Abstract
Smart healthcare systems that make use of abundant health data can improve access to healthcare services, reduce medical costs and provide consistently high-quality patient care. Medical dialogue systems that generate medically appropriate and human-like conversations have been developed using various pre-trained language models and a large-scale medical knowledge base based on Unified Medical Language System (UMLS). However, most of the knowledge-grounded dialogue models only use local structure in the observed triples, which suffer from knowledge graph incompleteness and hence cannot incorporate any information from dialogue history while creating entity embeddings. As a result, the performance of such models decreases significantly. To address this problem, we propose a general method to embed the triples in each graph into large-scalable models and thereby generate clinically correct responses based on the conversation history using the recently recently released MedDialog(EN) dataset. Given a set of triples, we first mask the head entities from the triples overlapping with the patient’s utterance and then compute the cross-entropy loss against the triples’ respective tail entities while predicting the masked entity. This process results in a representation of the medical concepts from a graph capable of learning contextual information from dialogues, which ultimately aids in leading to the gold response. We also fine-tune the proposed Masked Entity Dialogue (MED) model on smaller corpora which contain dialogues focusing only on the Covid-19 disease named as the Covid Dataset. In addition, since UMLS and other existing medical graphs lack data-specific medical information, we re-curate and perform plausible augmentation of knowledge graphs using our newly created Medical Entity Prediction (MEP) model. Empirical results on the MedDialog(EN) and Covid Dataset demonstrate that our proposed model outperforms the state-of-the-art methods in terms of both automatic and human evaluation metrics.
Similar content being viewed by others
Introduction
Health conversational assistants (or Chatbots) for patients act as a medical consultant, which usually provides simple and relevant measures to avoid infection from various diseases. They are designed to interact with patients in real time, inquiring about their medical issues and past records while also attempting to make useful suggestions. This enabled clinicians to use telemedicine, which is critical in pandemic circumstances where physical contact between patients and doctors is restricted. Chatbots are progressively being used to help people communicate in the open domain settings in order to exchange information1,2,3 and to assist professionals in completing a specific activity4,5. A medical dialogue system can serve as a physician’s assistant, inquiring about the patient’s medical, medication, social, personal, and family history, as well as a thorough review of symptoms and possibly a physical examination6,7,8. As a result, intelligent medical dialogue systems have the potential to reduce the workload of physicians.
More access to external sources of medical knowledge may aid in resolving the problem of limited semantic understanding during response generation. For example, to comprehend the utterance-response pair shown in Table 1, it is demonstrated how, rather than expecting a generic response, we might achieve the ground truth response by combining external medical information. Different weights should be assigned to different tail entities extracted from the triples acquired for the words dry eyes and cataract based on their presence in the gold response which have been linked using different relations. Tail entities, such as blurring of vision, cornea should certainly be given more importance due to their presence in the target response. Recently, Li et al.9 proposed a reasoning method over knowledge graphs for medical dialogue generation using a large-scale medical corpus to deliver appropriate medical responses, but they only learn local embeddings for the entities in different triples, ignoring the contextual information.
A medical knowledge graph is a multidimensional graph with nodes and edges, that represent the relationships between different medical concepts in several biomedical domains. However, the current medical graphs lack local information contained in medical dialogues i.e there is no representation of medical entities and relations with respect to the conversations. A team of subject-matter experts may be hired to map all of the connections between various treatments, illnesses, and other scientific concepts in order to address these issues. Unfortunately, the cost of hiring a group of medical experts to accomplish this task is rather high. In that instance, we may rapidly and simply extract those relations using NLP approaches.
In this study, we propose a novel method for medical dialogue generation called MED, to generate efficient responses by leveraging both the context information and relevant medical knowledge graphs using pre-trained language models. First, we build knowledge graphs using UMLS10 database and BERT-based entity prediction models as described in section “Construction of the medical entity graph”. Second, we employ our MED model to produce appropriate responses using these knowledge graphs. To be more specific, we mask the head entities from triples and use the masked sequence as the new input. The aim is to predict the tail entities that correspond to the masked head entities while also generating the response using the hidden states of the masked input sequence. In order to achieve this, we build models using the MedDialog(EN) corpus and the Covid Dataset for generating the appropriate responses. The proposed method performs well for knowledge graph enhanced medical dialogue generation.
Our current work makes the following contributions:
-
1.
We propose a new medical-knowledge-aware neural model, MED for medical dialogue generation that uses large-scale pre-trained language models to incorporate the triples from the knowledge graph for generating medically relevant dialogues. By greatly boosting the knowledge representation using a large amount of information from dialogue context, this method investigates the learning of contextual entity embeddings.
-
2.
We use the UMLS database to create a medical knowledge graph that is supplemented with several medical entities not discovered in the UMLS database and are extracted using BERT-based medical entity prediction models. This provides the language models with both global knowledge from UMLS as well as conversation-oriented medical entities from the medical datasets.
-
3.
We show that our model is capable of integrating graph contextualized information into large-scale pretrained language models which outperforms the strong baselines on the MedDialog(EN) and Covid Dataset using extensive qualitative and quantitative evaluation.
The rest of the article is organised as follows. Some of the earlier efforts in this field are summarised in the “Related work” section. Then, we describe our proposed methodology for generating clinically accurate response. The dataset used for the suggested generative model is briefly described in the next section. We give a detailed account of the experimental evaluation in the following part, followed by a quick review of how well the suggested generative model performed. We finish the article by describing the originality of the work and its future applications.
Related work
The first Chatbot, ELIZA11, used a keyword/pattern matching mechanism to find a pattern and the corresponding response for a given user text. Jia12 improved upon ELIZA by using textual knowledge and reasoning. For the task of multi-turn dialogue generation in open domain, several approaches13,14,15,16,17,18 for modeling hierarchy of the conversation using various frameworks and diversity-promoting objectives were proposed. Zhang et al.19 proposed a memory encoder which is trained with textual features to obtain dialogue representation, a decoder which is composed of an Recurrent Neural Networks (RNN) and a rule-memory network for response generation. Bidirectional Encoder Representations from Transformers (BERT)20 with around 340 million parameters successfully tackle a broad set of language understanding tasks. Denoising Sequence-to-Sequence Pre-training for Natural Language Generation, Translation, and Comprehension i.e BART21 with a bidirectional encoder20 and autoregressive decoder22 showed competitive performance on sequence classification as well as generation tasks. With 11 billion parameters, the T5 model23 is suggested to improve the performance for tasks requiring both natural language processing and generation. Generative Pre-trained Transformer (GPT)-324 having 175 billion parameters obtains exceptional performance on a variety of tasks in the few-shot and zero-shot settings. Various pre-trained language models1,2,3 have demonstrated compelling performance on generating responses for dialogue systems by leveraging the pre-train / fine-tune paradigms. MASS25 jointly trains the encoder which takes as input a sentence with a randomly masked fragment (several consecutive tokens) and decoder that attempts to predict this masked fragment for text generation.
Several approaches integrating commonsense knowledge graphs with dialogue systems were demonstrated in26,27,28,29,30,31. Zhou et al.27 incorporated commonsense knowledge using static and dynamic attention to generate a correct response for a given post. Wu et al.32 proposed ConKADI which improved the selection and effective integration of facts which were incredibly pertinent to the context of the generated response. Young et al.26 augmented the large scale commonsense knowledge and integrated it with an end-to-end neural dialogue model in the form of external memory. Wu et al.28 proposed a model which integrates both knowledge graph and topic specific knowledge to improve the existing post-response pair generation models using a teacher-student recommendation network. Liu et al.33 proposed to predict entities in a structured way by encoding knowledge triplets using a neural knowledge diffusion module. Similarly, for an open-domain dialogue system, Liu et al.34 integrated the knowledge graph with the dialogue pipeline using three modules viz. knowledge augmentation to augment a knowledge graph with texts, knowledge selector and knowledge aware response generator to perform graph reasoning. A knowledge infused model for capturing semantic relations and to model conversation structures was proposed by Varshney et al.35 using a multi-hop attention mechanism.
