Abstract
West Nile virus (WNV) is a Flavivirus, which can cause febrile illness in humans that may progress to encephalitis. Like any other obligate intracellular pathogens, Flaviviruses hijack cellular protein functions as a strategy for sustaining their life cycle. Many cellular proteins display globular domain known as PDZ domain that interacts with PDZ-Binding Motifs (PBM) identified in many viral proteins. Thus, cellular PDZ-containing proteins are common targets during viral infection. The non-structural protein 5 (NS5) from WNV provides both RNA cap methyltransferase and RNA polymerase activities and is involved in viral replication but its interactions with host proteins remain poorly known. In this study, we demonstrate that the C-terminal PBM of WNV NS5 recognizes several human PDZ-containing proteins using both in vitro and in cellulo high-throughput methods. Furthermore, we constructed and assayed in cell culture WNV replicons where the PBM within NS5 was mutated. Our results demonstrate that the PBM of WNV NS5 is important in WNV replication. Moreover, we show that knockdown of the PDZ-containing proteins TJP1, PARD3, ARHGAP21 or SHANK2 results in the decrease of WNV replication in cells. Altogether, our data reveal that interactions between the PBM of NS5 and PDZ-containing proteins affect West Nile virus replication.
Similar content being viewed by others
Introduction
Arboviruses include numerous human and animal pathogens that are important global health threats responsible for arboviroses. Some arboviroses are the most serious worldwide infectious risks to the human nervous system1. Among them, Flaviviruses from the Flaviviridae family are transmitted by the bite of arthropods (mosquitoes or ticks), and are able to replicate both in invertebrate and vertebrate organisms. Tick born encephalitis virus (TBEV), West Nile virus (WNV) and Japanese encephalitis virus (JEV) are neurotropic Flaviviruses causing febrile illness in humans that may progress to encephalitis and even death in some cases.
Hijacking of cellular protein functions is a widely used strategy for viruses as they are obligate intracellular pathogens. Each step of the viral life cycle from entry to transmission is orchestrated through interactions with cellular proteins. All families of viruses manipulate the cell proteome by targeting key proteins involved in the control of cell homeostasis. Interactions mediated by short linear motifs (SLiMs) are ubiquitous in eukaryotic proteome2 and the adaptation of viruses to their environment could involve the extensive use of SLiM mimicry to subvert host functionality. PDZ-Binding Motifs (PBMs) are short SLiMs that interact with a large family of protein–protein interaction domains found in prokaryotes and eukaryotes called PDZ (PSD-95/Dlg/ZO-1), and are usually located at the extreme carboxyl terminus of proteins, although a few internal binding motifs have also been identified3. PBMs play a central role in cell signaling by mediating protein–protein interactions in complex networks such as establishment and maintenance of cell polarity. The PDZ-PBM interactome is therefore one of the most prominent instances of a SLiM-mediated protein interaction network serving key cell signaling purposes. PBMs were identified in proteins of many viruses, responsible for acute to chronic infection, illustrating that cellular PDZ proteins are common targets during viral infection3. Cellular PDZ/viral PBM interactions were shown as directly involved in viral pathogenicity of severe acute respiratory syndrome coronavirus (SARS-CoV) and of rabies virus as well as in the oncogenicity of human papillomavirus (HPV16)4,5,6. During infection, the viral proteins compete with the endogenous protein ligands through the binding to the PDZ domain of the host protein targets but can also affect the catalytic activity of signaling proteins7. Functional perturbations of cellular processes due to interactions between viral PBMs and cellular PDZ-containing proteins may improve the virus life cycle in the host and the dissemination to new hosts. Thus, targeting the PBM-PDZ interface could lead to novel antiviral therapies.
Internal and C-terminal PBMs are present in the non-structural (NS) proteins NS5 of Flaviviruses which provides the RNA cap methyltransferase (MTase) and the RNA-dependent RNA polymerase (RdRp) activities as well as suppression of type 1 interferon signaling8. Their association with PDZ-containing proteins has been demonstrated for TBEV and dengue virus (DV)9,10. TBEV NS5 contains a PBM in both the MTase and RdRp regions. The binding of the internal PBM located in the MTase to a PDZ domain located in the Scribble cellular protein inhibits IFN-mediated JAK-STAT signaling, countering innate immunity to the virus11. Results with DV NS5 also suggest an internal binding mechanism to target PDZ-containing protein ZO-1 (also known as TJP1)9,11 that may contribute to changes at the tight junctions affecting the trans-endothelial permeability12. WNV NS5 displays a typical class I PDZ binding motif (S/T-X-L/V/I) at the C-terminal PBM of its RdRp region (-TVL) as does TBEV (-SII)10, while any internal PBM corresponding to that of TBEV MTase is rather elusive. To carry out viral genome replication, Flavivirus assembles a replication complex, NS5 protein is one major component of this complex. It is highly probable that the PBM of WNV NS5 protein plays a role during infection, in particular during replication by interacting with host partners.
Here, we decipher the role of PBM from WNV NS5 in the viral life cycle. We show both in vitro and in cellulo that this PBM is fully functional and binds to several PDZ-containing cellular proteins using high-throughput interactomic studies. Mutations in the PBM of NS5 result in significantly reduced WNV replication in vitro after cell transfection and depletion of some of these PDZ-containing proteins also affects WNV replication.
