr1+/+ and ngr1−/− mice (at clinical score 1 and 3) and then the immunoglobulin class was measured by Enzyme-linked immunosorbent assay (ELISA) with the supernatant diluted as follows: 1:10, 1:100 and 1:1000 for anti-IgG, -IgM and -IgD AP conjugated secondary antibodies (Southern Biotech). The 384-well assay plate (Corning) was coated with 20 µL/well of 2 µg/mL coating reagent (goat anti-mouse Ig (H + L)) (Southern Biotech) and incubated overnight at 4 °C. The plate was washed (2x) with wash buffer (0.1% Tween-20 (w/v), PBS) and (1x) with PBS. 100 µL/well of Block buffer (4% BSA (w/v), PBS) was added and incubated 30 min at room temperature, followed by two washes with wash buffer and one wash with PBS. 40 µL of diluted samples (supernatant collected from sorted and cultured cells) or control antibodies in a dilution buffer (0.5% (w/v) BSA, PBS) were added by serial dilution into the plate into serial dilution (1:10, 1:100 and 1:1000) for the supernatant samples and (1:2) for all control antibodies (Southern Biotech). At the same time, 20 µL of a dilution buffer was added to other wells. Then the plate was incubated for 30 min at room temperature on the plate shaker, followed by two washes with wash buffer and one wash with PBS. 20 µL/well of secondary antibody (diluted 1:2000 in a dilution buffer) (Southern Biotech) was added and incubated for 30 min at room temperature on the plate shaker. Then the plate was washed (4x) with wash buffer and (2x) with PBS. 20 µL/well of 1 mg/mL substrate solution (PNPP tablets dissolved in M.H2O; one 20 mg tablet per 20 mL M.H2O) (Sigma). Once colorimetric development was achieved, the absorbance measurement was performed at 405 nm using a microtitre plate reader (Bio-Rad Laboratories, CA, USA).
Statistical analysis
Statistical analysis was carried out using GraphPad Prism 8 software. Values were expressed as mean ± standard error of the mean (SEM). Normality was assessed by D’Agostino–Pearson test. Student’s t test and One-way ANOVA followed by Bonferroni post-hoc correction, were used for comparisons of 2 or more groups, respectively. In case of non-parametric data, Mann–Whitney or the Kruskall–Wallis test followed by Dunn’s multiple comparison were appropriately applied. Two-way repeated measures ANOVA was used to compare EAE progression in ngr1+/+ and ngr1−/− mice. p < 0.05 was the cut-off level of significance.
References
Dendrou, C. A., Fugger, L. & Friese, M. A. Immunopathology of multiple sclerosis. Nat. Rev. Immunol. 15, 545–558. https://doi.org/10.1038/nri3871 (2015).
Li, H., Hu, F., Zhang, Y. & Li, K. Comparative efficacy and acceptability of disease-modifying therapies in patients with relapsing-remitting multiple sclerosis: a systematic review and network meta-analysis. J. Neurol. https://doi.org/10.1007/s00415-019-09395-w (2019).
Kappos, L. et al. Ocrelizumab in relapsing-remitting multiple sclerosis: a phase 2, randomised, placebo-controlled, multicentre trial. Lancet 378, 1779–1787. https://doi.org/10.1016/S0140-6736(11)61649-8 (2011).
Hauser, S. L., Belachew, S. & Kappos, L. Ocrelizumab in primary progressive and relapsing multiple sclerosis. N. Engl. J. Med. 376, 1694. https://doi.org/10.1056/NEJMc1702076 (2017).
Montalban, X. et al. Ocrelizumab versus placebo in primary progressive multiple sclerosis. N. Engl. J. Med. 376, 209–220. https://doi.org/10.1056/NEJMoa1606468 (2017).
Fitzner, D. & Simons, M. Chronic progressive multiple sclerosis—pathogenesis of neurodegeneration and therapeutic strategies. Curr. Neuropharmacol. 8, 305–315 (2010).
Frischer, J. M. et al. The relation between inflammation and neurodegeneration in multiple sclerosis brains. Brain 132, 1175–1189. https://doi.org/10.1093/brain/awp070 (2009).
Palanichamy, A. et al. Immunoglobulin class-switched B cells form an active immune axis between CNS and periphery in multiple sclerosis. Sci. Transl. Med. 6, 248ra106. https://doi.org/10.1126/scitranslmed.3008930 (2014).
