Abstract
During mammalian brain development, neural progenitor cells proliferate extensively but can ensure the production of correct numbers of various types of mature cells by balancing symmetric proliferative versus asymmetric differentiative cell divisions. This process of cell fate determination may be harnessed for developing cancer therapy. Here, we test this idea by targeting KIF20A, a mitotic kinesin crucial for the control of cell division modes, in a genetic model of medulloblastoma (MB) and human MB cells. Inducible Kif20a knockout in both normal and MB-initiating granule neuron progenitors (GNPs) causes early cell cycle exit and precocious neuronal differentiation without causing cytokinesis failure and suppresses the development of Sonic Hedgehog (SHH)-activated MB. Inducible KIF20A knockdown in human MB cells inhibits proliferation both in cultures and in growing tumors. Our results indicate that targeting the fate specification process of nascent daughter cells presents a novel avenue for developing anti-proliferation treatment for malignant brain tumors.
Similar content being viewed by others
Introduction
During mammalian brain development, extensive expansion occurs in neural progenitor cells (NPCs); however, in contrast to the proliferating cells in a growing tumor, NPCs can produce correct numbers of diverse cell types without running a risk of excessive or dysregulated proliferation. NPCs achieve this feat through coordinated actions of growth-stimulating and -suppressing factors leading to symmetric or asymmetric cell divisions, which endow a proliferative or differentiative fate to daughter cells, so that an overall balance between proliferation and differentiation is ensured throughout brain morphogenesis. Disruption of the balance between symmetric and asymmetric cell divisions is expected to cause an imbalance between proliferation and differentiation, and may lead to tumorigenesis. Genetic studies in the Drosophila nervous system presented early evidence linking defects in cell division mode to tumorigenesis. In the Drosophila neuroblasts, the fly NPCs, mutations of multiple regulators of asymmetric cell division have been linked to hyperproliferation of neuroblasts in the embryos1,2,3,4,5,6 and tumor-like growth in recipient flies when mutant cells were transplanted1. These observations indicated that mis-regulation of the fate of an NPC’s daughter cells could lead to cancer7,8. These observations also suggested that the fate specification process of daughter cells might serve as a point of intervention for developing cancer therapy. However, key regulators that control symmetric proliferative vs. asymmetric differentiative divisions in the mammalian NPCs have remained largely elusive and, hence, the idea of targeting regulators of cell division mode for therapeutic intervention has not been directly tested in the mammalian systems.
We have recently identified KIF20A, a mitotic kinesin previously implicated in cytokinesis regulation in cancer cells, with a crucial role in balancing symmetric vs. asymmetric NPC divisions during cerebral cortical development9. Our data showed that inactivation of KIF20A through either short hairpin RNA (shRNA) knockdown or genetic knockout causes the affected cortical NPCs to switch from proliferative to differentiative mode of divisions. KIF20A exerts its function in the process of daughter cell-fate specification during NPC divisions in coordination with RGS39 and SEPT710. As tumor-initiating (stem/progenitor-like) cells share many regulatory mechanisms of cell proliferation with normal neural stem/progenitor cells11,12,13,14,15, it is conceivable that KIF20A is also crucial for the control of proliferation vs. differentiation of tumor-initiating cells.
In human cancers, KIF20A shows low mutation rate based on available TCGA database, which is consistent with the essential role of KIF20A in cell divisions of NPCs or other stem/progenitor cells (germline knockout of the Kif20a leads to embryonic/perinatal lethality). On the other hand, in silico studies of cancer expression data or expression analyses of clinical samples showed that KIF20A expression is positively correlated with poor prognosis of patients in different types of cancers16,17,18,19. Based on these observations, we anticipate that disruption of KIF20A function in brain tumor-initiating cells would similarly promote the affected cells to leave the cell cycle and become post mitotic, and as a result, this should lead to inhibition of brain tumor initiation and/or progression.