Results
The PBM within NS5 impacts West Nile virus replication
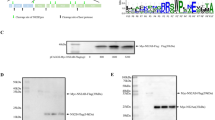
The NS5 protein of some Flaviviruses are known to contain an internal and/or C-terminus PBM. The WNV NS5 protein contains a −TVLCOOH motif at the C-terminal extremity, corresponding to a PBM of class I with the motif S/T-X-ΦCOOH where X is any amino-acid, and Φ is a hydrophobic amino-acid (Fig. 1A). As NS5 of Flaviviruses provides RNA cap MTase and RdRp activities, we focused our efforts to determine whether the PBM of WNV NS5 affects virus replication. To evaluate the contribution of the PBM to WNV replication, we constructed a DNA based sub-genomic WNV replicon expressing the 5′ and 3′ UTR, the genomic region encoding all the non-structural proteins of WNV as well as the reporter gene GLuc, thus generating a luciferase expressing replicon (Rep-IS98-Gluc-wt) (Fig. 1A). We also constructed a negative control replicon coding for an inactive RdRp with a single mutation in the replication “pocket” on WNV NS5 (Rep-IS98-Gluc-GVD) and a replicon with a deletion of the three residues -TVL identified as the PBM motif (Rep-IS98-Gluc-∆PBM) (Fig. 1A). The replicons were transfected into BHK-21 cells and their replication rates were monitored using luciferase luminescence at different time points over 72 h post-transfection (Fig. 1B). The deletion of the nucleotidic sequence coding for the PBM (∆PBM) resulted in a drastic effect with an absence of replication of the WNV replicon (Fig. 1B). In order to confirm the role of the PBM sequence in the replication of WNV, a mutagenized PBM in the replicon was also designed (Rep-IS98-Gluc-TVM: mPBM). We introduced suitable mutations into the replicon taking care not to alter the RNA structure. Indeed, the genome of WNV is an RNA molecule displaying a single ORF flanked by untranslated regions (UTRs). These UTRs are structural conserved parts in the viral genome and play important roles in viral translation and replication. Optimal viral RNA structure at the 3′ UTR is required for efficient polymerase activity during WNV replication leading to viral production in cultured cells13. Introducing a mutation in the RNA sequence encoding a PBM located at the hinge between translated and untranslated regions has proved challenging since cis-acting RNA structures in the 3′-genome must be considered in WNV replication in cells in addition to RdRp activity. We did identify amino-acids and nucleotides required to maintain replication competence and replication efficiency as some PBM mutations in replicons abrogated replication, due to modifications of secondary structures of the 3′UTR. A functional replicon that conserved a proper predicted RNA structure while altering the PBM was thereby obtained using the mutation Leu to Met at the C-terminus of NS5. Thus, we introduced a mutation in the PBM of NS5 that alters the PBM motif and hence potentially affecting the affinities and the pattern of cellular PDZ-containing proteins that could be recognized by the NS5 protein during the infection. Interestingly, the substitution of –TVLCOOH by –TVMCOOH (mPBM) resulted in a significant delay in replication and also in a significantly lower replication with a ten-fold decrease compared to the wt at 48 and 72 h (Fig. 1B) and a five-fold decrease of band intensity for mPBM NS5 compared to wt NS5 as monitored by western blot experiment (Fig. 1C). Collectively, these results show that WNV replication is affected by an amino-acid substitution that alters the PBM within NS5.
WNV replicon experiments. (A) Schematic construction of DNA based subgenomic WNV IS98 replicon models with mutations in the PBM of NS5. Rep-IS98-Gluc is a replicon containing the non-structural genes of WNV while the deleted structural genes were replaced by the non-secreted form of Gaussia luciferase (Gluc) reporter gene under the control of Sp6 promoter. The PBM sequence is indicated in green, a (−TVL) motif at the C-terminal extremity of NS5 (Rep-IS98-Gluc-wt). Three other replicons were constructed: a negative control replicon (Rep-IS98-Gluc-GVD), a replicon with a deletion of the three residues –TVL– (Rep-IS98-Gluc-∆PBM) and a mutagenized PBM in the replicon (Rep-IS98-Gluc-TVM: mPBM). (B). Replication of IS-98-Gluc replicons into BHK-21 cells. Representative replication curves after transfection into BHK-21 cells are plotted for replicons expressing different NS5 sequences: Rep-IS98-Gluc-wt (WT), Rep-IS98-Gluc-TVM (mBPM), Rep-IS98-Gluc-∆PBM (∆PBM) and a negative control replicon Rep-IS98-Gluc-GVD (GVD). The cells were lyzed for subsequent analysis post-transfection. Relative expression of luminescence was normalised to the luminescence obtained 4 h post-transfection. Average arbitrary luminescence units ± SEM is shown from 4 wells per group; experiments were done in triplicate. (C). Western blot showing NS5 protein levels in BHK-21 cells transfected with no replicon (medium only), wt replicon or mPMB replicon. Western blot of lysates from transfected BHK-21 cells at 48 h post electroporation of medium only (negative control/no replicon), mPBM replicon or wt replicon as indicated. The Western-blot is performed with the anti-NS5 antibody. Uncropped WB is shown in Supplementary Fig. 3.
Then, we have attempted to look at the knock-down on viral replication of wt WNV and mPBM WNV. We observed viral replication quantified by RTqCR (Suppl. Fig. 2A) and by FFU (Suppl. Fig. 2B). As we did not see any difference for both quantitation, we wondered if any reversions were occurring, permitting this replication. Indeed, after viral amplification, the last residue of the mPBM (Met, ATG) virus had reverted to a Leu (TTG) as the wild-type virus which has a Leu (CTG) (Suppl. Fig. 2C,D). These observations suggest that the Leu in the PBM motif if important for virus amplification.
West Nile virus NS5 recognizes several cellular PDZ-containing proteins
In order to quantify the binding activity of the NS5 PBM we used an in vitro automated high-throughput chromatography assay called holdup (Fig. 2A)14. A 13-mer peptide was synthetized encompassing the C-terminal PBM sequence of WNV NS5 protein linked to a biotinyl group. This peptide was used as bait to quantify the interaction between the WNV NS5 PBM peptide and a full human PDZ domain library expressed in Escherichia coli. The updated PDZ library called PDZome V.2 covers 97.3% of the human “PDZome” (259 over 266 PDZ domains identified in the human genome)13. The holdup approach displays a high sensitivity for low-to-medium affinity PDZ/PBM pairs and provides an affinity-based ranking of the identified binders corresponding to a specificity profile. Five replicate experiments were performed. This specificity profile for NS5 PBM with the mean values of binding intensities (BI) is reported in Fig. 2A with an affinity-based ranking. Highest values on the Y-axis indicate the PDZ domains recognized with the best affinities by our viral peptide bait. 29 PDZ NS5 binders displayed significant BI values higher than 0.2, a stringent threshold previously defined for significant binders (Fig. 2A, zoomed-in view, Table 1)14. We concluded that the PBM sequence of WNV NS5 protein is unambiguously functional in vitro as illustrated by its capacity to bind several PDZ domains in the 1–100 µM affinity range typically found for PDZ/PBM interactions. Thus, the holdup assay identified potential PDZ-domain partners from host that can be classified according to their affinity values for NS5 PBM: 5 PDZ NS5 binders with a BI > 0.6 (NHERF2_2, MAST2, SNX27, NHERF-1 and MAGI_2); 11 PDZ NS5 binders with a 0.3 < BI < 0.6 (MAGI1_2, MAST1, PARD3B_1, NHERF3_1, SHANK2, SHANK3, SHANK1, MAGI2_2, PDZRN3_1, SCRIB_3 and FRMPD4) and 13 PDZ NS5 binders with a 0.2 < BI < 0.3 (ARHGAP21, HTRA1, PARD3_3, PDZD7_3, SCRIB_1, ARHGEF12, DLG2_1, DLG4_3, TJP3_2, DLG1_2, SNTB1, SYNJ2BP and NHERF2_1) (Fig. 2A).