Using large scale pre-trained language model, BART36 incorporated knowledge graphs using multi-head graph attention for commonsense reasoning. There are also findings Peters et al.37, He et al.38 that focused on inserting prior knowledge by using entity and relation embedding into deep neural language models. In order to train the models to distinguish between the right entity mention and randomly selected ones, WKLM39 replaced entity mentions in the original texts with names of additional entities of the same type. To align global information and language representation into the same semantic space, KEPLER40 optimised the models with mask language model objective and extracts knowledge from text by encoding the entities from their corresponding descriptions. A word-knowledge graph was used by CoLAKE41 to combine the language context and the knowledge context, and the extended mask language model aim was used to jointly learn a context aware representation for both language and knowledge. Sun et al.42 proposed knowledge-aware pre-training tasks to incorporate knowledge graphs into language models for language understanding and generation. To forecast the relation in the triple, the model must recognise mentions of both the head and tail entities in the associated sentence and establish the semantic link that exists between them. In relation extraction tasks, the essence of this procedure is identical to the distant supervision algorithm. Our work, on the other hand, only masks the head entities and predicts the tails entities that correlate to them, rather than the head entities themselves. This is done so that the model can learn to use the graph attributes in dialogues as well as the contextual information.
As the number of biomedical documents grows, so does the importance of biomedical text mining. With advancements in natural language processing (NLP), researchers have become more interested in extracting valuable information from biomedical literature, and deep learning has aided in the development of effective biomedical text mining models. Domain-specific language representation model such as Bidirectional Encoder Representations from Transformers for Biomedical Text Mining (BioBERT)43 and ClinicalBERT44 are pretrained utilising large scale data from biomedical domains and have demonstrated high quality performance on various medical tasks. They outperform significantly on three representative biomedical text mining tasks: biomedical named entity recognition, biomedical relation extraction, and biomedical question answering. Rao et al.45 developed a system that integrates language models with knowledge graph embeddings to provide understandable answers to queries from biologists. Auti et al.46 used the GAN-BERT architecture to categorize pharmaceutical texts, replacing the BERT model with the BioBERT model to capture domain-specific information. Yang et al.47 introduced the Pathway2Text dataset, which consists of 2,367 pairs of biomedical pathways and corresponding textual descriptions, and proposed a graph-based text generation method called kNN-Graph2Text that uses descriptions of related graphs to generate new descriptions. Luo et al.48 developed the BioGPT language model, a domain-specific generative Transformer that was trained on a large volume of biological literature and tested on six biomedical natural language processing tasks. Blanc et al.49 used FlauBERT and CamemBERT to design a study to ascertain which language model and neural network architecture combination was best for intent and slot prediction by a chatbot using a French corpora of clinical cases.
Zeng et al.50 released a high-quality medical dialogue dataset in Chinese and English that covers more than 50 diseases. There are 3.4 million patient-doctor dialogues, 11.3 million utterances, and 660.2 million tokens in the Chinese dataset, while there are 0.26 million conversations, 0.51 million utterances, and 44.53 million tokens in the English dataset, which covers 96 illnesses categories. Liu et al.51 released another high-quality chinese medical dialogue dataset containing 12 types of common Gastrointestinal diseases named MedDG, with more than 17K conversations. They put forth two challenges, the first of which is to predict the subsequent entities based on dialogue contexts, and the second of which is to generate virtual doctor’s responses. On four pediatric disorders namely upper respiratory infection, functional dyspepsia, infantile diarrhea, bronchitis, the CMDD dataset52 containing 2,067 dialogues was released. This dataset did not handle the problem of data imbalance between diseases. A much larger medical dialogue dataset, namely Chunyu53 was proposed to improve upon the previous model, which now contains 15 diseases with comparatively distinct data ratios.
In the medical domain, on the MedDialog(CN) dataset, an end-to-end variational bayesian generative strategy9 was developed to generate medical dialogue by approximating posterior distributions over patient states and physician actions. In order to test the performance of their proposed VRBot model, they curate a large medical dialogue dataset with over 60,000 medical conversations having 5,682 entities (such as Asthma and Atropine). Liu et al.54 proposed an effective technique by auto encoding knowledge graphs for multimodal medical report generation using a knowledge-driven encoder and a knowledge-driven decoder. Similarly, Liang et al.55 presented a lightweight as well as a scalable mechanism using the transformer and BERT-GPT architecture to integrate the medical knowledge into different neural generative models on the MedDG and the MedDialog(CN) dialogue corpora. Lin et al.53 proposed a low-resource medical dialogue-generating system along with a Graph-Evolving Meta-Learning (GEML) framework that learns to evolve the commonsense graph for reasoning disease-symptom connections.
In this work, we exploit the largest conversational medical corpus, MedDialog(EN)50 containing 0.26 million English consultations between the patients and doctors, and demonstrate how external medical knowledge can improve the task of medical dialogue generation. We propose a knowledge-aware neural medical conversation model named MED to generate contextualized entity embeddings for incorporating relevant information from the knowledge graphs in accordance with the diverse conversation structures. Furthermore, we annotate the utterances with symptoms, diseases, tests, and other pertinent categories using semi-automated NLP techniques which are further used to enhance the knowledge graph retrieved using UMLS10 database. The efficacy of our proposed approach is demonstrated through our experiments on two medical dialogue corpus viz. MedDialog(EN) and Covid Dataset, a newly augmented and much larger corpora focusing mainly on Covid-19. Our method allows the use of a large-scale biomedical knowledge graph to facilitate the understanding of the clinical entities and relations present in the current dialogue and the generation of a response with clinically correct information.
Methodology
Problem definition: The objective of this research is to build a virtual doctor (i.e. conversational agent), which could assist the users (i.e. patients and/or other stakeholders) by providing an appropriate response. Given the current utterance and contextual conversations between patients and doctors, represented as a sequence of words, the task is to generate medically relevant responses. To generate more knowledgeable and engaging dialogue, we make our conversational agent knowledge grounded by leveraging an external knowledge graph.
Conversation: We have a set of conversations, each consisting of multiple turns, where the patient and the doctor take turns speaking. For each turn, we have the patient’s utterance and the doctor’s response, both represented as a sequence of words.
-
Consider a conversation \(C = \{c_i\}_{i=1}^K\) consisting of K turns, where each turn is represented by a tuple \((b_k, d_k)\). \(b_k\) denotes the patient’s utterance and \(d_k\) is the virtual doctor’s response at the k-th turn.
-
The patient’s utterance \(b_k\) is a sequence of \(\left( b_{k,1}, b_{k,2}, \ldots , b_{k,|b_k|}\right)\) words, where \(|b_k|\) denotes the total number of words in the utterance. Similarly, the doctor’s response \(d_k = \left( d_{k,1}, d_{k,2}, \ldots , d_{k,|d_k|}\right)\), comprising of \(|d_k|\) words.
Knowledge Graph: The augmented medical knowledge graph is denoted as: G = \(\left( g_{1},g_{2},\ldots ,g_{|b_k|}\right)\).
-
G is a set of graphs, each graph is a set of triples, and each triple is a tuple of head, relation, and tail entities.
-
Each graph, \(g_i\), contains a set of triples \(g_i = \left( {\tau }_1, {\tau }_2, \ldots , {\tau }_{|g_i|}\right)\).
-
Each triple \({\tau }_j\) has \(({\textbf {h}},{\textbf {r}},{\textbf {t}})\) denoting head, relation, and tail entity respectively, with \({|g_i|}\) indicating the total number of triples for every graph \(g_i\).
We create the augmented medical knowledge graph in the following ways:
-
1.
