Abstract
Use of menopausal hormone therapy (MHT) is associated with increased risk for breast cancer. However, the relevant mechanisms and its interaction with genetic variants are not fully understood. We conducted a genome-wide interaction analysis between MHT use and genetic variants for breast cancer risk in 27,585 cases and 34,785 controls from 26 observational studies. All women were post-menopausal and of European ancestry. Multivariable logistic regression models were used to test for multiplicative interactions between genetic variants and current MHT use. We considered interaction p-values < 5 × 10–8 as genome-wide significant, and p-values < 1 × 10–5 as suggestive. Linkage disequilibrium (LD)-based clumping was performed to identify independent candidate variants. None of the 9.7 million genetic variants tested for interactions with MHT use reached genome-wide significance. Only 213 variants, representing 18 independent loci, had p-values < 1 × 105. The strongest evidence was found for rs4674019 (p-value = 2.27 × 10–7), which showed genome-wide significant interaction (p-value = 3.8 × 10–8) with current MHT use when analysis was restricted to population-based studies only. Limiting the analyses to combined estrogen–progesterone MHT use only or to estrogen receptor (ER) positive cases did not identify any genome-wide significant evidence of interactions. In this large genome-wide SNP-MHT interaction study of breast cancer, we found no strong support for common genetic variants modifying the effect of MHT on breast cancer risk. These results suggest that common genetic variation has limited impact on the observed MHT–breast cancer risk association.
Similar content being viewed by others
Introduction
Breast cancer is one of the most common cancers in women. There were 268,600 new cases and 41,760 deaths due to breast cancer estimated in the U.S. in 20191. The use of menopausal hormone therapy (MHT) is associated with up to 23% increased risk of breast cancer. MHT use has been reduced among postmenopausal women since the report by the Women’s Health Initiative (WHI) clinical trial and observational study2,3 which has been subsequently confirmed by other studies and meta-analyses4,5. Breast cancer risk increases with longer duration of use6, and is higher for combined estrogen–progesterone MHT (EPT) use as compared with estrogen-only (ET) regimens4,5. Additionally, the association between MHT use and breast cancer may also differ by tumor molecular subtype. A prospective cohort study in UK found that current MHT use was associated with increased risk for estrogen receptor positive (ER+) breast cancers, but not with ER- breast cancers7. Several other observational studies also found that MHT use was associated with elevated risk of ER+ breast cancer8,9,10,11,12.
The biological mechanisms underlying the effect of MHT use on breast cancer risk is not fully understood. One proposed mechanism is that higher estrogen and progesterone levels increase the proliferation of breast epithelial cells, which results in accumulation of genetic mutations and insufficient DNA repair13,14, and therefore induces mutagenesis15,16. Genome-wide association studies (GWAS) have identified over 200 single nucleotide polymorphisms (SNPs) that are associated with invasive breast cancer risk17,18,19. Further analyses based on these GWAS findings have identified several genes that might interact with MHT use on breast cancer risk, including SNPs regulating the fibroblast growth factor receptor two (FGFR2) gene20, as well as SNPs close to the Kruppel like factor 4 (KLF4) gene and the insulin like growth-factor-binding protein 5 (IGFBP5) gene21,22,23. A meta-analysis of four genome-wide case-only interaction studies found suggestive evidence of interactions between MHT use and SNPs in genes related to transmembrane signaling and immune cell activation24. However, none of the findings reached genome-wide significance.
In the present study, we performed a comprehensive genome-wide interaction analysis of current MHT use by pooling individual-level data from 26 epidemiological studies. We also performed genome-wide interaction analysis of MHT use on ER+ breast cancer specifically.
Methods
Study population and data collection
Individual level data were pooled from 26 epidemiological studies, including eight population-based case–control studies, 13 nested studies from prospective cohort studies and five studies with mixed design from the Breast Cancer Association Consortium (BCAC) (Table S1). Data collection instruments for individual studies have been described previously19,23. Breast cancer cases were defined as incident invasive or in-situ breast tumors, confirmed by medical records, pathological reports or death certificates. Cases of benign breast disease or cases diagnosed more than five years before study enrollment were excluded.
Participants were excluded if they were male, pre-menopausal, of non-European ancestry, with unknown age at reference date, or missing information on MHT use. Reference date was defined as date of diagnosis for cases, and date of interview for controls. Menopausal status was reported at time of interview. For women with missing menopausal status, we assumed postmenopausal status for those who were > 54 years old. Only studies with information on MHT use in at least 150 breast cancer cases and 150 controls were included in the data analysis.
Ethnical approval and consent to participate
All participating studies were approved by the relevant ethics committees and informed consent was obtained from study participants.
Menopausal hormone therapy use definition
MHT use was defined as use for at least three months of any type of MHT, including EPT and ET. Current MHT use was defined as use at, or within the six months prior to the reference date. Former MHT use was defined as women who had a history of using MHT but had quit more than 6 months prior to the reference date.