PDZome binding profiles of WNV PBM by two high-throughput techniques: holdup assay (A) and Split-Nanoluciferase Protein Complementation Assay (N2H) (B). (A) Binding intensity profiles for the PDZome. PDZ domains revealed by holdup assay are ranked on the basis of the BIs of WNV NS5 PBM. Zoomed-in view (corresponding to the red box) shows the 29 PDZ NS5 binders which displayed significant BI values higher than 0.2. B. Frequency distribution for NLR(NS5) – NLR(NS5∆PBM) obtained by N2H method are ranked on their values. Zoomed-in view (corresponding to the red box) shows 28 PDZ NS5 binders displayed an NLR difference > 0.25 NLR.
In parallel, we monitored full-length NS5-PDZ domain interactions in a cellular context using the Split-Nanoluciferase Protein Complementation Assay (N2H)15. We used the full-length (FL) NS5 protein (wt PBM) instead of NS5 PBM peptide used for holdup assay. We constructed a FL NS5ΔPBM protein which contains –TVL base deletions (∆PBM), used as control. The FL NS5 proteins and the library of PDZ domains (257 over 266 PDZ domains identified in the human genome, as we failed to clone the two PDZ domains DLG5_3 and SNTB2) were cloned in mammalian expression vector for high-throughput protein–protein interaction (PPI) detection16. Our new PDZ library in the mammalian vector covers 96.6% of the human “PDZome”. Three replicate experiments were performed. Mammalian vectors containing FL NS5 and PDZome were transfected into 293HEK cells. We monitored PPIs by measuring interaction-mediated normalized luminescence ratio (NLR) for both NS5 and NS5∆PBM. The NLR obtained with FL NS5∆PBM protein, used as negative control, was subtracted from the NLR obtained with FL NS5 (wt PBM) protein and plotted in Fig. 2B. Highest values on the Y-axis indicate the highest intensity of PPI for the NS5 protein containing PBM sequence. 28 PDZ NS5 binders displayed an NLR difference > 0.25 NLR and among them 12 > 1.0 NLR difference (Fig. 2B). Thus, wt PBM in the context of full length NS5 protein is functional in cellulo, comforting our in vitro results with the holdup assay.
Together, these two high-throughput methods confirmed the functionality of WNV NS5 PBM to bind PDZ-containing proteins. To go further, we started assembling from the human ORFeome collection a subset of ORFs encoding full-length proteins containing PDZ domains to be transferred in the N2H system. To date, our bank covers 60.5% of the human PDZome with 91 full-length proteins out of the 152 human proteins known to contain PDZ domains (Table 1). 23 full-length proteins interacting with full-length NS5 protein displayed an NLR > 1.35 NLR (Suppl. Fig. 1A). We concluded that PBM of NS5 and these PDZ domains were unambiguously functional in this context of full-length proteins.
The three lists of PDZ proteins obtained by high-throughput holdup and N2H methods (PDZ domains and full-length PDZ proteins) were compared using a Venn diagram (Suppl. Fig. 1). Eleven proteins were identified in common by holdup assay and N2H method (PDZ domains), five were identified by holdup assay and N2H method (full length proteins) and four were identified by the two N2H methods (PDZ domains and full-length proteins).
To determine whether a particular ontology class or pathway could be established among the PDZ-containing proteins identified as WNV NS5 binders by our experimental approaches, we chose to use the protein annotation program PANTHER (protein annotation through evolutionary relationships). This overrepresentation test led us to analyze our large-scale experimental data against current annotated gene data set. PANTHER analysis was performed with the two lists of PDZ-containing proteins obtained from holdup (29 PDZ NS5 binders; 27 proteins) and from N2H (28 PDZ NS5 binders; 25 proteins) assays. Protein IDs were imported into the Gene Ontology (GO) enrichment analysis tool based on protein analysis through the evolutionary relationships classification system. The cellular component categories of GO having P values < 0.05 were considered to be statistically significant and then selected. PANTHER overrepresentation test based on the GO of molecular functions, cellular component, biological process and protein class (Suppl. Fig. 1B–D and Suppl. Table 1) showed that the majority of the 41 proteins have binding function, are involved in cellular process and more particularly the top five were annotated to membrane, cell periphery, plasma membrane, cell junction and cell projection. From these 41 proteins, some were already known to interact with viruses, and more particularly with Flaviviruses. Considering the results obtained with our high-throughput approaches, PANTHER analysis and the literature, we selected 10 PDZ-containing proteins for further analysis, which were at least in two lists of proteins identified (Suppl. Fig. 1) and which have at least been documented in interaction with another viral protein: ARHGAP21, CNKSR3, DLG1, HTRA1, MAGI1, PARD3, RIMS2, SHANK2, SNX27 and TJP1 (Table 1).
Depletion of cellular PDZ-containing proteins affects West Nile virus replication
To investigate whether the selected cellular PDZ proteins we identified to interact with the WNV NS5 PBM could have a functional role in viral replication, we decided to test depletion of these proteins from HEK-293T cells using siRNAs (Fig. 3). We first compared the replication rate of wt and mPBM replicons with the non-targeted control siRNA using a luciferase assay at different time point over 48 h post-transfection of HEK-293T cells (Fig. 3A). We obtained results similar to that presented in Fig. 1B using BHK-21 cells with a delay in replication for the mPBM (Fig. 3A) and a significant ten-fold decrease compared to wt at 24- and 48-h post-transfection (Fig. 3B). Then, we tested for depletion of the 10 selected cellular PDZ proteins, namely ARHGAP21, CNKSR3, DLG1, HTRA1, MAGI1, PARD3, RIMS2, SHANK2, SNX27 and TJP1 (Table 1). Depletion of the corresponding transcripts was assessed by real-time quantitative PCR and transcripts with a significant decrease in quantity are presented in Fig. 4A. The level of protein expression was checked by Western Blot after siRNA treatment (Fig. 4B). We obtained both a significant decrease in transcripts and a decrease in protein level ranging from 52.5 to 91.6% in HEK-293T cells at 48 h post-transfection (Fig. 4A,B).
Effect of control siRNA transfection into HEK-293T cells at 24- and 48 h post electroporation of Rep-IS98-Gluc wt and mPBM. HEK-293T cells were transfected with the non-targeting siRNA (control siRNA) 48 h before the electroporation with 10 μg of synthetized WNV replicon RNA. Cells were lysed at 24- and 48 h post electroporation. (A) Representative replication curves after electroporation into HEK-293T cells are plotted for Rep-IS98-Gluc-wt (WT) and Rep-iS98-Gluc mPBM (mPBM). (B) Statistical data analysis of Rep-IS98-Gluc wt and Rep-IS98-Gluc mPBM activities at 24- and 48 h post electroporation, respectively. Data show relative Gluc activity and error bars represent mean ± SD from three independent experiments in quadruplicate (n = 12). Asterisks indicate significant differences between replicon activities, **p < 0.01.