We first take as input an utterance, \(b_k\), and initialize the first word as the first node for the graph, \(g_i\). For every node, we extract different subgraphs using UMLS (section “Construction of the medical knowledge graph using UMLS”). Starting from the first node in \(g_i\), the agent follows a path in the graph and stops at a node that it finds in the doctor’s response.
-
2.
We extract medical entities from each utterance and use the four different annotated entity categories for defining relationships between them (section “Construction of the medical entity graph”). We add these new triples to the subgraphs extracted from UMLS.
Medical knowledge graph
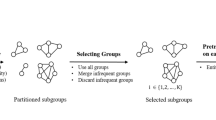
Utilizing the knowledge graph is driven by the fact that generating responses for the virtual doctor typically necessitates specialized medical knowledge. As a result, we can use the knowledge graph, which simulates the knowledge structure specific to the medical domain. Specifically, we build a global medical knowledge graph \(g_i = ({\tau }_1, {\tau }_2, \ldots , {\tau }_{|g_i|})\) where each triple covers the most common diseases and symptoms. In this section, we describe the steps to form a large augmented graph of disease-symptom relationships using medical concepts extracted from several sources. The process for preparing the medical knowledge graph is shown in Fig. 1 and explained in sections “Construction of the medical knowledge graph using UMLS” and “Construction of the medical entity graph”.
Medical Knowledge Graph: The patient-doctor dialogue as shown on the left is first passed through the Quick-UMLS tool, which is used to extract the medical concepts and relations from the UMLS knowledge base as shown in the box on top-middle. The box on the bottom-middle uses the MEP model to discover additional biomedical words present in the dialogues. Finally, the augmented knowledge graph used in the dialogue generation pipeline is shown in the right.
Construction of the medical knowledge graph using UMLS
The UMLS10 is a vast knowledge base that contains huge number of concept names and relationships between various health and biomedical words. The three knowledge sources available in UMLS are Metathesaurus, Semantic Network, and Lexical Tools. We use the Metathesaurus and the Semantic Network modules to implement our approach. Metathesaurus contains over 1 million biomedical concepts, and each concept is related to the other concept in some way. Furthermore, each concept is assigned with one or more semantic types, and their relationship with the semantic types is determined using a semantic relationship network. The semantic network contains 127 semantic types and 54 relationships between them. We use the QuickUMLS56 (https://github.com/Georgetown-IR-Lab/QuickUMLS) tool to extract the required medical concepts and construct the clinical graphs.
Quick-UMLS is used to extract biomedical entities from the UMLS Metathesaurus and associate them with unique identifiers (CUIs) and semantic type. It takes the utterances from the medical conversation as input, finds the closest match in the UMLS set of strings, and then returns the CUI, or list of semantic categories, for each concept in the utterance. Each unique identifier (CUI) in UMLS is treated as a node in our knowledge graph, and then using the semantic network module from UMLS appropriate relationship between each node is determined. Let us say, we have an input utterance as “Hello doctor, how to improve fetal growth and heart beat ?”. We utilise the Semantic Network to create a sub graph for the medical concept “heart beat” and form the following set of triples: (’heart beat’, ’affects’,’anxiety’),(’heart beat’, ’affects’, ’circulation’), (’heart beat’, ’affects’, ’collapsed lung’), (’heart beat’, ’process of’, ’unconscious’) and so on. This is shown in the top side of the Fig. 1.
The UMLS knowledge base overlooks some important medical information found in many medical dialogue datasets, such as medical tests, medication, and home remedies even after using all the 54 relationships and 127 semantic types from the UMLS semantic network. We use a neural BERT based medical entity prediction model to annotate the dialogues with symptoms, medical tests, medications, and home remedies in order to enrich the knowledge graph obtained from UMLS.
Construction of the medical entity graph
Medical Entity Annotation: We choose the following four different kinds of entities for annotation after consulting with domain experts: Symptom such as chest pain, thyroid dysfunction and so on; Medication such as analgesic, pantoprazole and so on; Home Remedy such as regular exercise, jogging, and so on; and Medical Test such as x-rays, blood tests and so on. The utterances of the conversation are labeled individually with four categories of entities using Named Entity Recognition (NER) method as the annotation technique, as illustrated in Table 3.
Four annotators with relevant backgrounds participated in the annotation process. The annotators have a mix of qualifications that includes linguistics with PhDs, medical experts with medical degrees, specifically masters in medicine. This combination of qualifications ensures that the annotators have the necessary linguistic skills, as well as the medical expertise to accurately understand and interpret the medical texts. The annotators are regular employees who are paid at a rate of $35,000 per month (in accordance with university standards). The annotators have been working on related projects for the past three years as members of our research team. They talk about making an annotation template first. Four blank columns are offered to the annotators so they can select the appropriate medical phrase for the various categories. For example in Table 3, for the first utterance, the relevant medical entities to be annotated are Symptom: Myalgia, costo-chondritis; Medication: analgesic cream, Ibuprofen; and so on. We present a few triples from the UMLS knowledge base, along with the corresponding utterances in the first column, to demonstrate the difference in the information obtained using different methods of graph creation. The triples are shown in the last column. Each participant annotates a small portion of the data and reports the confusing utterance. They summarize their observations and then revise the annotations once more.
To ensure that the guidelines are effective, a number of pilot studies were conducted with linguistics on around 200 test dialog samples. These pilot studies were used to test the annotation guidelines and identify any inconsistencies or issues that needed to be addressed. After the pilot studies, the annotation guidelines were discussed with medical experts, who verified the annotations. Any inconsistencies found in the pilot studies were removed and resolved in order to ensure consistency in the annotation process. This process guarantees that the final annotation guidelines are accurate and reliable, which is essential for the credibility of the research. Inconsistencies were errors, ambiguities, and sometimes a lack of clarity in the annotation guidelines. They were resolved by revising the guidelines or providing additional training for the annotators. Additionally, we took care to clearly define the concepts and entities that needed to be annotated.
We observe a Fleiss’ kappa57 score of 0.89 between annotators denoting great agreement between them for the entity annotation task. However, in addition to Fleiss kappa, we compute Krippendorff’s Alpha58: a measure of inter-rater variability. We obtain Krippendorff’s Alpha score of 0.62 for medical entity annotation indicating a moderate inner-annotator agreement. We annotate the first 6K utterances using the above method from the Covid Dataset and then using the algorithm described in the following paragraph, we annotate both the MedDialog Dataset and remaining conversation in the Covid Dataset.
Medical Entity Prediction (MEP) Model: To recognise medical entities from the dialogue context C, we use the MEP model described as follows. We first annotated the 6K samples manually tagged with entities as described previously using the Inside-Outside-Beginning (IOB)-tagging as the annotation scheme. Each tag indicates whether the corresponding token is inside, outside, or at the beginning of a specific named entity. We had four labels: Symptom, Medication, Medical test, and Remedy, for a total of nine tags: B-Medication, I-Medication, B-Symptom, I-Symptom, B-Medical test, I-Medical test, B-Remedy, I-Remedy, B-remedy, I-remedy, and O. We then fine-tune the pre-trained BERT base model (Uncased: hidden-768, heads-12, layer-12) for the medical entity prediction task. The input is in the form of a tokenized sentence, and the tokens are truncated or padded according to the maximum length of the model. The training was done on 80% of the total data (4978 sentences) while validation was done on the remaining 20% (1244 sentences). For evaluation, we calculate the precision, recall and F1 score for each label using seqeval.metrics (https://pypi.org/project/seqeval/). The scores from all the metrics are shown in Table 2 and the predictions from the MEP model are shown in Table 3. Using the trained model, we annotate all the utterances of the MedDialog Dataset and Covid Dataset. Finally, the medical entity graph is built with the extracted medical entities as nodes and edges for each pair of medical entities that appear in the four different categories.