To test this idea, we employed a genetic model of medulloblastoma (MB) induced by sustained activation of the Sonic Hedgehog (SHH) pathway. In this model system, inducible knockout of the Patched (Ptc) gene in cerebellar granule neuron progenitors (GNPs) (via expression of CreER under the control of the Atoh1 promoter) results in MB formation within a few months20. As KIF20A is a mitotic protein that should be expressed by NPCs present in other central nervous system areas beyond the cerebral cortex, we expected that KIF20A might play a similar role in regulating cell division mode, both in normal cerebellar GNPs during development and in tumor-initiating GNPs during tumor formation. To target KIF20A in the cerebellum, we established compound mice carrying Atoh1-CreER, Kif20afl/fl and Atoh1-CreER, Kif20afl/fl, Ptcfl/fl alleles. Deletion of the Kif20a gene alone or deletion of both the Ptc and Kif20a genes in cerebellar GNPs was investigated. We will present data obtained from these experiments showing that loss-of-function (LOF) of KIF20A resulted in early cell cycle exit and precocious neuronal differentiation in both normal and tumor-initiating GNPs, as well as data from targeting KIF20A function in human MB cells both in vitro and in vivo.
Results
Conditional knockout of Kif20a in cerebellar GNPs causes early cell cycle exit and precocious neuronal differentiation
In the mouse cerebellum, immunostaining showed that strong KIF20A expression is enriched within the external granular layer (EGL), where GNPs are located during early postnatal stages (Fig. 1a). To study the potential function of KIF20A in cerebellar development, we performed inducible deletion of the Kif20a gene in GNPs. Conditional Kif20a-knockout mice (littermates of Atoh1-CreER; Kif20afl/fl and control Kif20afl/fl mice) were treated with tamoxifen at postnatal day 4 (P4). Brain samples were then collected from P5 for analyses. Assessment of KIF20A expression in the P5 and P6 samples indicated that deletion of Kif20a was evident 1 day after tamoxifen induction (Supplementary Fig. 1a). Examination of Ki67 expression in the brain samples revealed a loss of Ki67+ cells in the mutant cerebellums following the induced deletion of Kif20a (Fig. 1b and Supplementary Fig. 1b). To address whether the loss of proliferating cells resulted from increased death of mutant GNPs (due to possible defect in cytokinesis after Kif20a knockout), we performed immunostaining for activated caspase 3. Our results from P6 and P7 brain samples showed that the apoptosis levels in the cerebellums were overall low and comparable between the Kif20a-knockout and wild-type littermate brains (Fig. 1c and Supplementary Fig. 1c). We previously found that knockout of Kif20a did not cause a noticeable failure of cytokinesis in neural progenitor divisions of the developing cerebral cortex9. To assess the status of cytokinesis process in the mutant cerebellums, we performed flow cytometry-based cell cycle analysis. Our results showed that the 4N DNA contents between the P6 mutant (Atoh1-CreER; Kif20afl/fl) and control (Kif20afl/fl) cerebellar cells were comparable (Fig. 1d), indicating that knockout of Kif20a did not cause an obvious defect in cytokinesis of GNP divisions, or a possible cytokinesis defect was largely compensated by other factors.
a KIF20A expression was mainly enriched in the proliferating cell zone in the external granular layer (EGL) of the early postnatal cerebellum. Scale bar represents 50 μm. b Tamoxifen (TM)-induced knockout of Kif20a in GNPs caused a reduction in the number of proliferating cells in the mutant cerebellums. Ki67+ cells within boxed region at the center of individual cerebellar sulcus were used for quantification. White arrowheads indicated the tips of sulci in the mutant cerebellums, which often had diminished Ki67 signal. Scale bar represents 50 μm. Data are mean ± SD. P = 0.0019 (P5), 6.65E − 05 (P6), 9.52E − 05 (P7) (Student’s t-test). c Knockout of Kif20a did not cause a noticeable increase of apoptosis level marked by cleaved caspase 3 in the P6 mutant cerebellums. Scale bar represents 50 μm. Data are mean ± SD. d Flow cytometry-based cell cycle analysis of P6 littermate of knockout and control cerebellums. The 4N DNA contents (representing G2/M phase cells or cells with two nuclei resulted from cytokinesis defect) were little changed between the Kif20a-knockout and control brains. n.s., not significant.