SiRNA knocked down ARHGAP21, CNKSR3, DLG1, HTRA1, MAGI1, PARD3, RIMS2, SHANK2, SNX27 and TJP1 expression. HEK-293T cells were transfected with two siRNAs against each protein: ARHGAP21, CNKSR3, DLG1, HTRA1, MAGI1, PARD3, RIMS2, SHANK2, SNX27 and TJP1 with 10 µM for each target siRNA in Lipofectamine RNAimax. After 48 h, the corresponding transcripts were measured by RTqPCR (A) and gene expression measured by Western blot assay (β-actin = loading control) (B). (A) Total RNA from transfected HEK-293T cells was extracted and used for real time quantitative PCR. Relative expression was normalised to the housekeeping gene GAPDH and is presented as the mean ± SD (*p < 0.05 by Mann Whitney t-test). (B) Western blot of a representative sample is showed for each protein. Protein expression was normalized to β actin or calnexin expression and indicated in each WB. Uncropped WB are shown in Supplementary Fig. 3.
We then monitored the Gluc activity of both wt and mPBM replicons over 48 h post-transfection upon siRNA treatment. Depletion of RIMS2 and SNX27 did not impact replication of either replicon in our conditions at 24- and 48 h post-transfection compared to control siRNA treatment (non-targeted siRNA) (Fig. 5A). RIMS2 and SNX27, although able to interact with WNV NS5 PBM through their PDZ domains in vitro and in cell, do not seem to play a role in the virus replication in our assay. Depletion of CNKSR3, DLG1, HTRA1 and MAGI1 affected replication of both wt and mPBM replicons in our conditions at 24- and 48 h post-transfection compared to control siRNA treatment, suggesting that these proteins impact WNV replication independently from the substitution we incorporated in the PBM sequence (Fig. 5B). In contrast, depletion of PARD3, ARHGAP21 and SHANK2 resulted in a significant 5.3-, 7.2- and 5.1-fold decrease respectively at 24 h and 2.7-, 19.7- and 14.0-fold decrease respectively at 48 h for the wt replicons post-transfection but not for mPBM replicons (Fig. 5C). Similarly, depletion of TJP1 resulted in a significant two-fold decrease at 48 h of replication for the wt replicons but not for the mPBM replicon. Thus, these results reveal a significant PBM-dependent effect of NS5 on WNV replication involving at least the four host PDZ-containing proteins PARD3, ARHGAP21, SHANK2 and TJP1.
Effect of ARHGAP21, CNKSR3, DLG1, HTRA1, MAGI1, PARD3, RIMS2, SHANK2, SNX27 and TJP1 siRNA transfection into HEK293T cells at 24 and 48 h post electroporation of Rep-IS98-Gluc wt and mPBM plasmids. HEK-293T cells were transfected with ARHGAP21, CNKSR3, DLG1, HTRA1, MAGI1, PARD3, RIMS2, SHANK2, SNX27 and TJP1 siRNA 48 h before the electroporation with 10 μg of synthetized WNV replicon RNA. Cells were lysed at 24- and 48 h post electroporation of Rep-IS98-Gluc wt and Rep-IS98-Gluc mPBM. (A) HEK293T cells were transfected with control siRNA, RIMS2 and SNX27 siRNAs showing no effect of the transfection. (B) HEK293T cells were transfected with control siRNA, CNKSR3, DLG1, HTRA1 and MAGI1 siRNAs showing an independent-PBM effect. (C) HEK-293T cells were transfected with control siRNA, TJP1, PARD3, ARHGAP21 and SHANK2 siRNAs showing a PBM-dependent effect. Data show relative Gluc activity and error bars represent mean ± SD from three independent experiments in quadruplicate (n = 12). Asterisks indicate significant differences between replicon activities, *p < 0.05, **p < 0.01 and ***p < 0.005 obtained with a Mann–Whitney unpaired t-test.
N2H method with full-length proteins (full-length NS5 and full-length human ORF protein) confirmed the interaction between WNV NS5 and four PDZ-containing proteins PARD3, ARHGAP21, SHANK2 and TJP1. Of note, TJP1, PARD3, SNX27 and DLG1 are not present in this bank. We performed a protein–protein interaction in cellular context using N2H method with this new bank and found strong interactions for ARHGAP21 and SHANK2 (NLRs values of 4.41 and 4.38; respectively (Table 1, second part)), in the top 4 over the 91 NLR obtained, we also found a weaker interaction with CNKSR3 (NLR of 1.9). By contrast, RIMS2, HTRA1, MAGI1 displayed low NLR values (Table 1, second part). Thus, we confirmed in cell the significant interaction of full length PDZ-containing proteins ARHGAP21 and SHANK2 with the FL NS5 protein.
Discussion
The viral NS5 protein is the largest and most conserved Flavivirus proteins. It plays a major role in viral replication through its MTase and RdRP domains and in IFN signaling11,17,18. Interactions between NS5 PBM and PDZ-containing cellular proteins have been associated with interferon antagonism, viral replication and disease pathogenesis19,20,21. The biological relevance of PDZ binding for TBEV is the most documented, especially regarding viral replication9,10,11. In this study, we showed that the PBM (–TVLCOOH) motif at the C-terminus of WNV NS5 protein interacts with human PDZ domains in vitro but is also capable of interacting with cellular PDZ-containing proteins in a cellular context. We established in this study the promiscuity of 41 different PDZ-containing proteins both in vitro and in cellulo (listed in Table 1) with the NS5 PBM of WNV by screening a PDZ bank which covers more than 96% of the human “PDZome”. Melik and co-workers previously observed such a broad repertoire of potential host PDZ proteins recognized by the NS5 PBM of WNV using a qualitative PDZ array partially covering the human PDZome10. We noted that the lists of partners selected from our two high-throughput screenings only partially overlapped. Several reasons may explain this observation; by contrast with their cellular expression in the N2H assay, the concentrations of partners are strictly controlled in vitro holdup assay. While the interactions involved viral PBM peptide and the PDZome in holdup assays, the NH2 screening involves full-length NS5 as bait, the PDZome but also full length PDZ-containing proteins in the case of ORFome screening. Thus, the expression environment and the type of constructs used could also have a strong impact on PPI detection, justifying our choice to combine results from holdup and N2H assays.