Stern, J. N. et al. B cells populating the multiple sclerosis brain mature in the draining cervical lymph nodes. Sci. Transl. Med. 6, 248ra107. https://doi.org/10.1126/scitranslmed.3008879 (2014).
Obermeier, B. et al. Related B cell clones that populate the CSF and CNS of patients with multiple sclerosis produce CSF immunoglobulin. J. Neuroimmunol. 233, 245–248 (2011).
Molnarfi, N. et al. MHC class II-dependent B cell APC function is required for induction of CNS autoimmunity independent of myelin-specific antibodies. J. Exp. Med. 210, 2921–2937 (2013).
Barr, T. A. et al. B cell depletion therapy ameliorates autoimmune disease through ablation of IL-6-producing B cells. J. Exp. Med. 209, 1001–1010 (2012).
Meinl, E., Krumbholz, M. & Hohlfeld, R. B lineage cells in the inflammatory central nervous system environment: migration, maintenance, local antibody production, and therapeutic modulation. Ann. Neurol. 59, 880–892 (2006).
Lisak, R. P. et al. Secretory products of multiple sclerosis B cells are cytotoxic to oligodendroglia in vitro. J. Neuroimmunol. 246, 85–95 (2012).
Kappos, L. et al. Atacicept in multiple sclerosis (ATAMS): a randomised, placebo-controlled, double-blind, phase 2 trial. Lancet Neurol. 13, 353–363. https://doi.org/10.1016/S1474-4422(14)70028-6 (2014).
Krumbholz, M. et al. BAFF is produced by astrocytes and up-regulated in multiple sclerosis lesions and primary central nervous system lymphoma. J. Exp. Med. 201, 195–200. https://doi.org/10.1084/jem.20041674 (2005).
Mackay, F. & Schneider, P. Cracking the BAFF code. Nat. Rev. Immunol. 9, 491–502 (2009).
Mackay, F. et al. Mice transgenic for BAFF develop lymphocytic disorders along with autoimmune manifestations. J. Exp. Med. 190, 1697–1710. https://doi.org/10.1084/jem.190.11.1697 (1999).
Schneider, P. et al. BAFF, a novel ligand of the tumor necrosis factor family, stimulates B cell growth. J. Exp. Med. 189, 1747–1756. https://doi.org/10.1084/jem.189.11.1747 (1999).
Thompson, J. S. et al. BAFF binds to the tumor necrosis factor receptor-like molecule B cell maturation antigen and is important for maintaining the peripheral B cell population. J. Exp. Med. 192, 129–135. https://doi.org/10.1084/jem.192.1.129 (2000).
Zhang, L. et al. Identification of BLyS (B Lymphocyte Stimulator), a non-myelin-associated protein, as a functional ligand for Nogo-66 receptor. J. Neurosci. 29, 6348–6352 (2009).
Baert, L. et al. A proliferation-inducing ligand-mediated anti-inflammatory response of astrocytes in multiple sclerosis. Ann. Neurol. 85, 406–420. https://doi.org/10.1002/ana.25415 (2019).
Vincent, F. B., Saulep-Easton, D., Figgett, W. A., Fairfax, K. A. & Mackay, F. The BAFF/APRIL system: emerging functions beyond B cell biology and autoimmunity. Cytokine Growth Factor Rev. 24, 203–215. https://doi.org/10.1016/j.cytogfr.2013.04.003 (2013).
Magliozzi, R. et al. Inflammatory intrathecal profiles and cortical damage in multiple sclerosis. Ann. Neurol. 83, 739–755. https://doi.org/10.1002/ana.25197 (2018).
Puthenparampil, M. et al. BAFF is decreased in the cerebrospinal fluid of multiple sclerosis at clinical onset. J. Neuroimmunol. 297, 63–67. https://doi.org/10.1016/j.jneuroim.2016.05.013 (2016).
Miyazaki, Y. et al. Fingolimod induces BAFF and expands circulating transitional B cells without activating memory B cells and plasma cells in multiple sclerosis. Clin. Immunol. 187, 95–101. https://doi.org/10.1016/j.clim.2017.10.009 (2018).
Kawakami, Y., Saito, Y., Nakagawa, R., Kurihara, Y. & Takei, K. The soluble form of LOTUS inhibits Nogo receptor type 1-mediated signaling induced by B lymphocyte stimulator and chondroitin sulfate proteoglycans. Neurosci. Lett. 683, 61–68. https://doi.org/10.1016/j.neulet.2018.06.045 (2018).