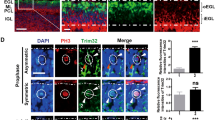
We next examined the cell cycle exit and re-entry in GNPs of the Kif20a-knockout cerebellum. In this experiment, conditional Kif20a mice were treated with tamoxifen at P4 followed by an injection of EdU at P5, and brain samples were then collected at P6 or P7 for analyses (Fig. 2). In the P6 brains, co-staining of EdU and Ki67 showed that within individual cerebellar sulci, there were relatively more EdU+Ki67− cells (cells having left the cell cycle after being labeled by EdU) in the mutant EGL than in the wild-type littermate EGL (Fig. 2a). Co-staining with neuronal marker NeuN further showed that more EdU+Ki67− cells in the Kif20a-knockout brains were positive for NeuN (Fig. 2b). These EdU+Ki67−NeuN+ cells were located outside the proliferating zone (Ki67+ cell zone) (Fig. 2b), reflecting developmental progression of differentiated granule neurons emanating from the EGL. This result also suggested that knockout of Kif20a did not compromise the completion of cell division (cytokinesis) of the mutant GNPs. In the P7 brains, within individual cerebellar sulci, significantly fewer EdU+Ki67+ cells (cells remaining in the cell cycle) could be seen in the mutant EGL than in the wild-type EGL (Fig. 2c). These data collectively suggested that KIF20A is essential for maintaining the proliferative state of GNPs and that LOF of KIF20A in GNPs causes cell cycle exit and leads to an early depletion of the progenitor population.
a After tamoxifen (TM) treatment at P4, animal pups were labeled with EdU at P5 and then brains were collected at P6. Co-staining of EdU and Ki67 showed that knockout of Kif20a resulted in relatively more EdU+Ki67− cells in the EdU+ cell population compared to the wild-type littermate brains. Scale bar represents 50 μm. Data are mean ± SD. P = 1.09E − 09 (Student’s t-test). b More EdU+Ki67− cells in the mutant cerebellums were positive for neuronal marker NeuN (white arrowheads). These cells formed a line outside the Ki67+ cells, reflecting they are differentiating and migrating out of the EGL. Scale bar represents 50 μm. Data are mean ± SD. P = 1.29E − 06 (Student’s t-test). c After tamoxifen treatment at P4, animal pups were labeled with EdU at P5 and then brains were collected at P7. Co-staining of EdU and Ki67 showed that knockout of Kif20a resulted in fewer EdU+Ki67+ proliferating cells in the EGL at this stage. EdU+Ki67+ cells within individual cerebellar sulcus were used for quantification. Scale bar represents 50 μm. Data are mean ± SD. P = 1.58E − 07 (Student’s t-test).
Blocking KIF20A function inhibits SHH-induced MB
To address whether KIF20A might be similarly crucial for maintaining the proliferation of brain tumor-initiating cells, we crossed the Kif20afl/fl floxed mice with Atoh1-CreER; Ptcfl/fl mice to generate a strain of compound mice carrying the Atoh1-CreER; Ptcfl/fl; Kif20afl/fl alleles. When tamoxifen was given to the pups at P4, Ptc-knockout mice with intact Kif20a (Atoh1-CreER; Ptcfl/fl) developed MB with 100% penetrance within about 5 months (Fig. 3a), as previously reported20. Ptc/Kif20a double-knockout mice (Atoh1-CreER; Ptcfl/fl; Kif20afl/fl), on the other hand, showed a longer survival time before symptoms occurred and some of the mice appeared normal even 1 year after tamoxifen induction (Fig. 3a). Among the mice that developed symptoms, the brains of the Ptc/Kif20a double-knockout mice had smaller tumors than those seen in Ptc-knockout mice with intact Kif20a (Fig. 3b–h). The brain samples from the Ptc/Kif20a double-knockout mice generally also retained more normal morphology than those Ptc single-knockout mice (Fig. 3b and Fig. 3f–h vs. 3d, e). These results suggest that KIF20A is crucial for the development of SHH-induced MB.