Our study supports a short-list of 10 PDZ-containing proteins selected from our crossed high-throughput screenings and for their functions in the cells and during viral infection. We tested the physiological relevance of these 10 interactors on WNV RNA replication using the siRNA technology and the replicon containing an altered PBM with the mutation Leu to Met at the C-terminus of NS5 allowing to conserve a proper predicted RNA structure. Eight of the ten selected PDZ-containing proteins significantly impacted WNV replicon replication: CNKSR3, DLG1, HTRA1, MAGI1, PARD3, ARHGAP21, SHANK2 and TJP1. We show that PARD3, ARHGAP21, SHANK2 and TJP1 displayed a significant PBM-dependent effect on WNV replication using the L-to-M PBM mutation as a gauge of the NS5 PBM influence. In contrast, CNKSR3, DLG1, HTRA1 and MAGI1 affected WNV replication regardless of the presence or absence of mutation in the PBM. We cannot exclude the presence of an unidentified internal PBM in NS5 that might interfere as other PBM motifs located internally rather than at the C-terminus of NS5 were shown to play a role the replication of TBEV and DV10. How CNKSR3, DLG1, HTRA1 and MAGI1 proteins are directly or indirectly connected to WNV replication should be documented in further studies. Finally, we identified RIMS2 and SNX27 as WNV NS5 PBM binders. They did not interfere with the viral replication assay, but they could participate in other steps of the viral cycle. Interestingly, RIMS2 has been shown to interact with TBEV NS5, potentially affecting the neuronal signalling of this neurotropic Flavivirus9.
Importantly, all our selected PDZ-containing proteins, except CNKSR3 and SHANK2, have been previously reported as host proteins targeted by viruses. MAGI1, PARD3, RIMS2 and TJP1 are targets of Flavivirus proteins (WNV, TBEV and DV)9,10,11. Others cellular PDZ proteins were also identified as target for other virus including DLG1, MAGI1 and ARHGAP21 for Influenza virus16,22,23,24; HTRA1, PARD3, SNX27, DLG1, MAGI1 for Human Papillomavirus11,25,26,27,28. The large number of cell proteins targeted by WNV could be related to the broad host repertoire of Flaviviruses replicating in very different cells and species necessary for its complex zoonotic transmission cycle. Our PANTHER analysis shows that these ten proteins were involved in a global perturbation of cellular homeostasis, architecture of the cells (membrane trafficking, cell junction, cellular polarity…), innate immune response (IFN mediated-JAK-STAT signalling) and neuronal cells (synapse). We report that NS5 of WNV can interact with TJP1 (also known as ZO-1), ARHGAP21 and PARD3 proteins through PBM-PDZ interactions. These three proteins are involved in cell polarity, trafficking, cell adhesion and cell junction. ARHGAP21 protein is involved in trafficking through the control of CDC42 activity29. Taye et al. performed a microarray analysis and obtained an overexpression of ARHGAP21 which negatively regulates the transport of Influenza A virus (H1N1) neuraminidase to the cell surface22,30. TJP1 was reported to interact with other Flavivirus NS59,10. Interestingly, we found both TJP1 and TJP3 as interactors of WNV NS5 PBM in our study (Fig. 2A and B and Table 1). TJP1, TJP2 and TJP3 are members of the membrane-associated guanylate kinase (MAGuK) family of proteins31 that play a particular role in tight-junctions (TJs) organization32. TJP1 can bind directly to TJP3 to form a TJP1/TJP3 complex that acts directly on the actin skeleton. Interestingly, it was demonstrated that actin filaments participate in WNV maturation process33. Moreover, TJs assembly and permeability are highly regulated by signalling pathways34. It was shown that TBEV NS5 interacts with GIPC and TJP2 through PDZ/PBM complex. The MAGuK TJP2 protein is an interactor of Scribble, a PDZ-containing protein involved in cell polarity and neuronal function. The complex NS5-TJP2-hScrib may shuttle the polymerase to the plasma membrane, regulating the flaviviral replication that occurs at ER membrane-associated complexes11. As TJP1, PARD3 displayed a significant NS5 PBM-dependent effect on WNV replication. Chen et al. showed that suppression of PARD3 expression by RNA interference (RNAi) caused a dramatic disruption of TJ assembly35. Of note, we also detected PARD3B, a human PARD3 homologue (Fig. 2A,B and Table 1), which also localizes at the TJs36. Moreover, PARD3 and TJP1 are recruited by JAM-2 to cell–cell contacts37. TJP1 is also associated with JAM-1 through PDZ domain 338. The association of TJP1 with these different proteins in TJs serves to cluster the integral membrane proteins at TJs and could be partially disrupted by WNV NS5 during infection resulting in a progressive disruption of TJ, which leads to leakage between adjacent cells, loss of barrier function, and infiltration of WNV virions37.
Suppression of host innate immunity is one of the multiple crucial functions of Flavivirus NS5 protein. The type I IFN signalling pathway included in the first lines of defence against Flavivirus infection of mammals. TBEV and WNV NS5 inhibit IFN-I signalling by suppressing surface expression of interferon alpha and beta receptor subunit 1 (IFNAR1)39. Indeed, TBEV may manipulate the IFN pathways through interaction with TJP1 as TBEV-NS5 (and DV-NS5) was shown to be present in the nucleus at the same moment as TJP19. To note, in WNV-infected cells, a high proportion of NS5 resides in the nucleus40 but it is also accurate for other Flavivirus NS5 (DV41; Yellow Fever42). A consequence of the NS5/PDZ-containing protein interactions could be a mislocalization of these PDZ-containing proteins which can alter their original function. For example, TJP2 and MAGI1 were reported to be sequestered by adenovirus E4-ORF1 in the cytoplasm of fibroblasts43 and this mislocalization disrupted both the tight junction barrier and the apicobasal polarity.
The PDZ-containing proteins interaction with NS5 protein could result in a less restrictive environment for viral replication and provide a new understanding on the mechanism of WNV immune evasion.
Several of the PDZ-containing cellular proteins identified in our crossed high-throughput screenings are involved in the development, function and architecture of neuronal cells (RIMS2, SHANK2, DLG1), highlighting the relevance of our findings as WNV is indeed a neurotropic virus. The nature of the assay we used, restricted the characterization of the functional significance of such PDZ dependent WNV-host interactions to viral replication. Further characterization of the mechanisms by which PBM/PDZ interactions may alter neuronal functions is therefore essential.
Materials and methods
Cell lines and viruses
Baby Hamster Kidney fibroblasts (BHK-21) cells and human embryonic kidney 293 T (HEK-293T) cells were grown and maintained in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 5% fetal bovine serum (FBS, Qualified, Gibco) and in DMEM supplemented with 10% FBS and 100 U ml−1 penicillin, 100 U ml−1 streptomycin, respectively. Both lineages were cultured at 37 °C in a humidified 5% CO2 incubator. The replicon Rep-IS98-Gluc/pCR2.1 has been previously described30.