Kawakami, Y. et al. The soluble form of LOTUS inhibits nogo receptor-mediated signaling by interfering with the interaction between nogo receptor type 1 and p75 neurotrophin receptor. J. Neurosci. 38, 2589–2604. https://doi.org/10.1523/JNEUROSCI.0953-17.2018 (2018).
Karnezis, T. et al. The neurite outgrowth inhibitor Nogo A is involved in autoimmune-mediated demyelination. Nat. Neurosci. 7, 736–744. https://doi.org/10.1038/nn1261 (2004).
Petratos, S. et al. Limiting multiple sclerosis related axonopathy by blocking Nogo receptor and CRMP-2 phosphorylation. Brain 135, 1794–1818. https://doi.org/10.1093/brain/aws100 (2012).
Lee, J. Y. et al. Limiting neuronal nogo receptor 1 signaling during experimental autoimmune encephalomyelitis preserves axonal transport and abrogates inflammatory demyelination. J. Neurosci. 39, 5562–5580. https://doi.org/10.1523/JNEUROSCI.1760-18.2019 (2019).
Tada, S. et al. BAFF controls neural cell survival through BAFF receptor. PLoS ONE 8, e70924. https://doi.org/10.1371/journal.pone.0070924 (2013).
Lee, J. Y. & Petratos, S. Multiple sclerosis: does Nogo play a role?. Neurosci. Rev. J. Bring. Neurobiol. Neurol. Psychiatry 19, 394–408. https://doi.org/10.1177/1073858413477207 (2013).
Baer, A. S. et al. Myelin-mediated inhibition of oligodendrocyte precursor differentiation can be overcome by pharmacological modulation of Fyn-RhoA and protein kinase C signalling. Brain 132, 465–481. https://doi.org/10.1093/brain/awn334 (2009).
Robinson, S. & Miller, R. H. Contact with central nervous system myelin inhibits oligodendrocyte progenitor maturation. Dev. Biol. 216, 359–368. https://doi.org/10.1006/dbio.1999.9466 (1999).
Syed, Y. A. et al. Inhibition of oligodendrocyte precursor cell differentiation by myelin-associated proteins. Neurosurg. Focus 24, E5. https://doi.org/10.3171/foc/2008/24/3-4/e4 (2008).
Niederöst, B., Oertle, T., Fritsche, J., McKinney, R. A. & Bandtlow, C. E. Nogo-A and myelin-associated glycoprotein mediate neurite growth inhibition by antagonistic regulation of RhoA and Rac1. J. Neurosci. Off. J. Soc. Neurosci. 22, 10368–10376. https://doi.org/10.1523/jneurosci.22-23-10368.2002 (2002).
Petratos, S., Azari, M. F., Ozturk, E., Papadopoulos, R. & Bernard, C. C. Novel therapeutic targets for axonal degeneration in multiple sclerosis. J. Neuropathol. Exp. Neurol. 69, 323–334. https://doi.org/10.1097/NEN.0b013e3181d60ddb (2010).
Meabon, J. S. et al. LINGO-1 protein interacts with the p75 neurotrophin receptor in intracellular membrane compartments. J. Biol. Chem. 290, 9511–9520. https://doi.org/10.1074/jbc.M114.608018 (2015).
Fournier, A. E., GrandPre, T. & Strittmatter, S. M. Identification of a receptor mediating Nogo-66 inhibition of axonal regeneration. Nature 409, 341–346. https://doi.org/10.1038/35053072 (2001).
Mi, S. et al. LINGO-1 is a component of the Nogo-66 receptor/p75 signaling complex. Nat. Neurosci. 7, 221–228. https://doi.org/10.1038/nn1188 (2004).
Wang, K. C., Kim, J. A., Sivasankaran, R., Segal, R. & He, Z. P75 interacts with the Nogo receptor as a co-receptor for Nogo, MAG and OMgp. Nature 420, 74–78. https://doi.org/10.1038/nature01176 (2002).
Lyons, J. A., Ramsbottom, M. J. & Cross, A. H. Critical role of antigen-specific antibody in experimental autoimmune encephalomyelitis induced by recombinant myelin oligodendrocyte glycoprotein. Eur. J. Immunol. 32, 1905–1913 (2002).