Genotyping
Samples were genotyped by the Illumina custom iSelect genotyping array (iCOGs)25,26 or the Illumina OncoArray 500K (OncoArray)19,27. Details on genotyping, imputation and quality-control checks have been published previously19,26. For these analyses, 9680 cases and 10,598 controls were genotyped using iCOGs, and 17,905 cases and 24,187 controls were genotyped using OncoArray. Both datasets were imputed to the 1000 Genomes Phase 3 release28. For samples that were genotyped on both iCOGs and OncoArray, OncoArray data was used. SNPs were excluded if imputation r2 < 0.5 for iCOGs, and r2 < 0.8 for OncoArray. A total of 9,661,037 genetic variants (SNPs and indels) were included for analysis in both datasets.
Statistical analysis
We used multivariable logistic regression models to test for interaction between each genetic variant and current MHT use (compared to never users) on breast cancer risk, adjusting for age at reference date, study, former MHT use, an indicator for study design (1 for population-based case–control or prospective studies, 0 for non-population or mixed case–control studies), an interaction term of study design indicator and current MHT use to account for different main effect of current MHT use by study design, and principal components to account for potential population stratification29, thus fitting a model of the form:
Each genetic variant was assessed as a continuous variable in a log-additive odds ratio model. For genetic variants that were not directly genotyped, the expected number of copies of the variant allele (“dosage”) was used30. OncoArray and iCOGs datasets were analyzed separately, and platform-specific interaction parameter estimates (\({\beta }_{gc}\)) were combined using METAL31 to obtain summary estimates for each SNP. Similar analyses were also performed for EPT use only and for ER+ breast cancer. Q-Q plots were used to assess whether the distribution of the p-values indicated genomic inflation. A p-value at 5 × 10–8 was used as the genome-wide significance level32.
For variants reaching suggestive evidence of interaction (p < 1 × 10–5), we performed linkage disequilibrium (LD)-based clumping to identify independent loci that might interact with MHT use on breast cancer risk (SWISS version 1.0.05b). SNPs in LD (r2 > 0.1 based on the build-in 1000G_2014-11_EUR) within 1 Mb from the most significantly associated SNP were removed so that independent SNPs remained in each region.
We also performed sensitivity analysis among patients from the population-based studies only. All analyses were performed using R version 3.6.1 unless otherwise specified.
Results
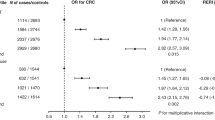
A total of 62,370 post-menopausal women from 26 studies (27,585 cases and 34,785 controls), were included in the analyses (Table S1). Cases were slightly older (mean age: 64 years) than controls (mean age: 63 years). Current use of MHT was more common among breast cancer cases (34%) than controls (28%), showing a suggestive increased breast cancer risk (OR = 1.16; 95% CI: 0.99, 1.36; Fig. 1A). A total of 20,131 cases and 22,601 controls from 18 studies also had information on current use of EPT. Current EPT use was more common among cases (19%) than controls (13%) and was associated with an estimated 48% risk increase of breast cancer, compared to non-EPT users (OR: 1.48; 95% CI: 1.29, 1.70; Fig. 1B).
A total of 9,661,271 SNPs and indels were successfully imputed from both the OncoArray and iCOGs genotyping platforms and were included in the combined analysis. We did not observe any interactions between variants and current MHT use at genome-wide significance level (p-value < 5 × 10–8, Fig. 2A). 213 SNPs had suggestive evidence of interaction with MHT use on breast cancer risk (p-value < 1 × 10–5). After LD-based clumping, 18 independent SNPs remained, none of which were in LD with currently known breast cancer risk GWAS loci (Table 1). The strongest evidence of interaction was for SNP rs4674019, located at chromosome 2q35 (p-value = 2.27 × 10–7). When restricting the analyses to population-based studies only (23,063 cases and 30,250 controls), this same SNP rs4674019 showed statistically significantly interaction with current MHT use on breast cancer risk (p-value = 3.75 × 10–8; Fig. S1).
Similarly, we did not observe any genome-wide significant interactions between SNPs and combined EPT use on breast cancer risk (Fig. 2B). There were 71 SNPs that reached suggestive significance level at p-value < 1 × 10–5. After LD-based clumping, 21 independent SNPs showed suggestive interactions (Table 2). The strongest evidence of interaction was for SNP rs4865075, located on chromosome 4q12 (p-value = 5.5 × 10–7). Sensitivity analysis using population-based studies only did not find statistically significant interactions.
Restricting our cases to those with ER+ breast cancer did not result in any genome-wide significant findings (Figs. S2 and S3). No genomic inflation was observed in primary or subgroup analyses (Figs. S1, S2 and S3).
Discussion
In this large genome-wide analysis of postmenopausal women of European ancestry, we did not identify any genetic variants that were strong modifiers of the association between current MHT use on breast cancer risk. Although the interaction between SNP rs4674019 and current MHT use was statistically significant among population-based studies only, the variate allele frequency is relatively rare (EAF = 5%) and needs further validation.