a Single (Atoh1-CreER; Ptcfl/fl)- and double (Atoh1-CreER; Ptcfl/fl; Kif20afl/fl)-knockout mice were treated with tamoxifen at P4 by gavage. Brain samples were collected for analyses when brain tumor symptoms were developed. Survival of mice was summarized in the Kaplan–Meier curve. b Representative whole brain samples from wild-type (Ptcfl/fl), Ptc single (Atoh1-CreER; Ptcfl/fl)-, or Ptc/Kif20a double (Atoh1-CreER; Ptcfl/fl; Kif20afl/fl)-knockout mice. Numbers in each panel indicate the days when brain samples were collected after TM injection (sections of these brains were shown in c–h). c Normal brain features from section of wild-type control (Ptcfl/fl) brain collected 145 days after TM injection. Arrowhead indicates the cerebellum. d, e Examples of sections from Ptc single-knockout mice displayed strong tumor growth and often disformed brain structures. Arrows indicate the tumor mass. f–h Examples of sections from Ptc/Kif20a double mice displayed varied tumor sizes. The overall brain structures were in general in better shape than the single-knockout mice.
LOF of KIF20A in MB-initiating GNPs causes early cell cycle exit
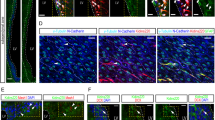
To study the possible mechanism of inhibition of SHH-induced MB by LOF of KIF20A, we looked at cell cycle exit and re-entry in tumor-initiating GNPs of the Ptc single- and Ptc/Kif20a double-knockout mice. The pups of Atoh1-CreER; Ptcfl/fl and Atoh1-CreER; Ptcfl/fl; Kif20afl/fl mice were treated with tamoxifen at P4 followed by EdU labeling at P5. Brain samples were then collected at P6 and P7 for analysis. In the P6 brains, data from Ki67 staining showed that there were overall fewer Ki67+ cells in the cerebellar sulci of the Ptc/Kif20a double-knockout brains than those of the Ptc single-knockout brains (Fig. 4a). Co-staining of EdU and Ki67 showed that there were more EdU+Ki67− cells in the Ptc/Kif20a double-knockout EGL than in the Ptc single-knockout EGL (Fig. 4a), suggesting that more GNPs in the Ptc/Kif20a double-knockout brains had left the cell cycle earlier. Co-staining with neuronal marker NeuN further showed that more EdU+Ki67− cells in the Ptc/Kif20a double-knockout brains were positive for NeuN and these EdU+Ki67−NeuN+ cells formed a line surrounding the proliferating zone, reflecting newly born granule neurons exiting the EGL (Fig. 4b). In the P7 brains, fewer proliferating cells (EdU+Ki67+ cells) remained in the EGL of the Ptc/Kif20a double-knockout mice than in the Ptc single-knockout EGL (Fig. 4c). These data collectively indicate that KIF20A is also essential for maintaining the proliferative state of tumor-initiating GNPs.
a After tamoxifen treatment at P4, animal pups were labeled with EdU at P5 and then brains were collected at P6. Co-staining of EdU and Ki67 revealed that knockout of Kif20a in MB-initiating GNPs (Ptc single-knockout GNPs) resulted in more EdU+Ki67− cells in the EdU+ cell population. White arrowheads indicate the EdU+Ki67− cells, many of which line outside the proliferating cell zone. Scale bar represents 50 μm. Quantifications from Fig. 2a (green and red columns) were plotted together in the graph. Data are mean ± SD. P = 1.82E − 05 (between the first two columns) (Student’s t-test). b More EdU+Ki67− cells in the Ptc/Kif20a double-knockout cerebellums were positive for neuronal marker NeuN (white arrowheads). Scale bar represents 50 μm. Data are mean ± SD. P = 0.00013 (Student’s t-test). c After tamoxifen treatment at P4, animal pups were labeled with EdU at P5 and then brains were collected at P7. Co-staining of EdU and Ki67 showed that knockout of Kif20a in MB-initiating GNPs resulted in fewer EdU+Ki67+ proliferating cells in the EGL. EdU+Ki67+ cells within individual cerebellar sulcus were used for quantification. Scale bar represents 50 μm. Data are mean ± SD. P = 0.00042 (Student’s t-test).