Production of recombinant WNV
We first produced a full-length infectious clone for wt WNV and mPBM WNV as previously described by Alsaleh et al.44; the DNA was purified and transcribed in vitro using the mMessage SP6 kit (Thermo Fisher Scientific). The resulting RNAs were electroporated in C6/36 cells (400 V, 25 µF, 800 Ω) in OPTI-MEM medium (Thermo Fisher Scientific). Cell culture supernatants were collected at 72 h post-electroporation and used to infect 107 C6/36 cells. At 3 days post-infection, viral supernatants were amplified by infecting 5 × 107 C6/36 cells for 3 days before collection and utilization as final viral stocks. The two stocks were used for genome quantitation by RTqPCR and for virus titers by FFU using Vero cells. Full-length viral genomes were sequenced by Sanger method from cDNA obtained by reverse transcription using the Superscript II reverse transcription kit (Invitrogen) and amplified by PCR using the Phusion high-fidelity kit (Thermo Fisher Scientific).
Cloning of human PDZ domains for Holdup assay
The full PDZome clone collection (266 clones), previously cloned by Vincentelli et al.; PDZome V.216. These constructs were initially cloned into pDONRzeo for the 266 clones. All the PDZ (266 clones) were sequenced in the destination vector.
Holdup assay with human PDZ domains
All PDZ domains (259/266 PDZ) were expressed following the high-throughput protocol previously described45. The peptides biotinylated WNV NS5 PBM (peptide sequence-RYEDTTLVEDTVL) was synthesized in solid phase using Fmoc strategy (Proteogenix) and resuspended in H2O. The holdup assay was carried out against the biotinylated WNV NS5 PBM in quadruplicates as previously described14,45 with minor modifications. We measured WNV NS5 PBM interactions against 259 human PDZ domains. The minimal BI threshold value is 0.2 to define a significant interaction as previously reported14.
Cloning all PDZ domains and ORFs into gateway-compatible expression plasmids for split-nanoluciferase protein complementation assay (N2H)
All PDZ domains (257/266 PDZ) and ORFs containing PDZ domains (91 full-length proteins out of the 152 human proteins) were introduced into Gateway destination vector pYN2H-LEU-N145 whereas NS5 and NS5∆PBM (NS5 with a deletion of the three residues –TVL–) were introduced into Gateway destination vector pYN2H-TRP-N2, via LR clonase-mediated Gateway reaction (Life Technologies), following the protocol previously described46. Briefly, LR reaction products were subsequently transformed into DH5α competent bacterial cells and grown for 24 h at 37 °C at 900 rpm in carbenicillin-containing TB liquid medium. Plasmid DNA was extracted using a NucleoSpin 96 Plasmid kit from Macherey–Nagel, following manufacturer's instructions. All DNA were sequenced using plasmid-specific primers.
Split-nanoluciferase protein complementation assay (N2H) into HEK cells
HEK-293T cells were seeded at 5.4 × 104 cells per well in 96-well, flat-bottom, cell culture microplates (Greiner Bio-One, #655083), and cultured in Dulbeccoʼs modified Eagleʼs medium (DMEM) supplemented with 10% fetal calf serum at 37 °C and 5% CO2. Twenty-four hour later, cells were transfected with 100 ng of pYN2H-LEU-N bait and 100 ng of pYN2H-TRP-N2 prey using linear polyethylenimine (PEI) to co-express the peptide/protein pairs or the protein/protein pairs fused with complementary NanoLuc fragments, N1 and N2, as previously described by Choi et al.46. Twenty four hours after DNA transfection, the cell culture medium was removed and 50 µL of mix (100 mM MES pH 6.0, 1 mM CDTA, 0.5% (v/v) Tergitol, 0.05% (v/v), Mazu DF 204, 150 mM KCl, 1 mM DTT, and 35 mM thiourea; Furimazine 20 µg/ml) were added to each well, plates were incubated for 3 min at room temperature, and luminescence output was measured using a Centro XS LB 960 luminometer (Berthold; 2 s integration time). All binary PPI experiments were independently performed three times. Control experiments were performed similarly where NS5/NS5∆PBM protein fused to the NanoLuc fragment N1 was co-expressed with the matching NanoLuc fragment alone. For each peptide/protein pair –N1/–N2, we calculated a normalized luminescence ratio (NLR) corresponding to the raw luminescence value of the tested pair (–N1/–N2) divided by the maximum luminescence value from the control, as previously described by Cassonnet et al.15. One NLR was obtained for the NS5/PDZ and one for NS5∆PBM/PDZ. We decided to subtract the NLR calculated with NS5∆PBM protein to the NLR calculated with NS5 protein, all the values are listed in the Table 1 with mean ± SD.
Plasmid DNA constructs
All recombinant DNA techniques and cloning procedures were carried out by standard procedures47. We modified the replicons by site-directed mutagenesis methods on the PBM of NS5 proteins using Phusion polymerase the following primers respectively: Forward mPBM: 5′GGTTGAGGACACAGTAATGTAGATATTT3′, Reverse mPBM: 5′AAATATCTACATTACTGTGTCCTCAACC3′.
In vitro transcription
The RNA was transcribed from linearized reconstituted full-length plasmids using mMESSAGE MEGAscript Sp6 Kit (Ambion-Life technologies) according to the manufacturer’s instructions as previously described44. Briefly, after 4 h of incubation at 37 °C, the DNA template was digested by DNase treatment. Synthesized RNA was precipitated by LiCl and purified according to manufacturer’s instructions (Ambion-Life Technologies). The RNA was quantified by Nanodrop and used for transfection.
Plasmid transfection into BHK-21 and HEK-293T cells and Luciferase assays
4 × 106 BHK-21 cells were electroporated with 10 μg of synthetized RNAs using the following settings: 1 pulse of 140 V, 25 µF and ∞ Ω. Cells were resuspended in DMEM-2% FBS and seeded at 1 × 105 cells/well in a 24 well plate.
4 × 106 HEK-293T cells electroporated with the different siRNAs were electroporated with 10 μg of synthetized RNAs using the following settings: 1 pulse of 220 V, 960 µF and 25 ms. Cells were resuspended in DMEM-10% FBS and seeded at 2 × 105 cells/well in a 24 well plate.