For example, for the input sequence “Hi, from history it seems that you might be having myalgia or costo-chondritis. Apply analgesic cream locally. Take Ibuprofen tablet if require. Do regular exercise like running, jogging, swimming. To remove doubt about cardiac problem, go for EKG. Ok and take care” medical concepts such as analgesic, tablet, exercise, swimming, cardiac, care and so on and triples such as (analgesic, diagnoses, heart diseases), (cardiac, location of, heart sounds), (swimming, isa, relaxation) and so on are extracted from the UMLS database. As can be clearly seen, UMLS fails to incorporate medical concepts such as myalgia, costo-chondritis and ibuprofen. We make an attempt to extract these concepts using our MEP model. Examples of the extracted medical concepts can be seen in the Table 3. Using the extracted medical concepts from entity prediction model, we form an entity graph having the following nodes and relations: (Myalgia, medication, analgesic cream), (Myalgia, medication, ibuprofen), (Myalgia, medical test, EKG), (Myalgia, medication, ibuprofen), (Myalgia, remedies, regular exercise), and so on. This is finally appended with the triples obtained from the UMLS database to form the new augmented medical graph and finally used in section “Model description” for building knowledge-aware medical dialogue systems.
Model description
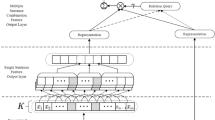
In this part, we outline our suggested method for having the virtual doctor produce responses to patient inquiries that are appropriate from a medical standpoint. Figure 2 depicts the detailed model architecture proposed for the task of medical dialogue generation. We use the BioBERT BASE43 as the pre-trained language model to dynamically build contextualised representations for the input sequences using graphical knowledge. With the conversation sequence C in hand, we first mask the tokens for which we acquire a medical graph \(g_i\) in accordance with section “Construction of the medical entity graph”, and instead of predicting the token itself, we infer the tail entities extracted from the corresponding graphs. We call this as the MED model. For example, given the triples: ’(cough, treats, eosinophilia)’, ’(blockage, occurs in, pulmonologist)’, we mask the tokens cough and blockage in the dialogue context, as “i am continuously suffering from [MASK] due to feeling of some [MASK] in upper wind pipe”; the model then predicts the tail entity corresponding to the masked tokens cough and blockage i.e eosinophilia and pulmonologist. We choose the tail token from the first triple in the sequence \(({\tau }_1, {\tau }_2, \ldots , {\tau }_{|g_i|})\), associated with the m-th masked token and denote it as \({\textbf {T}}_m\). Given the input sequence, I (Each token in I is first passed through a series of three embedding layers (Token, Segment, and Position). The resulting embeddings are concatenated and denoted as \(E_{b_{k,i}}\)), we first attempt to predict \(M_L\) by using BERT based token classification model which returns the hidden states, \(H_{k,i}\), for the input sequence which is usually the representation of the [CLS] token. While predicting any masked token, information from nearby words is utilized. Our approach directly utilizes the dialogue context, (I) , simultaneously incorporating the knowledge triples.
We first form the flattened token sequence for the input utterance with the masked entities:
The corresponding ground truth label, \(M_L\), for token classification is shown as follows:
where the [CLS] token is inserted at the beginning of the sequence as an indicator of the start of the sentence. The [SEP] token distinguishes one sequence from the next and indicates the end of a sentence.
The BioBERT-based decoder generates text by predicting one word at a time, using the hidden state \(H_{k,i}\) at each time step. During training, the decoder is provided with the actual next word in the sequence, taken from the set \(R_L\), as input. However, during inference, the decoder uses the word it has previously predicted as input. To start the decoding process, the first input to the decoder is taken from the first token in \(R_L\).
The BioBERT model uses its hidden state from the top layer, passed through a linear layer, to predict the next token in the target (output) sequence.
where \(W_1\) is a learnable weight matrix and \(b_1\) is the bias.
The decoder loss is the cross-entropy between the output distribution \(P(y_{k,j})\) and the reference distribution, \(d_j\), denoted as \(Loss = - {\sum }{d_j}log(P(y_{k,j}))\).
MED architecture: The proposed architecture first encodes the conversation history using a BioBERT encoder as shown in the left. We first attempt to predict the sequence \(M_L\) using the token classification task that provides the hidden states, \(H_{k,i}\), for the input sequence. This input sequence is then fed into a BioBERT decoder to generate the response.
Ethical declaration
The public can access all of the datasets used in this study freely. By crawling additional data from the sources cited in the datasets’ respective papers, we also add to the dataset new patient-doctor conversations. A committed group of full-time employees were employed for annotation purposes. All experiments were conducted in conformity with the applicable rules and guidelines. Medical dialogue systems might be employed in the real world to give patients advice and gather information for diagnoses. Doctors will eventually take control of the procedure, even if the agent makes a few minor errors along the way.
Datasets and experimental setup
In this section, we describe the datasets that we use for our experiments, implementation details, baseline and the evaluation metrics. The train, test and validation split details for both the MedDialog and Covid Dataset are mentioned in Table 4. More details of the dataset containing information on the number of utterances and so forth are mentioned in Table 5.
MedDialog(EN) dataset
The MedDialog(EN)50 dataset is the largest available dataset which comprises of 0.26 million patient-doctor conversations. Each conversation is composed of two sections: a comprehensive description of the patient’s medical issue, including symptoms, duration of the illness and other relevant information, as well as the physician’s recommended course of treatment and suggested remedies. The dataset includes 96 distinct medical specialities, such as andrology, cardiology, nephrology, and pharmacology, as well as 51 different categories of illnesses, such as diabetes, geriatric concerns and pain management. The total number of words in the corpora is 32,855,023, and the total number of utterances is 396,562. The average, maximum, and minimum number of tokens in an utterance are 83.0, 3,677, and 1, respectively. The average, maximum, and minimum number of utterances are 2.0, 2.0, and 2.0, respectively. Sample conversation from the dataset is shown in Table 1.
Covid dataset
We enhance the CovidDialog dataset59 having 603 conversations with the diseases which are the symptoms of Covid-19 such as fever, cough, allergies, and so forth. The reason for this is because dialogues regarding fever, cough, cold, and other Covid-19 symptoms can be helpful in response generation on the Covid-19 disease. The dataset now includes over 10,000 English conversations about Covid-19 and symptoms associated with Covid, which helps in the creation of a resourceful medical dialogue system. Each session starts with a brief overview of the patient’s health issues, which is followed by a chat between the patient and the doctor. The total number of utterances and tokens in the dataset is 26,658 and 2,735,112, respectively. The average, maximum, and minimum number of utterances are 3.0, 42, and 2, respectively. The average, maximum, and minimum number of tokens in an utterance are 103.0, 1509, and 1, respectively.
Implementation details
All the experiments are implemented using Pytorch. We chose the hidden size of 512 for all our model. The number of layers is set to 2 for BERT and BART based models, and 6 for the BioBERT and MED model. We use the cased models for every pre-trained model. We use the ADAM optimizer whose learning rate is fixed to 0.0005 and set the beam size to 1, while decoding the responses. The input utterances are truncated to a max token count of 400 and output utterances to 100. We choose the best model when the loss on the validation set does not decrease. We use the GeForce GTX 1080 Ti as the computing infrastructure. Each model is trained up to 30 epochs.
Baselines
We use the following baseline models: CCM27 and ConKADI32 for incorporating knowledge graphs for dialogue generation. Pre-trained language models, such as \({\textbf {DialogGPT}}_{{{\varvec{finetune}}}}\)2, BERT20, BART21 and BioBERT43 for dialogue generation.
-
1.
CCM27: Many tasks involving natural language processing require commonsense knowledge. To illustrate how extensive commonsense knowledge can aid language interpretation and generation, this model first forms an appropriate knowledge graph from a knowledge base. The graph is then encoded using a static graph attention technique, which improves the post’s semantic content and makes it easier to understand. The model then reads the extracted knowledge graphs and knowledge triples while generating each word of the response using a dynamic graph attention strategy within each graph and thus aids in improving the whole dialogue generation process. We replicated the results of CCM on our dataset using the codes in this link : https://github.com/thu-coai/ccm.