LOF of KIF20A in mouse tumor cells leads to inhibition of proliferation by inducing differentiation
To further examine the mechanisms of KIF20A function in MB, we sought to isolate proliferating cells from MBs derived from both the Atoh1-CreER; Ptcfl/fl and Atoh1-CreER; Ptcfl/fl; Kif20afl/fl knockout mice. Some tumor cells could expand in culture and we were able to establish two cell lines from each tumor type. We first examined CreER-mediated gene deletion by genotyping PCR on genomic DNAs (Supplementary Fig. 2a). Our results showed that targeted homozygous deletion of the Ptc gene was evident in all four tumor cell lines, consistent with the occurrence of MB in these mice (Supplementary Fig. 2b; results of one cell line from each tumor type were shown). Surprisingly, however, in the cell lines derived from the Ptc/Kif20a double-knockout mice, the Kif20a gene was in a heterozygous state, with one allele having the intact floxed Kif20a gene and the other allele having the floxed exons removed (Supplementary Fig. 2c). Further quantitative PCR on RNAs isolated from these cells showed that a Kif20a transcript was expressed, albeit in a reduced level (Supplementary Fig. 2d). In theory, tamoxifen-induced activation of CreER should have led to simultaneous homozygous deletion of both the Ptc and Kif20a genes in GNPs carrying the Atoh1-CreER; Ptcfl/fl; Kif20afl/fl alleles; however, our results suggested that simultaneous deletion of the Kif20a and Ptc genes was faulty in some GNPs of the double-knockout mice, such that although the Ptc gene was deleted, the Kif20a gene was left half intact. Thus, the cell lines derived from tumors of the Ptc single and Ptc/Kif20a double-knockout mice have the genotype of Ptc−/−; Kif20a+/+ and Ptc−/−; Kif20afl/−, respectively.
We next performed analyses on cell biological properties of these two types of tumor cells. Our results showed that tumor cells derived from the Ptc/Kif20a double-knockout mice had a slower proliferation rate (Fig. 5a), consistent with the overall longer survival time of the double-knockout mice (Fig. 3a). These tumor cells also showed fewer Ki67+ proliferating cells and were prone to exiting the cell cycle (Fig. 5b), compared to tumor cells derived from the Ptc single-knockout mice. The two types of tumor cells displayed comparable 4N DNA contents in cell cycle analysis (Supplementary Fig. 3), suggesting that deletion of Kif20a in SHH-MB cells did not alter the cytokinesis status. To further test the effect of Kif20a deletion, we next introduced Cre enzyme (Cre-2A-GFP), or control green fluorescent protein (GFP), into the tumor cells derived from the Ptc/Kif20a double-knockout mice, using a lentiviral expression system. The infected cells were next labeled with EdU for 24 h before being fixed for immunostaining. Our results showed that expression of Cre in these heterozygous Kif20a tumor cells caused an increase of EdU+Ki67− cells compared to cells expressing GFP control alone (Fig. 5c), suggesting more cells exited the cell cycle after Cre expression. In addition, cell cycle analysis of these tumor cells showed that the cells expressing Cre or control GFP displayed comparable 4N DNA contents (Fig. 5d). These results thus collectively indicated that knocking out the remaining Kif20a allele in the tumor cells derived from the Ptc/Kif20a double-knockout mice did not cause an obvious failure in cell division but led to increased exit of the cell cycle in daughter cells. Therefore, the slower growth rate of these tumor cells (Fig. 5a) might be attributed to their tendency for becoming post-mitotic cells due to the half dosage of Kif20a expression, which eventually leads to fewer proliferating cells in the population.