Cells were lysed at 4-, 24-, 48- and 72 h post electroporation with Renilla Luciferase Assay Lysis Buffer according to the manufacturer’s instruction (Promega). Mean values of Renilla-normalized firefly Luciferase expression were determined from quadruplicate wells. At least three independent transfections in cells were performed for each experiment.
siRNA
Two siRNAs against each protein: ARHGAP21, CNKSR3, DLG1, HTRA1, MAGI1, PARD3, RIMS2, SHANK2, SNX27 and TJP1 (Suppl. Table 2) were purchased from Life Technologies (10 μM for each target siRNA and 20 μM for control untargeted siRNA) were transfected into HEK-293T cells (24 well cultures dishes) using Lipofectamine RNAimax according to the manufacturer's reverse-transfection protocol. After 48 h, the cells were used for electroporation of RNA replicons as described above. Data were analyzed from two experiments in quadruplicate wells.
RTqPCR
Total RNA from infected or transfected HEK-293T cells was extracted using Nucleospin RNA kit (Machery Nagel) according to the manufacturer's protocol and reverse transcribed into cDNA using Moloney Murine Leukemia Virus Reverse Transcriptase (Invitrogen, Life Technologies). A SYBR Green-based real-time PCR assay (QuantiTect SYBR Green kit, Qiagen) for relative quantification of transcripts was performed on 384-well plate QuantStudio 6 Flex real-time PCR system (Applied Biosystems) according to protocol as previously described19,20. The relative quantities were determined by using raw Cp values as input for qBase, a flexible and open source program for qPCR data management and analysis48; the primers used in this manuscript were listed in Suppl. Table 2.
Western Blot
Protein lysates were prepared by cell lysis in radioimmunoprecipitation assay (RIPA) buffer (BioBasic Canada) containing protease inhibitors (Roche), then the cells were frozen at −20 °C. Five μl of NuPAGE LDS sample Buffer (Thermo Fisher Scientific) was added to 20 μl of lysate and incubated at 95 °C for 5 min. Samples were separated on NuPAGE 4–12% Bis–Tris Protein Gels and transferred to a polyvinylidene difluoride (PVDF) membrane (Life Technologies). The membrane was washed for 1 h at room temperature in PBS-Tween (PBS-T) plus 5% milk, then incubated overnight at 4 °C with the different antibodies (Suppl. Table 2). After washing in PBS-T the membrane was incubated for 1 h at room temperature with HRP-conjugated antibody (BioRad). After washes in PBS-T, the membrane was developed with enhanced chemiluminescence reagent (Pierce) and exposed to film.
Statistical analysis
The p values were determined with GraphPad Prism software version 8.0 (https://www.graphpad.com/scientific-software/prism/) using a Mann–Whitney unpaired t-test (*p < 0.05, **p < 0.01, ***p < 0.005).
References
Wasay, M., Khatri, I. A. & Abd-Allah, F. Arbovirus infections of the nervous system: current trends and future threats. Neurology 84, 421–423 (2015).
Via, A., Uyar, B., Brun, C. & Zanzoni, A. How pathogens use linear motifs to perturb host cell networks. Trends Biochem. Sci. 40, 36–48 (2015).
Javier, R. T. & Rice, A. P. Emerging theme: Cellular pdz proteins as common targets of pathogenic viruses. J. Virol. 85, 11544–11556 (2011).
Préhaud, C. et al. Attenuation of rabies virulence: Takeover by the cytoplasmic domain of its envelope protein. Sci. Signal. 3, 5 (2010).
Spanos, W. C. et al. Deletion of the PDZ motif of HPV16 E6 preventing immortalization and anchorage-independent growth in human tonsil epithelial cells. Head Neck 30, 139–147 (2008).
Teoh, K.-T. et al. The SARS coronavirus E protein interacts with PALS1 and alters tight junction formation and epithelial morphogenesis. MBoC 21, 3838–3852 (2010).
Maisonneuve, P. et al. Regulation of the catalytic activity of the human phosphatase PTPN4 by its PDZ domain. FEBS J. 281, 4852–4865 (2014).
Thurmond, S., Wang, B., Song, J. & Hai, R. Suppression of type I interferon signaling by flavivirus NS5. Viruses 10, 272 (2018).
Ellencrona, K., Syed, A. & Johansson, M. Flavivirus NS5 associates with host-cell proteins zonula occludens-1 (ZO-1) and regulating synaptic membrane exocytosis-2 (RIMS2) via an internal PDZ binding mechanism. Biol. Chem. 390, 319–323 (2009).
Melik, W. et al. Two PDZ binding motifs within NS5 have roles in Tick-borne encephalitis virus replication. Virus Res. 169, 54–62 (2012).
Werme, K., Wigerius, M. & Johansson, M. Tick-borne encephalitis virus NS5 associates with membrane protein scribble and impairs interferon-stimulated JAK-STAT signalling. Cell. Microbiol. 10, 696–712 (2008).
Talavera, D., Castillo, A. M., Dominguez, M. C., Gutierrez, A. E. & Meza, I. IL8 release, tight junction and cytoskeleton dynamic reorganization conducive to permeability increase are induced by dengue virus infection of microvascular endothelial monolayers. J. Gen. Virol. 85, 1801–1813 (2004).
de Borba, L. et al. RNA structure duplication in the dengue virus 3′ UTR: Redundancy or host specificity?. mBio 10, e02506-e2518 (2019).
Vincentelli, R. et al. Quantifying domain-ligand affinities and specificities by high-throughput holdup assay. Nat. Methods 12, 787–793 (2015).
Cassonnet, P. et al. Benchmarking a luciferase complementation assay for detecting protein complexes. Nat. Methods 8, 990–992 (2011).
Duhoo, Y. et al. High-throughput production of a new library of human single and tandem PDZ domains allows quantitative PDZ-peptide interaction screening through high-throughput holdup assay. in High-Throughput Protein Production and Purification: Methods and Protocols (ed. Vincentelli, R.) 439–476 (Springer, New York, 2019).
Laurent-Rolle, M. et al. The interferon signaling antagonist function of yellow fever virus NS5 protein is activated by type I interferon. Cell Host Microbe 16, 314–327 (2014).
Grant, A. et al. Zika virus targets human STAT2 to inhibit type i interferon signaling. Cell Host Microbe 19, 882–890 (2016).
Johansson, M. A. et al. Assessing the risk of international spread of yellow fever virus: A mathematical analysis of an urban outbreak in asunción, 2008. Am. J. Trop. Med. Hyg. 86, 349–358 (2012).
Johansson, M. A., Dominici, F. & Glass, G. E. Local and global effects of climate on dengue transmission in Puerto Rico. PLoS Negl. Trop. Dis. 3, e382 (2009).
Johansson, M. A., Arana-Vizcarrondo, N., Biggerstaff, B. J. & Staples, J. E. Incubation periods of yellow fever virus. Am. J. Trop. Med. Hyg. 83, 183–188 (2010).