-
2.
ConKADI32: To make the dialogue models aware of the context when using the knowledge, ConKADI provides a Felicitous Fact mechanism to help the model focus on knowledge facts that are extremely relevant to the context in order to help the dialogue models be aware of the context when integerating the knowledge. Context-Knowledge Fusion and Flexible Mode Fusion are two further strategies that aid in combining the knowledge information for the model. Additionally, ConKADI employs trainee responses as posterior knowledge. The codes for generating the results on the dataset can be found in this link: https://github.com/pku-sixing/ACL2020-ConKADI
-
3.
\({\textbf {DialogGPT}}_{{{\varvec{finetune}}}}\)2: Dialogue generative pre-trained transformer (DialogGPT), a large scale fine-tuned neural conversational response generation model, was constructed using the OpenAI GPT-2 architecture and trained with 147 million Reddit conversations. Additionally, the model that performed the best was selected with 345M parameters. The GPT-2 transformer model uses a stack of masked multi-head self attention layers and the generic transformer language model to train on the large amount of web-text data. The codes for replicating the results can be found in the link as follows: https://github.com/UCSD-AI4H/COVID-Dialogue/tree/master/src/DialogGPT/GPT2-chitchat-master
-
4.
BERT20: Transformer, which makes use of an attention mechanism to learn contextual relationships between words (or, sub-words) in a text, is used by BERT. They simply mask a portion of the input tokens at random, then predict those masked tokens in order to train a deep bidirectional representation. Using the BERT model as both an encoder and a decoder, the input utterances from the conversations are encoded, and the pertinent output is generated. Codes to obtain results is possible using the codes in this link: https://github.com/UCSD-AI4H/COVID-Dialogue/tree/master/src/Transformer
-
5.
BART21: BART comprises of a denoising autoencoder utilized for pre-training sequence-to-sequence models. BART is trained by altering documents and then optimizing the cross-entropy loss between the original document and the decoder’s output. It uses a sequence-to-sequence approach for emcoding the corrupted text with a bidirectional encoder and a autoregressive decoder works from left to right. For pre-training purposes, the negative log likelihood of the original document was optimized. For our datasets, the sentences in the conversations are encoded using the bidirectional encoder, and the relevant response is generated using autoregressive decoding. The codes for implementing the BART model is available in the link: https://github.com/UCSD-AI4H/COVID-Dialogue/tree/master/src/Bert-GPT
-
6.
BioBERT43: The BioBERT model adopts the previously described BERT architecture except that it has been pre-trained on a large-scale biomedical corpora (PMC full-text articles and PubMed abstracts). In a number of biomedical text mining tasks (NER, RE and QA), it has shown to outperform BERT and other state-of-the-art models. In order to adapt the BioBERT model, we encode the dialogue history using the BioBERT model as an encoder. Finally, the decoding process is completed by utilizing a BioBERT model as a decoder. Codes to replicate this model can be found on the link: https://github.com/deekshaVarshney/MED
Evaluation metrics
Automatic evaluation: We evaluate our models on the test set of both the MedDialog(EN) and Covid Dataset, using the following standard metrics. We use one of the most popular metrics for evaluating sequence like BLEU60, F1-score, Perplexity (PPL)61 and n-gram diversity (Div.)62 to automatically evaluate the quality of generated responses.
-
1.
Perplexity: We make use of perplexity as one of the automatic evaluation metric for the medical dialogue generation task. It is a measurement of how well a model can predict human responses. Generally, lower perplexity indicates better generation performance. It is defined in Equation 5. Our various models are tested on the generation ability by computing perplexity on the test data.
$$\begin{aligned} PPL = \text {exp} \{{\frac{-1}{N} \sum _{i=1}^{N} log(p(y|U))}\} \end{aligned}$$(5)where N is the total number of samples in the test set.
-
2.
BLEU (Bilingual Evaluation Understudy Score): We measure the accuracy of the generated responses by using BLEU, a word-based metric which measures n-gram overlaps with the gold response. First, sentence by sentence n-gram matches are calculated. Then the total number of candidate n-grams in the test corpus are multiplied by the sum of all the clipped n-gram counts for the candidate sentences to arrive at a modified precision score, for the full test corpus. The BLEU metric is in the range of 0 to 1. Unless they are exact replicas of a reference sentence, few sentencess will receive a score of 1.
-
3.
F1-score (https://github.com/facebookresearch/ParlAI/blob/master/parlai/core/metrics.py): We also compute unigram F1-score between the predicted sentences and the ground truth sentences. We first compute the precision using the number of overlapping unigram words between the gold and the generated sentence divided by the total number of words in the gold response. Second, we compute the recall using the count of unigram overlap divided by the count of words in generated text. Finally, the F1 score is computed as defined below:
$$\begin{aligned} F1 = \frac{2 \cdot recall \cdot precision}{recall + precision} \end{aligned}$$(6)
Embedding-based metrics63, such as Greedy Matching, Vector Extrema and Embedding Average are an alternative to word-matching-based metrics. Code to implement the above metrics: https://github.com/Maluuba/nlg-eval. They are defined as follows:
-
1.
Greedy matching: Given two sequences q and \({\hat{q}}\), the total score is calculated by greedily matching each token w \(\in\) q with a token \({\hat{w}}\) \(\in\) \({\hat{q}}\) based on their word embeddings’(\(e_w\)) cosine similarity and finally averaged across all words. It is mathematically defined as follows:
$$\begin{aligned} G(q, {\hat{q}}) = \frac{\sum _{w \in q;} max_{{\hat{w}} \in {\hat{q}}} cos\_sim(e_w, e_{{\hat{w}}})}{|q|} \end{aligned}$$(7)Due to the asymmetry of this algorithm, we must average the greedy matching scores G in both directions.
$$\begin{aligned} GM(q, {\hat{q}}) = \frac{G(q, {\hat{q}}) + G({\hat{q}},q)}{2} \end{aligned}$$(8) -
2.
Embedding Average: The mean of the word embeddings of each token in a sentence q is known as the embedding average (\(\bar{e}\)) or the sentence embeddings.
$$\begin{aligned} \bar{e}_q = \frac{\sum _{w \in q}e_w}{|\sum _{w' \in q}e_{w'}|} \end{aligned}$$(9)Then the cosine similarity between two sentence level embeddings, one representing the ground truth response q and the other representing the retrieved / generated response \({\hat{q}}\) is calculated as described below:
$$\begin{aligned} EA = cos(\bar{e}_q, \bar{e}_{{\hat{q}}}). \end{aligned}$$(10) -
3.
Vector Extrema: For each dimension of the word vectors, take the most extreme value amongst all word vectors in the sentence, and use that value in obtaining the sentence-level embedding:
$$\begin{aligned} e_{qd} = {\left\{ \begin{array}{ll} max_{w \in q} e_{wd} &{} \text {if }e_{wd} > |min_{w' \in r} e_{w'd}| \\ min_{w \in q} e_{wd} &{} \text {otherwise} \end{array}\right. } \end{aligned}$$(11)where d denotes a vector’s dimensions and \(e_{wd}\) denotes the embedding at the \(d-th\) dimension. We again compute the cosine similarity between the sentence-level vectors to obtain VE.
Human evaluation: To evaluate the quality of generated responses from a human perspective, we randomly select 50 dialogues from each model developed using the MedDialog and Covid datasets and analyse the predicted responses with the assistance of three human evaluators. Human raters are post-graduates in science and linguistics with annotation experience for text mining tasks especially in the medical domain. The annotators are regular employees and receive a monthly salary of approximately $35,000. (in accordance with university standards). As a part of our research team, the annotators have been working on related projects for the previous three years. We also had a physician with a postgraduate degree in medicine validate our model’s results. The responses were confirmed to have maintained the crucial medical information. To assess the accuracy of our model predictions, we employ the following metrics:
-
1.