a Two tumor cell lines were derived from the Ptc single-knockout mice and these cells carried a genotype of Ptc−/−; Kif20a+/+. Two tumor cell lines from the Ptc/Kif20a double-knockout mice were also established in culture and these cells showed a genotype of Ptc−/−; Kif20afl/−. The latter two tumor lines displayed slower proliferation rates. Individual data points represented replicates of cell samples. Data are mean ± SD. P = 9.46E − 05 (two-way ANOVA). b Proliferating tumor cells were labeled with EdU for 24 h and were then stained for EdU and Ki67. There were fewer Ki67+ cells and a relatively high percentage of EdU+Ki67− cells (in the total population of EdU+ cells) in the tumor line derived from the Ptc/Kif20a double-knockout mice. Scale bar represents 50 μm. Data are mean ± SD. P = 5.45E − 08; 1.75E − 07 (Student’s t-test). c Tumor cells (having a genotype of Ptc−/−; Kif20afl/−) derived from the Ptc/Kif20a double-knockout mice were infected with lentivirus expressing Cre-2A-GFP or control GFP and were next labeled with EdU for 24 h in culture. Knockout of the remaining Kif20a allele in these cells resulted in cell cycle exit. Scale bar represents 50 μm. Data are mean ± SD. P = 4.12E − 05 (Student’s t-test). d Tumor cells derived from the Ptc/Kif20a double-knockout mice were infected with lentivirus expressing Cre-2A-GFP or control GFP. After labeling with propidium iodide, the cells were examined for their DNA contents by flow cytometry analysis. PI, propidium iodide; 2N, cells in G1 phase; 4N, cells in G2/M phase or bi-nucleated cells. n.s., not significant.
Inducible knockdown of KIF20A in human MB cells inhibits proliferation in culture and tumor growth in xenograft
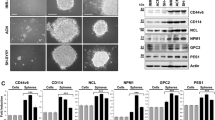
We next examined the expression status of KIF20A in relation to human MB cells using available expression data of patient tumor samples. Our results revealed that strong KIF20A expression is positively correlated with Ki67 level in different subgroups or subtypes of MBs (Fig. 6a–c), indicating that KIF20A is positively associated with proliferation of human MB cells. To address the function of KIF20A in human MB cell proliferation, we generated multiple shRNAs targeting human KIF20A transcript (Supplementary Fig. 4a). One such shRNA (shKIF20A726) was cloned into a Tet-on-inducible shRNA expression vector21. The inducible shKIF20A726 expression cassette and a GFP/firefly luciferase (ffluc) expression cassette were then stably integrated into Daoy human MB cells. Three puromycin-resistant cell clones (stably integrated with Tet-on shKIF20A726 and GFP/ffluc) were isolated. Our data showed that doxycycline treatment could induce significant knockdown of KIF20A expression in all three cell clones (Supplementary Fig. 4b). Doxycycline induction was also seen to significantly inhibit the proliferation of all three clones (Supplementary Fig. 4c). This inhibition was not due to cytotoxicity of doxycycline, because parental Daoy cells were not affected by doxycycline treatment (Supplementary Fig. 4c). Among the three clones, Clone#2 was chosen for further in vivo studies, as it showed comparable growth rate to the parental cells (indicating minimal alteration of cell properties from lentiviral integration and/or minimal leakage of the Tet-on shKIF20A cassette).
a Different subgroups of MB patient cells display strong expression of KIF20A, which is positively correlated with proliferating cell marker Ki67. b KIF20A and Ki67 expressions are also positively correlated in different subtypes of human MB cells. c Comparison between KIF20A expression and Ki67 expression in subgroups of human MB cells. Expression of the two factors are highly correlated. d Daoy cells stably integrated with Tet-on shKIF20A726 and firefly luciferase were intracranially injected into the cerebellar areas of recipient NSG mice (105 cells per mouse). Tumor growths were monitored by bioluminescence imaging. Doxycycline treatment started at 4 weeks after the initial cell implantation (indicated with a red font and line). The treatment group of mice was first given doxycycline by gavage for 2 consecutive days and was then fed with doxycycline-containing food for the entire duration of the experiment. e Survival data were analyzed by Kaplan–Meier plot.
To examine the function of KIF20A in tumor formation by Daoy cells, we injected 105 Clone#2 cells into the cerebellum of each recipient NOD scid gamma (NSG) mouse using a method previously described22. Growth of tumor cells was then monitored by bioluminescence imaging. Four weeks after the initial cell injection, tumor growths in the brains of recipient mice were evident by the increase of imaging signal (Fig. 6d). We then separated the injected mice into two groups: one group was treated with doxycycline and the other group was untreated, as control. For the treatment group, the mice were first fed with doxycycline solution (10 mg/ml, 0.2 ml) by gavage daily for 2 consecutive days. The mice were then kept on doxycycline-containing food continuously and were monitored for tumor growth by bioluminescence imaging. Our results revealed that, in comparison to the control group, the group of mice that received treatment showed significantly slower growth of brain tumors (Fig. 6d) and better overall survival (Fig. 6e), indicating that inhibition of KIF20A expression could suppress cell proliferation in a growing tumor.