Consistent with previous literature2,3,33, we found that current use of MHT, and in particular current EPT use, was associated with an increased risk of breast cancer for postmenopausal women. The mechanisms underlying this association are not fully understood. It has been hypothesized that estrogen stimulates cell proliferation through ERα-mediated hormone activity and increases mutation rates through a cytochrome P450-mediated metabolic activation that results in DNA damage34. In addition, the risk associated with ER+ breast cancer is substantially higher than for ER- breast cancer, particularly for EPT use, suggesting an ER-dependent pathway5. In vitro and in vivo studies found that estradiol and 4-OH-estradiol, metabolites of estrogen, may induce mutations and damage DNA by forming DNA adducts to bind to adenine and guanine on the DNA backbone35,36. The role of progestogens in human breast carcinogenesis is less clear, although it has been suggested that synthetic progestogens are pro-proliferative and may thus promote cancer cell growth37,38.
Although MHT use has been found to be associated with increased breast cancer risk in both epidemiologic and experimental studies, no published studies to date have identified genome-wide significant interactions for breast cancer risk between candidate single variants and MHT use among postmenopausal women39,40. In a previous two-stage GWAS interaction analysis among ~ 2700 cases and ~ 2700 controls, five SNPs had suggestive evidence of interaction with current MHT use; but none of them reached genome-wide significance41. A meta-analysis of genome-wide case-only studies in 2920 cases also found no statistically significant interactions between SNPs and MHT use on breast cancer overall or by subtype24. Although our study had a larger sample size and increased statistical power than previous genome-wide analyses, we similarly did not find any genome-wide statistically significant interactions between genetic variants and MHT use in this study, and we further did not replicate previously suggested SNPs (data not shown).
The region for which the strongest evidence of interaction with current MHT use on breast cancer risk was observed (lead SNP rs4674019), was also implicated in the analysis restricted to combined EPT use only (p-value = 4.5 × 10–6). The rs4674019 SNP is an intronic variant in the coding region for the long intergenic non-protein coding RNA 607 (LINC00607). Although the functionality of long non-coding RNAs is still not clear, it has been recently recognized that abnormal expression of long non-coding RNAs may play an important role in cell cycle control and cell differentiation, which is related to cancer and neurodegenerative disease42,43,44. Expression levels of LINC00607 were found to be significantly downregulated among lung adenocarcinoma tissues, compared to adjacent tissues45. Other GWAS have shown genetic variants in the LINC00607 gene to be associated with height in people of European ancestry46. Previous evidence for long noncoding RNAs in relation to breast cancer risk is limited; but it is possible that changes in exogenous hormone levels due to MHT use result in differential expression that eventually leads to tumorigenesis.
We also observed suggestive evidence of interaction between current use of both MHT and EPT and rs146251672. SNP rs146251672 is located in the intronic region for the prickle planar cell polarity protein 2 (PRICKLE2) gene on chromosome 3. PRICKLE2 encodes a non-canonical Wnt signaling protein that mediates feedback amplification to generate asymmetric planar cell polarity (PCP) signaling47. The Wnt pathway has been found to be activated in more than half of breast tumors, and is associated with lower overall survival for breast cancer patients48. In particular, the upregulation of the Wnt/PCP pathway has been suggested to be associated with more malignant phenotypes, such as abnormal tissue polarity, invasion and metastasis49. Exposure to estrogen has been associated with accelerated tumor formation in ER-knockout/Wnt-1 mice36. It is plausible that MHT acts partially through the alternative Wnt pathway rather than ER-dependent pathways to promote breast tumor development.
This study constitutes the largest genome-wide interaction analysis for current MHT use and breast cancer risk in postmenopausal women to date. We analyzed data from more than 62,000 women for whom we had both MHT use and genotypes from more than 9.6 million genetic variants. We controlled our analysis for potential confounding by population stratification by adjusting for principal components. We performed LD-based clumping, which accounted for correlations between genotypes to identify the strongest signal in each independent region, providing more targeted variants and regions for future investigation.
There are some limitations to our study. We used a single binary definition of current MHT use within 6 months prior to reference date and could not evaluate other measures such as age at MHT initiation or duration of MHT use. This could lead to some exposure misclassification, particularly for the non-population based studies, where it is possible that those cases had stopped their MHT use at time of recruitment and were classified as non-current users. Such misclassification would have attenuated the main effect of MHT and reduced our statistical power to detect any interactions. In our sensitivity analysis using population-based studies only, we found stronger interactions between the lead SNPs and MHT use. However, given a smaller sample size in the sensitivity analysis, it is possible that we did not have sufficient statistical power to detect any other potential interactions. We assumed no significant interactions between SNPs and former MHT use, and only adjusted for potential confounding from the main effect of former MHT use in the model based on previous evidence. It is possible that comparing the interaction effect of current MHT use to a combined reference group of never and former users may attenuate the point estimates of the interaction term. The use of estrogen only hormone therapy (ET) was also not available among the study participants, although the statistical power might be further limited since the main association of ET and breast cancer risk is much smaller than EPT use5. Although the impact may be small given our case–control study design and large sample size, it is still possible that the observed SNP-MHT interaction was due to the interaction between SNPs and potential uncontrolled confounders of MHT use that were not available in our study50, such as tolerance of menopausal symptoms or socioeconomic status. In addition, our study sample only included women of European ancestry, and thus, our findings may not be generalizable to other race/ethnicity groups.