Fluency: This is a measure of sentence grammatical accuracy (i.e no fragments or missing parts) and that the text is easy to understand.
-
2.
Adequacy: This metric is used to determine whether the generated response is meaningful and relevant to the conversation history.
-
3.
Entity Relevance: This metric is used to determine whether or not a response contains the correct medical entities which are mentioned / referred in the conversation.
The scale runs from 1 to 5. The higher the number, the better. The ratings for the fluency metric are incoherent, disfluent, non-native, satisfactory, and perfect English, respectively. For the adequacy metric, these correspond to none, little meaning, much meaning, most meaning, and all meaning, respectively. Similarly, for Entity Relevance, we give the higher score if the response contains all the entities mentioned in the ground truth dialogue. As the final results, the ratings from the various annotators are averaged. To assess inter-annotator agreement, we compute the Fleiss’ kappa score57.
Results and analysis
Tables 6 and 7 show the automatic and human evaluation results for the baselines and the proposed models.
Automatic evaluation
The results of using automatic evaluation metrics on both the dataset are shown in Table 6. On most metrics, we see that MED outperforms the baseline models, demonstrating the efficiency of introducing global and data specific medical information generated using the MEP model for the task of medical dialogue generation. Our proposed model outperforms the knowledge graph based models, CCM27 and ConKADI32 on all the automatic evaluation metrics as well as the embedding-based metrics, demonstrating the importance of masking and predicting relevant medical concepts in generating relevant responses. When compared to the strongest baseline, BioBERT, MED yields a significant performance improvement of around 5.2 F1-score points and 8.3 BLEU-4 points on the test-set of the MedDialog corpora. On the Covid Dataset also, MED outperforms the BioBERT baseline on F1 and BLEU-4 evaluation metrics by 8.5 and 9.3 points, respectively.
Human evaluation results
Table 7 displays the results of human evaluation. We only compare to the best models, BERT, BART, and BioBERT, on both MedDialog(EN) and Covid Dataset, because manual evaluation is costly. On fluency, adequacy, and medical knowledge-related criteria viz. entity relevance, MED outperforms the baseline models, demonstrating consistency with the results of automatic evaluation. All of the kappa values are greater than 0.75, indicating that the annotators agree with each other.
We present a few example conversations predicted by the proposed (MED) and baseline models (BERT, BART, and BioBERT) on the test set from MedDialog(EN) corpus and Covid Dataset in Table 10. As seen in the first example, MED correctly decodes the response by using context information as well as the correct triplets, such as (headaches, associated with, migraines), (headaches, co-occurs with, blurring of vision), (headaches, co-occurs with, tingling) as well as triples formed using the MEP model such as (pain, medication, ibugesic), (migraine, medication, analgesics), (pain, medication, omeparazole) as opposed to the BioBERT’s prediction which clearly fails to predict many medical entities. This shows the relevance of enhancing the graph with medications, medical tests and home remedies. The reason for correctly predicting medicines names, such as omeparazole and ibugesic is the association between headache and migraines with pain as inferred from the triples viz.(headache, evaluation of, pain), (migraine, associated with, pain) which is learned by using idea of masking to predict the tail entity i.e pain.
Overall, during evaluation, we found that the presence of medical entities in the predicted response by our proposed MED model especially belonging to the medicine, medical test and home remedies increased to 5% and 10% as opposed to the baseline (BioBERT) predictions on MedDialog(EN) and Covid dataset respectively.
We conduct statistical significance tests on our models using statistical hypothesis testing (t-test) at a 5% (0.05) significance level. We used the implementation provided at this link: https://github.com/rtmdrr/testSignificanceNLP to perform the t-test. The test compares the performance of two approaches, A and B, on a given metric M. P-value scores are reported in Table 8 for automatic evaluation and in Table 9 for human evaluation. All the p-values reported in Tables 8 and 9 are less than 0.05 (5% significance level). Hence it shows that our approach is statistically significant. We found that the improvement in the proposed model (MED) over the state-of-the-art models BioBERT, BART, and BERT on the MedDialog(EN) dataset and the Covid dataset, as well as on human evaluation metrics such as Fluency, Adequacy, Emotional Content, and Knowledge Relevance, was statistically significant with 95% confidence (i.e., p-value < 0.05) on both datasets.
Ablation study
To analyze the effectiveness of external medical knowledge for medical dialogue generation, we provide an ablation study for our proposed methodology. The results are reported in Tables 6 and 7.
-
1.
BioBERT: This setup solely uses the BioBERT-based conversation encoder and decoder to highlight the importance of using augmented knowledge graphs for generating medical dialogue. Table 6 shows that this results in a 5.1 percent decrease in F1 score and a similar drop in human evaluation metrics (Table 7) can also be noted, proving the efficiency of our knowledge assimilation technique in MED.
-
2.
MED-UMLS: In this configuration, the triples prepared using the UMLS knowledge base (section “Construction of the medical knowledge graph using UMLS”) are not used; only the triples obtained from the MEP model (section “Construction of the medical entity graph”) are used. Table 6 shows that this results in a 2.2% decrease in F1 score on the MedDialog(EN) dataset, signifying the importance of the triples formed using the entities obtained by the MEP model depicting the importance of local information in order to generate relevant response by the doctor.
-
3.
MED-MEP: This ablation model uses only the triples formed using the UMLS knowledge base for medical dialogue generation. Table 6 shows that using only UMLS triples leads to a significant decrease of 13.6% in F1 score on the MedDialog(EN) dataset, which demonstrates that the UMLS knowledge base may overlook some important medical information found in many medical dialogue datasets and instead adds noise into the model and thus supports our motivation to augment the UMLS knowledge graph with more relevant triples.
-
4.
MED-H: This experiment is designed to test the effectiveness of our approach for predicting the tail entity. In this module, we try to predict the head entities instead of the tail entities from the augmented knowledge graph for the masked head tokens. The results in Table 6 shows that this leads to a 4.5% decrease in the F1 score and results in a similar performance to the BioBERT model indicating predicting head entities is similar to performing fine-tuning on the dialog dataset and thus demonstrating the effectiveness of our approach of predicting the tail entity given the masked head entities.
Error analysis
Using the generated responses, we conduct a thorough examination of our proposed model and classify the errors it encounters into the following categories:
-
1.
Non-clinical Response: We observed around 1% cases where responses as “hello & welcome to healthcaremagic!” appear multiple times without any text with medical information. This is due to the fact that using a simple decoder in conjunction with complex context vectors can easily result in non-clinical and generic responses.
-
2.
Non-Fluency: We observe around 5% cases of non-fluent outputs. For example, “hi, i understand your concern to your history and your history. yes, i can help you better, the only way to find your particular diagnosis that you might be suffering from benign tumor of left breast. ... if it is just a simple cyst of the small and in the future i”. The reason for this is that input and output sequences are truncated because the maximum sequence length for pre-trained models is limited to a fixed length.
-
3.
Knowledge irrelevancy: We observe around 5% cases of knowledge irrelevancy. For example, Patient: “my baby has fever even after giving paracetamol. please advise . hello doctor, my four month old baby weighs 7 kg. he has a 100-degree fever since today noon. i had given 4 ml of 125g paracetamol syrup. but the temperature is still at 100.”; Doctor: “you can continue giving the 4ml paracetamol syrup, every four to six hours if there is a fever. it will take an hour or two for the temperature to normalize. if fever is nearing 102f, give ibuprofen 70mg. it is most likely to be a viral fever. so, if the baby is active, you can wait for two days. if you notice any new symptoms, or if the fever persists beyond 48 hours, then consult his previous doctor.”; Predicted Response: “don’t worry. with the help of medicine, it is fine. please don’t worry, as such nothing is required. if his symptoms not responding to him, wait and watch.”. We discovered that, while our proposed model produced an adequate response, it missed many clinical entities that were important. This might be attributed to the fact that since a head entity can have multiple tail entities associated with it and selection of relevant tail entities is still an open problem.