Discussion
In mouse spontaneous brain tumor models, accumulating evidence from genetic studies of the MB20,23,24,25 or glioma26,27,28,29,30 have demonstrated that brain tumors originate from dysregulated stem or progenitor cells. Gene expression profiling analyses have also revealed that brain tumor cell transcriptomes display molecular characteristics resembling neural stem/progenitor cell types31,32,33,34,35,36,37. These data thus suggest that targeting the proliferation or differentiation programs employed by stem/progenitor cells is expected to effectively interfere with brain tumor initiation or growth. Along these lines, the daughter cells’ fate specification process during stem/progenitor cell divisions presents a particularly good opportunity for therapeutic intervention, because the process is thought to occur during the late stages of mitosis and is expected to lie downstream of many proliferation-promoting (oncogenic) signaling pathways. In this report, our genetic analyses of KIF20A in both normal and cancer-initiating cerebellar GNPs presents an experimental validation of a cell division mode regulator as a potential target for brain tumor inhibition.
One interesting observation from our genetic analyses was that MB cells isolated from the Atoh1-CreER; Ptcfl/fl; Kif20afl/fl double-knockout mice were homozygous null for the Ptc gene but heterozygous for the Kif20a gene. This result suggested that in the Atoh1-CreER; Ptcfl/fl; Kif20afl/fl knockout mice, some GNPs responded to tamoxifen induction with one Kif20a allele being deleted but leaving the other Kif20a allele intact. One possible reason for this phenomenon might be that Cre recombinase had lower recombination efficiency at the Kif20a locus than at the Ptc locus. This could be due to some structural hindrance of the Kif20a gene; for instance, ten exons were floxed in the Kif20a gene but only one exon was floxed in the Ptc gene (Supplementary Fig. 2a). Another reason might be that Cre-mediated recombination could generate mosaic patterns of recombination products between the Ptc and Kif20a genes, such as cells with genotypes of Ptc−/−; Kif20afl/− or Ptcfl/−; Kif20a−/−, or other combinations. The GNPs of the Ptc−/−; Kif20afl/− genotype were detected in tumor cells, because they should have had an advantage in proliferation over GNPs of other combinations of genotypes and, as a result, they eventually grew out into a tumor over the time. Regardless of how the Ptc−/−; Kif20afl/− GNPs were produced, our results would indicate that the prolonged survival time in the tamoxifen-induced Atoh1-CreER; Ptcfl/fl; Kif20afl/fl mice (Fig. 3a) might be underestimated, due to the incomplete deletion of the Kif20a gene in some of the cancer-initialing progenitor cells. In support of this thought, further deletion of the remaining allele of the Kif20a gene in tumor cells derived from the Ptc/Kif20a double-knockout mice (cells having a genotype of Ptc−/−; Kif20afl/−) accelerated cell cycle exit (Fig. 5c).
Our data obtained from Atoh1-CreER; Kif20afl/fl single and Atoh1-CreER; Ptcfl/fl; Kif20afl/fl double-knockout mice together showed that KIF20A functions similarly in normal and cancer-initiating GNPs to maintain their proliferative potential. We previously found that blocking KIF20A function in NPCs of the developing cerebral cortex did not compromise the divisions of parental progenitor cells but drive the daughter cells to adopt a differentiative fate9. Similarly, data from this study indicated that GNPs with LOF of KIF20A were able to complete cell divisions but the nascent daughter cells became differentiated into neurons, leading to early depletion of proliferating cells in the developing cerebellum and in MB. These results thus indicate that targeting KIF20A, or other regulators of cell division mode control in a broad sense, is distinct from conventional anti-mitotic inhibitors, which act primarily to induce mitotic arrest or cytokinesis defect. Blocking the function of a cell division mode regulator does not disrupt a progenitor cell’s division process per se but promotes the daughter cells to take a differentiative path. Targeting the process of daughter cell-fate specification may thus present a new strategy for expanding the repertoire of anti-proliferation chemotherapy for malignant brain tumors.