It is important to note that the lack of statistical interaction, on the log-scale, does not necessarily imply a lack of biological interaction. The results are consistent with a model in which the effects of genetic variants and MHT use combine multiplicatively on risk, which could still indicate important interactions at a functional level. Overall, our results suggest that it is not necessary to include interaction variables for G × MHT use in development of breast cancer risk prediction models. Although our results suggested that potential interaction effect between SNP rs4674019 and current MHT, further validation is needed. Several suggestive interactions also warrant further investigations in independent studies.
Conclusion
In this large genome-wide SNP-MHT interaction study of breast cancer, we found no strong support for common genetic variants modifying the effect of MHT on breast cancer risk. These results suggest that common genetic variation has limited impact on the observed MHT–breast cancer risk association.
Data availability
The data that support the findings of this study are available from the Breast Cancer Association Consortium but restrictions apply to the availability of these data, which were used under license for the current study, and so are not publicly available. Data are however available from the authors upon reasonable request and with permission of the Breast Cancer Association Consortium.
Abbreviations
- MHT:
-
Menopausal hormone therapy
- LD:
-
Linkage disequilibrium
- SNP:
-
Single nucleotide polymorphism
- EPT:
-
Estrogen–progesterone menopausal hormone therapy
- ET:
-
Estrogen-only menopausal hormone therapy
- ER+:
-
Estrogen receptor positive
- GWAS:
-
Genome-wide association studies
- BCAC:
-
Breast Cancer Association Consortium
- iCOGs:
-
Samples genotyped using the Illumina custom iSelect genotyping array
- OncoArray:
-
Samples genotyped using the Illumina 500K array
References
Siegel, R. L., Miller, K. D. & Jemal, A. Cancer statistics, 2019. CA Cancer J. Clin. 69, 7–34 (2019).
Chlebowski, R. T. et al. Breast cancer after use of estrogen plus progestin in postmenopausal women. N. Engl. J. Med. 360, 573–587 (2009).
Prentice, R. L. et al. Conjugated equine estrogens and breast cancer risk in the Women’s Health Initiative clinical trial and observational study. Am. J. Epidemiol. 167, 1407–1415 (2008).
Anothaisintawee, T. et al. Risk factors of breast cancer: A systematic review and meta-analysis. Asia Pac. J. Public Health 25, 368–387 (2013).
Collaborative Group on Hormonal Factors in Breast C. Type and timing of menopausal hormone therapy and breast cancer risk: Individual participant meta-analysis of the worldwide epidemiological evidence. Lancet. 394, 1159–1168 (2019).
Breast cancer and hormone replacement therapy: Collaborative reanalysis of data from 51 epidemiological studies of 52,705 women with breast cancer and 108,411 women without breast cancer. Collaborative Group on Hormonal Factors in Breast Cancer. Lancet 350, 1047–1059 (1997).
Beral, V., Reeves, G., Bull, D. & Green, J. Million Women Study C. Breast cancer risk in relation to the interval between menopause and starting hormone therapy. J. Natl. Cancer Inst. 103, 296–305 (2011).
Chen, W. Y. et al. Association of hormone replacement therapy to estrogen and progesterone receptor status in invasive breast carcinoma. Cancer 101, 1490–1500 (2004).
Gertig, D. M. et al. Hormone therapy and breast cancer: What factors modify the association?. Menopause 13, 178–184 (2006).
Salagame, U., Banks, E., O’Connell, D. L., Egger, S. & Canfell, K. Menopausal hormone therapy use and breast cancer risk by receptor subtypes: Results from the New South Wales Cancer Lifestyle and EvaluAtion of Risk (CLEAR) study. PLoS ONE 13, e0205034 (2018).
Saxena, T. et al. Menopausal hormone therapy and subsequent risk of specific invasive breast cancer subtypes in the California Teachers Study. Cancer Epidemiol. Biomarkers Prevent. 19, 2366–2378 (2010).
Setiawan, V. W. et al. Breast cancer risk factors defined by estrogen and progesterone receptor status: The multiethnic cohort study. Am. J. Epidemiol. 169, 1251–1259 (2009).
Mello, M. L., Vidal, B. C., Russo, I. H., Lareef, M. H. & Russo, J. DNA content and chromatin texture of human breast epithelial cells transformed with 17-beta-estradiol and the estrogen antagonist ICI 182,780 as assessed by image analysis. Mutat. Res. 617, 1–7 (2007).
Saeed, M. et al. Formation of depurinating N3Adenine and N7Guanine adducts by MCF-10F cells cultured in the presence of 4-hydroxyestradiol. Int. J. Cancer 120, 1821–1824 (2007).
Persson, I. Estrogens in the causation of breast, endometrial and ovarian cancers—Evidence and hypotheses from epidemiological findings. J. Steroid Biochem. Mol. Biol. 74, 357–364 (2000).
Yager, J. D. & Liehr, J. G. Molecular mechanisms of estrogen carcinogenesis. Annu. Rev. Pharmacol. Toxicol. 36, 203–232 (1996).
Easton, D. F. et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature 447, 1087–1093 (2007).
Hunter, D. J. et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat. Genet. 39, 870–874 (2007).
Michailidou, K. et al. Association analysis identifies 65 new breast cancer risk loci. Nature 551, 92–94 (2017).