Conclusion and future works
A virtual doctor uses relevant knowledge graphs to generate clinically correct responses to patient queries. This is critical for the development of a robust and effective medical dialogue system. Previous approaches to integrating knowledge graphs only considered attentive methods to encode them into the current utterance. We proposed a knowledge-driven neural conversational model for medical dialogue generation in this paper. Unlike previous methods, we incorporate the knowledge without using any attention mechanism and by encoding the augmented knowledge graphs alongside the patient’s utterances using the large-scale pretrained language models. We use knowledge from the UMLS database and improve it using medical entity annotation in a semi-supervised fashion to facilitate effective conversation understanding and generation. On the MedDialog testset, our proposed method, MED results in a significant performance improvement of around 5.2 F1 score points and 8.3 BLEU-4 points. On F1 and BLEU-4, MED outperforms the BioBERT baseline by 8.5 and 9.3 points, respectively, on the Covid Dataset. The results on two benchmark medical dataset show masking relevant tokens by jointly utilizing the dialogues and graph based knowledge can successfully enrich entity embeddings based on the dialogue context and hence produce clinically correct and instructive responses. Aside from these, the model’s small misrepresentations are discussed in detail in the error analysis section.
In the future, we aim to make use of commonsense knowledge graphs as well as introduce reward functions to tackle the non-clinical response and non fluency errors for the model. The codes and dataset used to replicate our findings are available at https://github.com/deekshaVarshney/MED.git
Data availability
The MedDialog(EN) and CovidDialog dataset analysed in this work are included in these published articles by Zeng et al.50 and Yang et al.59 and can be downloaded from the links: https://github.com/UCSD-AI4H/Medical-Dialogue-System and https://github.com/UCSD-AI4H/COVID-Dialogue respectively. The extended CovidDialog dataset renamed as the Covid Dataset analysed / generated during the current study as explained in section “Covid dataset” are available in the https://github.com/deekshaVarshney/MED.git repository. We have crawled this data from the same websites (https://www.icliniq.com/ and https://www.healthcaremagic.com/) which is an online platform of healthcare services and all the rights reserve to them. The website completely anonymizes the patient’s identity.
References
Zhao, Y., Wu, W. & Xu, C. Are pre-trained language models knowledgeable to ground open domain dialogues? arXiv:2011.09708 (2020).
Zhang, Y. et al. Dialogpt: Large-scale generative pre-training for conversational response generation. In Proceedings of the 58th Annual Meeting of the Association for Computational Linguistics: System Demonstrations 270–278 (2020).
Zhao, X. et al. Knowledge-grounded dialogue generation with pre-trained language models. In Proceedings of the 2020 Conference on Empirical Methods in Natural Language Processing (EMNLP) 3377–3390. https://doi.org/10.18653/v1/2020.emnlp-main.272 (Association for Computational Linguistics, Online, 2020).
Reddy, R. G., Contractor, D., Raghu, D. & Joshi, S. Multi-level memory for task oriented dialogs. In Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Volume 1 (Long and Short Papers) 3744–3754 (2019).
Wang, J. et al. Dual dynamic memory network for end-to-end multi-turn task-oriented dialog systems. In Proceedings of the 28th International Conference on Computational Linguistics 4100–4110. https://doi.org/10.18653/v1/2020.coling-main.362 (International Committee on Computational Linguistics, Barcelona, Spain (Online), 2020).
Wei, Z. et al. Task-oriented dialogue system for automatic diagnosis. In Proceedings of the 56th Annual Meeting of the Association for Computational Linguistics (Volume 2: Short Papers) 201–207 (2018).
Xia, Y., Zhou, J., Shi, Z., Lu, C. & Huang, H. Generative adversarial regularized mutual information policy gradient framework for automatic diagnosis. Proc. AAAI Conf. Artif. Intell. 34, 1062–1069 (2020).
Xu, L. et al. End-to-end knowledge-routed relational dialogue system for automatic diagnosis. Proc. AAAI Conf. Artif. Intell. 33, 7346–7353 (2019).
Li, D. et al. Semi-supervised variational reasoning for medical dialogue generation. In Proceedings of the 44th International ACM SIGIR Conference on Research and Development in Information Retrieval 544–554 (2021).
Bodenreider, O. The unified medical language system (umls): Integrating biomedical terminology. Nucleic Acids Res. 32, D267–D270 (2004).
Weizenbaum, J. Eliza-a computer program for the study of natural language communication between man and machine. Commun. ACM 9, 36–45 (1966).
Jia, J. Csiec: A computer assisted english learning chatbot based on textual knowledge and reasoning. Knowl.-Based Syst. 22, 249–255 (2009).
Sutskever, I., Vinyals, O. & Le, Q. V. Sequence to sequence learning with neural networks. Adv. Neural. Inf. Process. Syst. 2014, 3104–3112 (2014).
Serban, I. V. et al. A hierarchical latent variable encoder-decoder model for generating dialogues. In Proceedings of the Thirty-First AAAI Conference on Artificial Intelligence 3295–3301 (2017).
Xing, C. et al. Topic aware neural response generation. In Proceedings of the Thirty-First AAAI Conference on Artificial Intelligence 3351–3357 (2017).
Li, J., Galley, M., Brockett, C., Gao, J. & Dolan, W. B. A diversity-promoting objective function for neural conversation models. In Proceedings of the 2016 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies 110–119 (2016).
Zhao, T., Zhao, R. & Eskenazi, M. Learning discourse-level diversity for neural dialog models using conditional variational autoencoders. In Proceedings of the 55th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers) 654–664 (2017).
Sun, B., Feng, S., Li, Y., Liu, J. & Li, K. Think: A novel conversation model for generating grammatically correct and coherent responses. Knowl. Based Syst. 2022, 108376 (2022).
Zhang, B. et al. A memory network based end-to-end personalized task-oriented dialogue generation. Knowl.-Based Syst. 207, 106398 (2020).
Devlin, J., Chang, M.-W., Lee, K. & Toutanova, K. BERT: Pre-training of deep bidirectional transformers for language understanding. In Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Volume 1 (Long and Short Papers) 4171–4186, https://doi.org/10.18653/v1/N19-1423 (Association for Computational Linguistics, Minneapolis, Minnesota, 2019).
Lewis, M. et al. Bart: Denoising sequence-to-sequence pre-training for natural language generation, translation, and comprehension. In Proceedings of the 58th Annual Meeting of the Association for Computational Linguistics 7871–7880 (2020).
Radford, A. et al. Language models are unsupervised multitask learners. OpenAI blog 1, 9 (2019).
Raffel, C. et al. Exploring the limits of transfer learning with a unified text-to-text transformer. J. Mach. Learn. Res. 21, 1–67 (2020).
Brown, T. et al. Language models are few-shot learners. Adv. Neural Inf. Process. Syst. 33, 1877–1901 (2020).
Song, K., Tan, X., Qin, T., Lu, J. & Liu, T.-Y. Mass: Masked sequence to sequence pre-training for language generation. In International Conference on Machine Learning 5926–5936 (PMLR, 2019).
Young, T. et al. Augmenting end-to-end dialogue systems with commonsense knowledge. In Proceedings of the Thirty-Second AAAI Conference on Artificial Intelligence and Thirtieth Innovative Applications of Artificial Intelligence Conference and Eighth AAAI Symposium on Educational Advances in Artificial Intelligence 4970–4977 (2018).
Zhou, H. et al. Commonsense knowledge aware conversation generation with graph attention. In Proceedings of the 27th International Joint Conference on Artificial Intelligence 4623–4629 (2018).