Oliver, A. R., Lyon, G. M. & Ruddle, N. H. Rat and human myelin oligodendrocyte glycoproteins induce experimental autoimmune encephalomyelitis by different mechanisms in C57BL/6 mice. J. Immunol. 171, 462–468. https://doi.org/10.4049/jimmunol.171.1.462 (2003).
Marta, C. B., Oliver, A. R., Sweet, R. A., Pfeiffer, S. E. & Ruddle, N. H. Pathogenic myelin oligodendrocyte glycoprotein antibodies recognize glycosylated epitopes and perturb oligodendrocyte physiology. Proc. Natl. Acad. Sci. U. S. A. 102, 13992–13997. https://doi.org/10.1073/pnas.0504979102 (2005).
Howell, O. W. et al. Meningeal inflammation is widespread and linked to cortical pathology in multiple sclerosis. Brain 134, 2755–2771. https://doi.org/10.1093/brain/awr182 (2011).
Wicken, C., Nguyen, J., Karna, R. & Bhargava, P. Leptomeningeal inflammation in multiple sclerosis: Insights from animal and human studies. Mult. Scler. Relat. Disord. 26, 173–182. https://doi.org/10.1016/j.msard.2018.09.025 (2018).
Bhargava, P. et al. Trial of intrathecal rituximab in progressive multiple sclerosis patients with evidence of leptomeningeal contrast enhancement. Mult. Scler. Relat. Disord. 30, 136–140. https://doi.org/10.1016/j.msard.2019.02.013 (2019).
Silk, M. & Nantz, E. Efficacy and Safety of Tabalumab in Patients with Relapsing-Remitting Multiple Sclerosis: A Randomized, Double-Blind, Placebo-Controlled Study (P3.397). Neurology 90 3.397 (2018).
Baker, D., Pryce, G., James, L. K., Schmierer, K. & Giovannoni, G. Failed B cell survival factor trials support the importance of memory B cells in multiple sclerosis. Eur. J. Neurol. 27, 221–228. https://doi.org/10.1111/ene.14105 (2020).
Dickendesher, T. L. et al. NgR1 and NgR3 are receptors for chondroitin sulfate proteoglycans. Nat. Neurosci. 15, 703–712. https://doi.org/10.1038/nn.3070 (2012).
Karlsson, T. E., Wellfelt, K. & Olson, L. Spatiotemporal and long lasting modulation of 11 key nogo signaling genes in response to strong neuroexcitation. Front. Mol. Neurosci. 10, 94. https://doi.org/10.3389/fnmol.2017.00094 (2017).
Zhu, J. W., Brdicka, T., Katsumoto, T. R., Lin, J. & Weiss, A. Structurally distinct phosphatases CD45 and CD148 both regulate B cell and macrophage immunoreceptor signaling. Immunity 28, 183–196. https://doi.org/10.1016/j.immuni.2007.11.024 (2008).
Levitt, D. & Olmstead, L. B-cell stimulation by T-cell-secreted proteoglycan. Cell. Immunol. 110, 425–430. https://doi.org/10.1016/0008-8749(87)90134-1 (1987).
Ciechomska, M. et al. Antigen-specific B lymphocytes acquire proteoglycan aggrecan from cartilage extracellular matrix resulting in antigen presentation and CD4+ T-cell activation. Immunology 141, 70–78. https://doi.org/10.1111/imm.12169 (2014).
Serafini, B., Rosicarelli, B., Magliozzi, R., Stigliano, E. & Aloisi, F. Detection of ectopic B-cell follicles with germinal centers in the meninges of patients with secondary progressive multiple sclerosis. Brain Pathol. 14, 164–174 (2004).
Magliozzi, R. et al. Meningeal B-cell follicles in secondary progressive multiple sclerosis associate with early onset of disease and severe cortical pathology. Brain 130, 1089–1104. https://doi.org/10.1093/brain/awm038 (2007).
Hauser, S. L. et al. B-cell depletion with rituximab in relapsing-remitting multiple sclerosis. N. Engl. J. Med. 358, 676–688. https://doi.org/10.1056/NEJMoa0706383 (2008).
Rojas, O. L. et al. Recirculating Intestinal IgA-Producing Cells Regulate Neuroinflammation via IL-10. Cell 176, 610-624.e618. https://doi.org/10.1016/j.cell.2018.11.035 (2019).