Prentice, R. L. et al. Variation in the FGFR2 gene and the effects of postmenopausal hormone therapy on invasive breast cancer. Cancer Epidemiol. Biomarkers Prevent. 18, 3079–3085 (2009).
Ghoussaini, M. et al. Evidence that breast cancer risk at the 2q35 locus is mediated through IGFBP5 regulation. Nat. Commun. 4, 4999 (2014).
Nickels, S. et al. Evidence of gene-environment interactions between common breast cancer susceptibility loci and established environmental risk factors. PLoS Genet. 9, e1003284 (2013).
Kapoor, P. M. et al. Assessment of interactions between 205 breast cancer susceptibility loci and 13 established risk factors in relation to breast cancer risk, in the Breast Cancer Association Consortium. Int. J. Epidemiol. 49, 216–232 (2020).
Rudolph, A. et al. Genetic modifiers of menopausal hormone replacement therapy and breast cancer risk: A genome-wide interaction study. Endocr. Relat. Cancer 20, 875–887 (2013).
Michailidou, K. et al. Genome-wide association analysis of more than 120,000 individuals identifies 15 new susceptibility loci for breast cancer. Nat. Genet. 47, 373–380 (2015).
Michailidou, K. et al. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat Genet 45, 353-361e1-2 (2013).
Amos, C. I. et al. The OncoArray Consortium: A network for understanding the genetic architecture of common cancers. Cancer Epidemiol. Biomarkers Prevent. 26, 126–135 (2017).
Genomes Project C et al. A global reference for human genetic variation. Nature 526, 68–74 (2015).
Price, A. L. et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904–909 (2006).
Jiao, S., Hsu, L., Hutter, C. M. & Peters, U. The use of imputed values in the meta-analysis of genome-wide association studies. Genet. Epidemiol. 35, 597–605 (2011).
Willer, C. J., Li, Y. & Abecasis, G. R. METAL: Fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26, 2190–2191 (2010).
Dudbridge, F. & Gusnanto, A. Estimation of significance thresholds for genomewide association scans. Genet. Epidemiol. 32, 227–234 (2008).
Rossouw, J. E. et al. Risks and benefits of estrogen plus progestin in healthy postmenopausal women: Principal results From the Women’s Health Initiative randomized controlled trial. JAMA 288, 321–333 (2002).
Russo, J. & Russo, I. H. The role of estrogen in the initiation of breast cancer. J. Steroid Biochem. Mol. Biol. 102, 89–96 (2006).
Cavalieri, E. et al. Catechol estrogen quinones as initiators of breast and other human cancers: Implications for biomarkers of susceptibility and cancer prevention. Biochim. Biophys. Acta 1766, 63–78 (2006).
Yue, W. et al. Effects of estrogen on breast cancer development: Role of estrogen receptor independent mechanisms. Int. J. Cancer 127, 1748–1757 (2010).
Horwitz, K. B. & Sartorius, C. A. Progestins in hormone replacement therapies reactivate cancer stem cells in women with preexisting breast cancers: A hypothesis. J. Clin. Endocrinol. Metab. 93, 3295–3298 (2008).
Santen, R. J. Risk of breast cancer with progestins: Critical assessment of current data. Steroids 68, 953–964 (2003).
Harlid, S. et al. Interactive effect of genetic susceptibility with height, body mass index, and hormone replacement therapy on the risk of breast cancer. BMC Womens Health 12, 17 (2012).
Travis, R. C. et al. Million Women Study C. Gene-environment interactions in 7610 women with breast cancer: Prospective evidence from the Million Women Study. Lancet 375, 2143–2151 (2010).
Hein, R. et al. A genome-wide association study to identify genetic susceptibility loci that modify ductal and lobular postmenopausal breast cancer risk associated with menopausal hormone therapy use: A two-stage design with replication. Breast Cancer Res. Treat. 138, 529–542 (2013).
Dey, B. K., Mueller, A. C. & Dutta, A. Long non-coding RNAs as emerging regulators of differentiation, development, and disease. Transcription. 5, e944014 (2014).
Ricciuti, B. et al. Long noncoding RNAs: New insights into non-small cell lung cancer biology, diagnosis and therapy. Med. Oncol. 33, 18 (2016).
Zhao, W., An, Y., Liang, Y. & Xie, X. W. Role of HOTAIR long noncoding RNA in metastatic progression of lung cancer. Eur. Rev. Med. Pharmacol. Sci. 18, 1930–1936 (2014).
Zhao, B. et al. Expression profiles of long noncoding RNAs in lung adenocarcinoma. Onco Targets Ther. 11, 5383–5390 (2018).
Kichaev, G. et al. Leveraging polygenic functional enrichment to improve GWAS power. Am. J. Hum. Genet. 104, 65–75 (2019).
Tree, D. R. et al. Prickle mediates feedback amplification to generate asymmetric planar cell polarity signaling. Cell 109, 371–381 (2002).
Khramtsov, A. I. et al. Wnt/beta-catenin pathway activation is enriched in basal-like breast cancers and predicts poor outcome. Am. J. Pathol. 176, 2911–2920 (2010).