Wang, S. et al. Transport of influenza virus neuraminidase (NA) to host cell surface is regulated by ARHGAP21 and Cdc42 proteins. J. Biol. Chem. 287, 9804–9816 (2012).
Awad, A. et al. SHIP2 regulates epithelial cell polarity through its lipid product, which binds to Dlg1, a pathway subverted by hepatitis C virus core protein. MBoC 24, 2171–2185 (2013).
Golebiewski, L., Liu, H., Javier, R. T. & Rice, A. P. The avian influenza virus NS1 ESEV PDZ binding motif associates with Dlg1 and scribble to disrupt cellular tight junctions. J. Virol. 85, 10639–10648 (2011).
Stuqui, B. et al. The differential role of HTRA1 in HPV-positive and HPV-negative cervical cell line proliferation. BMC Cancer 16, 840 (2016).
Facciuto, F. et al. Human papillomavirus (HPV)-18 E6 oncoprotein interferes with the epithelial cell polarity Par3 protein. Mol. Oncol. 8, 533–543 (2014).
Pim, D., Broniarczyk, J., Bergant, M., Playford, M. P. & Banks, L. A novel PDZ domain interaction mediates the binding between human papillomavirus 16 L2 and sorting Nexin 27 and modulates virion trafficking. J. Virol. 89, 10145–10155 (2015).
Ganti, K. et al. Interaction of the human papillomavirus e6 oncoprotein with sorting nexin 27 modulates endocytic cargo transport pathways. PLoS Pathog. 12, e1005854 (2016).
Chi, X., Wang, S., Huang, Y. & Chen, J. Mechanism underlying the anterograde transport of the influenza A virus transmembrane proteins and genome in host cytoplasm. Sheng Wu Gong Cheng Xue Bao 28, 1021–1030 (2012).
Taye, B. et al. Inter-species host gene expression differences in response to human and avian influenza A virus strains. Int. J. Mol. Sci. 18, 2295 (2017).
Haskins, J., Gu, L., Wittchen, E. S., Hibbard, J. & Stevenson, B. R. ZO-3, a novel member of the MAGUK protein family found at the tight junction, interacts with ZO-1 and occludin. J. Cell. Biol. 141, 199–208 (1998).
González-Mariscal, L., Betanzos, A. & Ávila-Flores, A. MAGUK proteins: Structure and role in the tight junction. Semin. Cell Dev. Biol. 11, 315–324 (2000).
Chu, J. J. H., Choo, B. G. H., Lee, J. W. M. & Ng, M. L. Actin filaments participate in West Nile (Sarafend) virus maturation process. J. Med. Virol. 71, 463–472 (2003).
Matter, K. & Balda, M. S. Signalling to and from tight junctions. Nat. Rev. Mol. Cell Biol. 4, 225–237 (2003).
Chen, X. & Macara, I. G. Par-3 controls tight junction assembly through the Rac exchange factor Tiam1. Nat. Cell Biol. 7, 262–269 (2005).
Kohjima, M. et al. PAR3β, a novel homologue of the cell polarity protein PAR3, localizes to tight junctions. Biochem. Biophys. Res. Commun. 299, 641–646 (2002).
Ebnet, K. et al. The junctional adhesion molecule (JAM) family members JAM-2 and JAM-3 associate with the cell polarity protein PAR-3: A possible role for JAMs in endothelial cell polarity. J. Cell. Sci. 116, 3879–3891 (2003).
Itoh, M. et al. Junctional adhesion molecule (JAM) binds to PAR-3 a possible mechanism for the recruitment of PAR-3 to tight junctions. J. Cell Biol. 154, 491–498 (2001).
Lubick, K. J. et al. Flavivirus antagonism of type I interferon signaling reveals prolidase as a regulator of IFNAR1 surface expression. Cell Host Microbe 18, 61–74 (2015).
Lopez-Denman, A. J. et al. Nucleocytoplasmic shuttling of the West Nile virus RNA-dependent RNA polymerase NS5 is critical to infection. Cell. Microbiol. 20, e12848 (2018).
Kapoor, M. et al. Association between NS3 and NS5 proteins of dengue virus type 2 in the putative RNA replicase is linked to differential phosphorylation of NS5. J. Biol. Chem. 270, 19100–19106 (1995).
Buckley, A., Gaidamovich, S., Turchinskaya, A. & Gould, E. A. Monoclonal antibodies identify the NS5 yellow fever virus non-structural protein in the nuclei of infected cells. J. Gen. Virol. 73(Pt 5), 1125–1130 (1992).
Latorre, I. J. et al. Viral oncoprotein-induced mislocalization of select PDZ proteins disrupts tight junctions and causes polarity defects in epithelial cells. J. Cell Sci. 118, 4283–4293 (2005).
Alsaleh, K. et al. The E glycoprotein plays an essential role in the high pathogenicity of European-Mediterranean IS98 strain of West Nile virus. Virology 492, 53–65 (2016).
Duhoo, Y. et al. High-throughput production of a new library of human single and tandem PDZ domains allows quantitative PDZ-peptide interaction screening through high-throughput holdup assay. Methods Mol. Biol. 2025, 439–476 (2019).
Choi, S. G. et al. Maximizing binary interactome mapping with a minimal number of assays. Nat. Commun. 10, 1–13 (2019).
Sambrook. Molecular cloning: A laboratory manual. Second edition. Volumes 1, 2, and 3. Current protocols in molecular biology. Cell 61, 17–18 (1989).
Hellemans, J., Mortier, G., De Paepe, A., Speleman, F. & Vandesompele, J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 8, R19 (2007).
Acknowledgements
This work was supported by the Institut Pasteur’s Transverse Research Programs PTR-N° 10-2016.
Author information
Authors and Affiliations
Contributions
Conceptualization, E.G., Y.J., N.P., N.W.; Formal analysis, E.G., C.C-S., Investigation, E.G., C.O., C.C-S., N.Z., C.K., J.C.; Funding acquisition, Y.J., N.P., N.W.; Methodology, E.G., C.C-S., Y.J., N.P., N.W.; Software, E.G., C.C-S.; Visualization, E.G.; Resources, Y.J., N.P., N.W.; Supervision, Y.J., N.P., N.W.; Writing-original draft, E.G., N.W.; Writing-review/editing, C.C-S., N.P.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Giraud, E., del Val, C.O., Caillet-Saguy, C. et al. Role of PDZ-binding motif from West Nile virus NS5 protein on viral replication. Sci Rep 11, 3266 (2021). https://doi.org/10.1038/s41598-021-82751-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-82751-x
This article is cited by
-
Quantitative fragmentomics allow affinity mapping of interactomes
Nature Communications (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.