Magliozzi, R., Columba-Cabezas, S., Serafini, B. & Aloisi, F. Intracerebral expression of CXCL13 and BAFF is accompanied by formation of lymphoid follicle-like structures in the meninges of mice with relapsing experimental autoimmune encephalomyelitis. J Neuroimmunol 148, 11–23. https://doi.org/10.1016/j.jneuroim.2003.10.056 (2004).
Kim, J. E., Liu, B. P., Park, J. H. & Strittmatter, S. M. Nogo-66 receptor prevents raphespinal and rubrospinal axon regeneration and limits functional recovery from spinal cord injury. Neuron 44, 439–451. https://doi.org/10.1016/j.neuron.2004.10.015 (2004).
Theotokis, P. et al. Time course and spatial profile of Nogo-A expression in experimental autoimmune encephalomyelitis in C57BL/6 mice. J. Neuropathol. Exp. Neurol. 71, 907–920. https://doi.org/10.1097/NEN.0b013e31826caebe (2012).
Theotokis, P. et al. Nogo receptor complex expression dynamics in the inflammatory foci of central nervous system experimental autoimmune demyelination. J. Neuroinflammation 13, 265. https://doi.org/10.1186/s12974-016-0730-4 (2016).
Theotokis, P. et al. Connexin43 and connexin47 alterations after neural precursor cells transplantation in experimental autoimmune encephalomyelitis. Glia 63, 1772–1783. https://doi.org/10.1002/glia.22843 (2015).
Geczy, C. L., Roberts, I. M., Meyer, P. & Bernard, C. C. Susceptibility and resistance to experimental autoimmune encephalomyelitis and neuritis in the guinea pig correlate with the induction of procoagulant and anticoagulant activities. J. Immunol. 133, 3026–3036 (1984).
Acknowledgements
JYL supported by Multiple Sclerosis Research Australia and Trish Multiple Sclerosis Research Foundation Postgraduate Scholarship; SP supported by National Multiple Sclerosis Society Project Grant #RG4398A1/1, International Progressive Multiple Sclerosis Alliance Challenge Award #PA0065, Multiple Sclerosis Research Australia and Trish Multiple Sclerosis Research Foundation #15-022 and Bethlehem Griffiths Research Foundation #BGRF1706. S.M.S supported by N.I.H. The authors thank Stephen Cody from Monash Micro Imaging, Central Clinical School, Monash University, Melbourne, Australia.
Author information
Authors and Affiliations
Contributions
Conceptualization: S.P.; Methodology: M.M.B., P.T., J.Y.L., A.A.A., P.M.A., W.A.F., M.F.A., J.P.A., F.M., C.S., F.A., S.M.S., N.G., S.P.; Formal analysis and investigation: M.M.B., P.T., J.Y.L., A.A.A., P.M.A., W.A.F., M.F.A., J.P.A., F.M., C.S., F.A., S.M.S., N.G., S.P.; Writing—original draft: M.M.B., P.T., S.P.; Writing—review and editing: P.T., S.P.; Visualization: P.T., J.Y.L.; Data curation: P.T., J.Y.L., S.P.; Resources: S.M.S., N.G., S.P.; Funding acquisition: S.M.S., N.G., S.P.; Supervision: S.P.
Corresponding author
Ethics declarations
Competing interests
The authors declare that S.M.S. is a co-founder and consultant for ReNetX Bio, which seeks to develop NgR1-based therapies for neural repair. All other authors listed do not have competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Bakhuraysah, M.M., Theotokis, P., Lee, J.Y. et al. B-cells expressing NgR1 and NgR3 are localized to EAE-induced inflammatory infiltrates and are stimulated by BAFF. Sci Rep 11, 2890 (2021). https://doi.org/10.1038/s41598-021-82346-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-82346-6
This article is cited by
-
BAFF blockade in experimental autoimmune encephalomyelitis reduces inflammation in the meninges and synaptic and neuronal loss in adjacent brain regions
Journal of Neuroinflammation (2023)
-
Microglia and meningeal macrophages depletion delays the onset of experimental autoimmune encephalomyelitis
Cell Death & Disease (2023)
-
Distinct binding mode of BAFF antagonist antibodies belimumab and tabalumab, analyzed by computer simulation
Journal of Molecular Modeling (2022)
-
Evolution of Anti-B Cell Therapeutics in Autoimmune Neurological Diseases
Neurotherapeutics (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.