Katoh, M. WNT/PCP signaling pathway and human cancer (review). Oncol. Rep. 14, 1583–1588 (2005).
Vanderweele, T. J., Ko, Y. A. & Mukherjee, B. Environmental confounding in gene-environment interaction studies. Am. J. Epidemiol. 178, 144–152 (2013).
Acknowledgements
We thank all the individuals who took part in these studies and all the researchers, clinicians, technicians and administrative staff who have enabled this work to be carried out. The COGS study would not have been possible without the contributions of the following: Andrew Lee, and Ed Dicks, Craig Luccarini and the staff of the Centre for Genetic Epidemiology Laboratory, Javier Benitez, Anna Gonzalez-Neira and the staff of the CNIO genotyping unit, Jacques Simard and Daniel C. Tessier, Francois Bacot, Daniel Vincent, Sylvie LaBoissière and Frederic Robidoux and the staff of the McGill University and Génome Québec Innovation Centre, Stig E. Bojesen, Sune F. Nielsen, Borge G. Nordestgaard, and the staff of the Copenhagen DNA laboratory, and Julie M. Cunningham, Sharon A. Windebank, Christopher A. Hilker, Jeffrey Meyer and the staff of Mayo Clinic Genotyping Core Facility. ABCFS thank Maggie Angelakos, Judi Maskiell, Gillian Dite. CBCS thanks study participants, co-investigators, collaborators and staff of the Canadian Breast Cancer Study, and project coordinators Agnes Lai and Celine Morissette. Investigators from the CPS-II cohort thank the participants and Study Management Group for their invaluable contributions to this research. They also acknowledge the contribution to this study from central cancer registries supported through the Centers for Disease Control and Prevention National Program of Cancer Registries, as well as cancer registries supported by the National Cancer Institute Surveillance Epidemiology and End Results program. We thank the participants and the investigators of EPIC (European Prospective Investigation into Cancer and Nutrition). The GENICA Network: Dr. Margarete Fischer-Bosch-Institute of Clinical Pharmacology, Stuttgart, and University of Tübingen, Germany [Hiltrud Brauch, WYL, RH], German Cancer Consortium (DKTK) and German Cancer Research Center (DKFZ), Partner Site Tübingen [Hiltrud Brauch], Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany's Excellence Strategy—EXC 2180—390900677 [Hiltrud Brauch], Department of Internal Medicine, Evangelische Kliniken Bonn gGmbH, Johanniter Krankenhaus, Bonn, Germany [Yon-Dschun Ko, Christian Baisch], Institute of Pathology, University of Bonn, Germany [Hans-Peter Fischer], Molecular Genetics of Breast Cancer, Deutsches Krebsforschungszentrum (DKFZ), Heidelberg, Germany [UH], Institute for Prevention and Occupational Medicine of the German Social Accident Insurance, Institute of the Ruhr University Bochum (IPA), Bochum, Germany [TB, Beate Pesch, Sylvia Rabstein, Anne Lotz]; and Institute of Occupational Medicine and Maritime Medicine, University Medical Center Hamburg-Eppendorf, Germany [Volker Harth]. KARMA and SASBAC thank the Swedish Medical Research Counsel. MARIE thanks Petra Seibold, Dieter Flesch-Janys, Judith Heinz, Nadia Obi, Alina Vrieling, Sabine Behrens, Ursula Eilber, Muhabbet Celik, Til Olchers and Stefan Nickels. The MCCS was made possible by the contribution of many people, including the original investigators, the teams that recruited the participants and continue working on follow-up, and the many thousands of Melbourne residents who continue to participate in the study. We thank the coordinators, the research staff and especially the MMHS participants for their continued collaboration on research studies in breast cancer. MSKCC thanks Marina Corines, Lauren Jacobs. `For NHS and NHS2 the study protocol was approved by the institutional review boards of the Brigham and Women’s Hospital and Harvard T.H. Chan School of Public Health, and those of participating registries as required. We would like to thank the participants and staff of the NHS and NHS2 for their valuable contributions as well as the following state cancer registries for their help: AL, AZ, AR, CA, CO, CT, DE, FL, GA, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, WY. The authors assume full responsibility for analyses and interpretation of these data. The OFBCR thanks Teresa Selander, Nayana Weerasooriya and Steve Gallinger. ORIGO thanks E. Krol-Warmerdam, and J. Blom for patient accrual, administering questionnaires, and managing clinical information. The LUMC survival data were retrieved from the Leiden hospital-based cancer registry system (ONCDOC) with the help of Dr. J. Molenaar. PBCS thanks Louise Brinton, Mark Sherman, Neonila Szeszenia-Dabrowska, Beata Peplonska, Witold Zatonski, Pei Chao, Michael Stagner. PROCAS thanks NIHR for funding. UCIBCS thanks Irene Masunaka. UKBGS thanks Breast Cancer Now and the Institute of Cancer Research for support and funding of the Generations Study, and the study participants, study staff, and the doctors, nurses and other health care providers and health information sources who have contributed to the study. We acknowledge NHS funding to the Royal Marsden/ICR NIHR Biomedical Research Centre. We acknowledge funding to the Manchester NIHR Biomedical Research Centre (IS-BRC-1215-20007). The authors thank the WHI investigators and staff for their dedication and the study participants for making the program possible.