Wu, S., Li, Y., Zhang, D., Zhou, Y. & Wu, Z. Topicka: Generating commonsense knowledge-aware dialogue responses towards the recommended topic fact. In Proceedings of the Twenty-Ninth International Joint Conference on Artificial Intelligence, IJCAI 3766–3772 (2020).
Jiang, B. et al. Knowledge augmented dialogue generation with divergent facts selection. Knowl.-Based Syst. 210, 106479 (2020).
He, W. et al. Multi-goal multi-agent learning for task-oriented dialogue with bidirectional teacher-student learning. Knowl.-Based Syst. 213, 106667 (2021).
Liu, Q. et al. Heterogeneous relational graph neural networks with adaptive objective for end-to-end task-oriented dialogue. Knowl.-Based Syst. 227, 107186 (2021).
Wu, S., Li, Y., Zhang, D., Zhou, Y. & Wu, Z. Diverse and informative dialogue generation with context-specific commonsense knowledge awareness. In Proceedings of the 58th Annual Meeting of the Association for Computational Linguistics 5811–5820.https://doi.org/10.18653/v1/2020.acl-main.515 (Association for Computational Linguistics, Online, 2020).
Liu, S. et al. Knowledge diffusion for neural dialogue generation. In Proceedings of the 56th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers) 1489–1498 (2018).
Liu, Z., Niu, Z.-Y., Wu, H. & Wang, H. Knowledge aware conversation generation with explainable reasoning over augmented graphs. In Proceedings of the 2019 Conference on Empirical Methods in Natural Language Processing and the 9th International Joint Conference on Natural Language Processing (EMNLP-IJCNLP) 1782–1792 (2019).
Varshney, D., Prabhakar, A. & Ekbal, A. Commonsense and named entity aware knowledge grounded dialogue generation. In Proceedings of the 2022 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies 1322–1335, https://doi.org/10.18653/v1/2022.naacl-main.95 (Association for Computational Linguistics, Seattle, United States, 2022).
Liu, Y., Wan, Y., He, L., Peng, H. & Yu, P. S. Kg-bart: Knowledge graph-augmented bart for generative commonsense reasoning. Proc. AAAI Conf. Artif. Intell. 35, 6418–6425 (2021).
Matthew, E. P. et al. Knowledge Enhanced Contextual Word Representations. In Proceedings of the 2019 Conference on Empirical Methods in Natural Language Processing and the 9th International Joint Conference on Natural Language Processing (EMNLP-IJCNLP). 43–54 (2019).
He, B. et al BERT-MK: Integrating Graph Contextualized Knowledge into Pre-trained Language Models. In Findings of the Association for Computational Linguistics: EMNLP 2020. 2281–2290 (2020).
Xiong, W., Du, J., Wang, W. Y. & Stoyanov, V. Pretrained encyclopedia: Weakly supervised knowledge-pretrained language model. In International Conference on Learning Representations (2019).
Wang, X. et al. Kepler: A unified model for knowledge embedding and pre-trained language representation. Trans. Assoc. Comput. Linguist. 9, 176–194 (2021).
Sun, T. et al. Colake: Contextualized language and knowledge embedding. In Proceedings of the 28th International Conference on Computational Linguistics 3660–3670 (2020).
Sun, Y. et al. Ernie 3.0: Large-scale knowledge enhanced pre-training for language understanding and generation. arXiv:2107.02137 (2021).
Lee, J. et al. Biobert: A pre-trained biomedical language representation model for biomedical text mining. Bioinformatics 36, 1234–1240 (2020).
Alsentzer, E. et al. Publicly available clinical bert embeddings. In Proceedings of the 2nd Clinical Natural Language Processing Workshop 72–78 (2019).
Rao, D. J., Mane, S. S. & Paliwal, M. A. Biomedical multi-hop question answering using knowledge graph embeddings and language models. arXiv:2211.05351 (2022).
Auti, T. et al. Towards classification of legal pharmaceutical text using gan-bert. In Proceedings of the First Computing Social Responsibility Workshop within the 13th Language Resources and Evaluation Conference 52–57 (2022).
Yang, J., Liu, Z., Zhang, M. & Wang, S. Pathway2text: Dataset and method for biomedical pathway description generation. Find. Assoc. Comput. Linguist.: NAACL 2022, 1441–1454 (2022).
Luo, R. et al. Biogpt: Generative pre-trained transformer for biomedical text generation and mining. Brief. Bioinform. 2022, 409 (2022).
Blanc, C. et al. Flaubert vs. camembert: Understanding patient’s answers by a french medical chatbot. Artif. Intell. Med. 2022, 102264 (2022).
Zeng, G. et al. Meddialog: A large-scale medical dialogue dataset. In Proceedings of the 2020 Conference on Empirical Methods in Natural Language Processing (EMNLP) 9241–9250 (2020).
Liu, W. et al. Meddg: A large-scale medical consultation dataset for building medical dialogue system. arXiv:2010.07497 (2020).
Lin, X. et al. Enhancing dialogue symptom diagnosis with global attention and symptom graph. In Proceedings of the 2019 Conference on Empirical Methods in Natural Language Processing and the 9th International Joint Conference on Natural Language Processing (EMNLP-IJCNLP) 5033–5042 (2019).
Lin, S. et al. Graph-evolving meta-learning for low-resource medical dialogue generation. In Proceedings of the 35th AAAI Conference on Artificial Intelligence 13362–13370 (AAAI Press, 2021).
Liu, F. et al. Auto-encoding knowledge graph for unsupervised medical report generation. Adv. Neural. Inf. Process. Syst. 34, 16266–16279 (2021).
Liang, K., Wu, S. & Gu, J. Mka: A scalable medical knowledge-assisted mechanism for generative models on medical conversation tasks. Comput. Math. Methods Med. 2021, 5294627–5294627 (2021).
Soldaini, L. & Goharian, N. Quickumls: A fast, unsupervised approach for medical concept extraction. In andMedIR workshop, sigir 1–4 (2016).
Fleiss, J. L. Measuring nominal scale agreement among many raters. Psychol. Bull. 76, 378 (1971).
Hayes, A. F. & Krippendorff, K. Answering the call for a standard reliability measure for coding data. Commun. Methods Meas. 1, 77–89 (2007).
Yang, W. et al. On the generation of medical dialogues for covid-19. arXiv:2005.05442 (2020).
Papineni, K., Roukos, S., Ward, T. & Zhu, W.-J. Bleu: A method for automatic evaluation of machine translation. In Proceedings of the 40th annual meeting on association for computational linguistics 311–318 (Association for Computational Linguistics, 2002).
Vinyals, O. & Le, Q. A neural conversational model. arXiv:1506.05869 (2015).
Gopalakrishnan, K. et al. Topical-chat: Towards knowledge-grounded open-domain conversations. In INTERSPEECH 1891–1895 (2019).
Liu, C.-W. et al. How NOT to evaluate your dialogue system: An empirical study of unsupervised evaluation metrics for dialogue response generation. In Proceedings of the 2016 Conference on Empirical Methods in Natural Language Processing 2122–2132. https://doi.org/10.18653/v1/D16-1230 (Association for Computational Linguistics, Austin, Texas, 2016).
Acknowledgements
Authors gratefully acknowledge the support from the projects “Percuro-A Holistic Solution for Text Mining“, sponsored by Wipro Ltd; and “Sevak-An Intelligent Indian Language Chabot“, sponsored by Imprint 2, SERB, Government of India.
Author information
Authors and Affiliations
Contributions
This study's idea was created by D.V. and A.E. The experiment(s) were carried out by D.V., A.Z., and N.B. All authors then analyzed the results. The manuscript was written by D.V., A.Z., and N.B. with helpful suggestions from A.E. The manuscript was reviewed by all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Varshney, D., Zafar, A., Behera, N.K. et al. Knowledge grounded medical dialogue generation using augmented graphs. Sci Rep 13, 3310 (2023). https://doi.org/10.1038/s41598-023-29213-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-29213-8
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.