Disclaimer
Where authors are identified as personnel of the International Agency for Research on Cancer/World Health Organization, the authors alone are responsible for the views expressed in this article and they do not necessarily represent the decisions, policy or views of the International Agency for Research on Cancer/World Health.
Funding
BCAC is funded by Cancer Research UK [C1287/A16563, C1287/A10118], the European Union's Horizon 2020 Research and Innovation Programme (grant numbers 634935 and 633784 for BRIDGES and B-CAST respectively), and by the European Community´s Seventh Framework Programme under grant agreement number 223175 (grant number HEALTH-F2-2009-223175) (COGS). The EU Horizon 2020 Research and Innovation Programme funding source had no role in study design, data collection, data analysis, data interpretation or writing of the report. Genotyping of the OncoArray was funded by the NIH Grant U19 CA148065, and Cancer UK Grant C1287/A16563 and the PERSPECTIVE project supported by the Government of Canada through Genome Canada and the Canadian Institutes of Health Research (grant GPH-129344) and, the Ministère de l’Économie, Science et Innovation du Québec through Genome Québec and the PSRSIIRI-701 grant, and the Quebec Breast Cancer Foundation. Funding for the iCOGS infrastructure came from: the European Community's Seventh Framework Programme under grant agreement no. 223175 (HEALTH-F2-2009-223175) (COGS), Cancer Research UK (C1287/A10118, C1287/A10710, C12292/A11174, C1281/A12014, C5047/A8384, C5047/A15007, C5047/A10692, C8197/A16565), the National Institutes of Health (CA128978) and Post-Cancer GWAS initiative (1U19 CA148537, 1U19 CA148065 and 1U19 CA148112—the GAME-ON initiative), the Department of Defense (W81XWH-10-1-0341), the Canadian Institutes of Health Research (CIHR) for the CIHR Team in Familial Risks of Breast Cancer, and Komen Foundation for the Cure, the Breast Cancer Research Foundation, and the Ovarian Cancer Research Fund. The DRIVE Consortium was funded by U19 CA148065. CBCS is funded by the Canadian Cancer Society (grant # 313404) and the Canadian Institutes of Health Research. The CECILE study was supported by Fondation de France, Institut National du Cancer (INCa), Ligue Nationale contre le Cancer, Agence Nationale de Sécurité Sanitaire, de l'Alimentation, de l'Environnement et du Travail (ANSES), Agence Nationale de la Recherche (ANR). The American Cancer Society funds the creation, maintenance, and updating of the CPS-II cohort. The coordination of EPIC is financially supported by International Agency for Research on Cancer (IARC) and also by the Department of Epidemiology and Biostatistics, School of Public Health, Imperial College London which has additional infrastructure support provided by the NIHR Imperial Biomedical Research Centre (BRC). The national cohorts are supported by: Danish Cancer Society (Denmark); Ligue Contre le Cancer, Institut Gustave Roussy, Mutuelle Générale de l’Education Nationale, Institut National de la Santé et de la Recherche Médicale (INSERM) (France); German Cancer Aid, German Cancer Research Center (DKFZ), German Institute of Human Nutrition Potsdam- Rehbruecke (DIfE), Federal Ministry of Education and Research (BMBF) (Germany); Associazione Italiana per la Ricerca sul Cancro-AIRC-Italy, Compagnia di SanPaolo and National Research Council (Italy); Dutch Ministry of Public Health, Welfare and Sports (VWS), Netherlands Cancer Registry (NKR), LK Research Funds, Dutch Prevention Funds, Dutch ZON (Zorg Onderzoek Nederland), World Cancer Research Fund (WCRF), Statistics Netherlands (The Netherlands); Health Research Fund (FIS)—Instituto de Salud Carlos III (ISCIII), Regional Governments of Andalucía, Asturias, Basque Country, Murcia and Navarra, and the Catalan Institute of Oncology—ICO (Spain); Swedish Cancer Society, Swedish Research Council and County Councils of Skåne and Västerbotten (Sweden); Cancer Research UK (14136 to EPIC-Norfolk; C8221/A29017 to EPIC-Oxford), Medical Research Council (1000143 to EPIC-Norfolk; MR/M012190/1 to EPIC-Oxford). (United Kingdom).The GENICA was funded by the Federal Ministry of Education and Research (BMBF) Germany grants 01KW9975/5, 01KW9976/8, 01KW9977/0 and 01KW0114, the Robert Bosch Foundation, Stuttgart, Deutsches Krebsforschungszentrum (DKFZ), Heidelberg, the Institute for Prevention and Occupational Medicine of the German Social Accident Insurance, Institute of the Ruhr University Bochum (IPA), Bochum, as well as the Department of Internal Medicine, Evangelische Kliniken Bonn gGmbH, Johanniter Krankenhaus, Bonn, Germany. The KARMA study was supported by Märit and Hans Rausings Initiative Against Breast Cancer. The MARIE study was supported by the Deutsche Krebshilfe e.V. [70-2892-BR I, 106332, 108253, 108419, 110826, 110828], the Hamburg Cancer Society, the German Cancer Research Center (DKFZ) and the Federal Ministry of Education and Research (BMBF) Germany [01KH0402]. The MCBCS was supported by the NIH grants CA192393, CA116167, CA176785 an NIH Specialized Program of Research Excellence (SPORE) in Breast Cancer [CA116201], and the Breast Cancer Research Foundation and a generous gift from the David F. and Margaret T. Grohne Family Foundation. The Melbourne Collaborative Cohort Study (MCCS) cohort recruitment was funded by VicHealth and Cancer Council Victoria. The MCCS was further augmented by Australian National Health and Medical Research Council grants 209057, 396414 and 1074383 and by infrastructure provided by Cancer Council Victoria. Cases and their vital status were ascertained through the Victorian Cancer Registry and the Australian Institute of Health and Welfare, including the National Death Index and the Australian Cancer Database. The MEC was supported by NIH grants CA63464, CA54281, CA098758, CA132839 and CA164973. The MISS study is supported by funding from ERC-2011-294576 Advanced grant, Swedish Cancer Society, Swedish Research Council, Local hospital funds, Berta Kamprad Foundation, Gunnar Nilsson. The MMHS study was supported by NIH grants CA97396, CA128931, CA116201, CA140286 and CA177150. The Carolina Breast Cancer Study (NCBCS) was funded by Komen Foundation, the National Cancer Institute (P50 CA058223, U54 CA156733, U01 CA179715), and the North Carolina University Cancer Research Fund. The NHS was supported by NIH grants P01 CA87969, UM1 CA186107, and U19 CA148065. The NHS2 was supported by NIH grants UM1 CA176726 and U19 CA148065. The PBCS was funded by Intramural Research Funds of the National Cancer Institute, Department of Health and Human Services, USA. Genotyping for PLCO was supported by the Intramural Research Program of the National Institutes of Health, NCI, Division of Cancer Epidemiology and Genetics. The PLCO is supported by the Intramural Research Program of the Division of Cancer Epidemiology and Genetics and supported by contracts from the Division of Cancer Prevention, National Cancer Institute, National Institutes of Health. PROCAS is funded from NIHR grant PGfAR 0707-10031. The SASBAC study was supported by funding from the Agency for Science, Technology and Research of Singapore (A*STAR), the US National Institute of Health (NIH) and the Susan G. Komen Breast Cancer Foundation. The SMC is funded by the Swedish Cancer Foundation and the Swedish Research Council (VR 2017-00644) grant for the Swedish Infrastructure for Medical Population-based Life-course Environmental Research (SIMPLER). The UCIBCS component of this research was supported by the NIH [CA58860, CA92044] and the Lon V Smith Foundation [LVS39420]. The UKBGS is funded by Breast Cancer Now and the Institute of Cancer Research (ICR), London. ICR acknowledges NHS funding to the NIHR Biomedical Research Centre. The USRT Study was funded by Intramural Research Funds of the National Cancer Institute, Department of Health and Human Services, USA. The WHI program is funded by the National Heart, Lung, and Blood Institute, the US National Institutes of Health and the US Department of Health and Human Services (HHSN268201100046C, HHSN268201100001C, HHSN268201100002C, HHSN268201100003C, HHSN268201100004C and HHSN271201100004C). This work was also funded by NCI U19 CA148065-01.
Author information
Authors and Affiliations
Contributions
X.W. and S.L. formed the research proposal, analyzed and interpreted the data. X.W., S.L., P.M.K., P.L.A., R.L.M., R.K. and J.C.C. were major contributors in writing the manuscript. J.D., A.M.D., Q.W., M.L., K.M., M.K.B., K.J.A., R.A.M., A.B., D.G.L., E.C., P.G., T.T., C.M., L.R.T., A.V.P., L.D., R.K., R.H., W.Y.L., T.B., U.H., K.C., M.G., P.H., M.K., A.J., H.B., F.J.C., N.L.L., J.E.O., K.J.R., G.G.G., R.J.M., M.C.S., L.L.M., L.R.W., C.A.H., H.O., A.A., U.K., P.W., C.S., S.J.W., C.M.V., C.M.P., A.F.O., M.A.T., D.J.H., A.H.E., R.M.T., K.B., I.L.A., J.F., S.J.C., T.U.A., M.G.C., D.G.E., W.G.N., E.M.V., A.H., A.W., N.H., H.A.C., A.Z., M.E.J., N.O., M.J.S., A.J.S., C.M.K., M.L., R.L.P., D.F.E. collected and provided genotyping and epidemiologic data, and provided comments on the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wang, X., Kapoor, P.M., Auer, P.L. et al. Genome-wide interaction analysis of menopausal hormone therapy use and breast cancer risk among 62,370 women. Sci Rep 12, 6199 (2022). https://doi.org/10.1038/s41598-022-10121-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-10121-2
This article is cited by
-
The 100 top-cited articles in menopausal syndrome: a bibliometric analysis
Reproductive Health (2024)
-
Polygenic scores in cancer
Nature Reviews Cancer (2023)
-
A genome-wide gene-environment interaction study of breast cancer risk for women of European ancestry
Breast Cancer Research (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.