Methods
Antibodies
The following antibodies were used: KIF20A(L-13) (Santa Cruz, sc-104954), KIF20A (OriGene, AP01361PU-N), Alpha-tubulin(TU-01) (ThermoFisher, MA1-19162), Ki67(SP6) (Abcam, Ab16667), Active caspase 3 (BD Biosciences, 559565), and NeuN (Millipore, MAB377). Secondary antibodies were purchased from Jackson ImmunoResearch Laboratories (Cy3, Cy5, and Cy2 AffiniPure conjugated) and Click-iT®EdU Alexa Fluor®594 image kit (ThermoFisher Scientific) was used.
DNA constructs and lentivirus production
Lentiviral transfer plasmids used in this study include the following: Hiv7CMV-Cre-myc-2A-GFP (Addgene #117148), Hiv7CMV-GFP, Hiv7CMV-GFP-IRES-Luciferase, and Tet-on-inducible shRNA plasmids Tet-pLKO-shKIF20A726 or Tet-pLKO-shKIF20A181-Puro (shRNA sequences were cloned into pLKO-Tet-On plasmid, Addgene # 21915 at AgeI/EcoRI sites). The plasmids used for shRNA Luciferase in vitro screening include the following: pNUTS-shScramble9, pNUTS-shKIF20A726 or pNUTS-shKIF20A181, and shRNA template plasmid pSiCheck2.2-hKIF20A-CDS. Lentiviruses were generated by co-transfecting 293T cells with 15 μg Lentiviral transfer plasmids, 15 μg second-generation lentiviral packaging plasmid psPAX2 (Addgene #12260), and 5 μg lentiviral envelope plasmid pCMV-G in 10 cm tissue culture plate using calcium phosphate cell transfection reagents. Growth media was exchanged around 6 h later and lentivirus-containing supernatant was collected 48 h later.
Mice
Kif20afl/fl mice were deposited in Mutant Mouse Resources and Research Centers (MMRRC) (strain ID: MMRRC_050513-UCD). Atoh1-CreER; Ptcfl/fl mice were obtained from Dr. Robert Wechsler-Reya’s group. The two strains were crossed to generate Atoh1-CreER; Kif20afl/fl and Atoh1-CreER; Ptcfl/fl; Kif20afl/fl mice. For analyses of normal and MB-initiating GNPs in knockout animals, all mice were treated with a single dose of tamoxifen by gavage at P4, as described in the SHH-MB model20. Animals were group housed and maintained in the temperature range and environmental conditions recommended by Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC). Animal procedures were approved by the Institutional Animal Care and Use Committee of Beckman Research Institute of the City of Hope and were carried out in accordance with NIH guideline and the Guide for the Care and Use of Laboratory Animals.
shRNA design and screening
Potential shRNA sequences were selected using web-based design tool (https://rnaidesigner.thermofisher.com/rnaiexpress/design.do). shRNAs were expressed under the control of a mouse U6 promoter in pNUTs vector, which additionally contains a ubiquitin promoter-EGFP expression cassette. Candidate shRNAs in pNUTs and cDNA target in psi-CHECK were co-transfected into HEK293 cells in triplicates; 48 h later, the firefly and Renilla Luciferase values were determined with Promega’s Dual-Luciferase® Reporter Assay System (Promega, E1910). The final inhibition unit (% luciferase activity) was the normalized value (Renilla/Firefly). The sequences of 19mer human KIF20A shRNAs (the loop of shRNA is underlined) are as below:
Scrambled shRNA, 5′-CGGCTGAAACAAGAGTTGG-TTCAAGAGA-CCAACTCTTGTTTCAGCCG-3′;
shKIF20A726, 5′-GAGGAGTGTCTACATCGAA-TTCAAGAGA-TTCGATGTAGACACTCCTC-3′;
shKIF20A181, 5′-AGTATGGAGAAGGTGAAAG-TTCAAGAGA-CTTTCACCTTCTCCATACT-3′.
Western blotting
Daoy cell clones stably integrated with Tet-on shKIF20A726 were cultured with or without Doxycycline induction for 3 days. These Daoy cells were washed with phosphate-buffered saline (PBS), lysed with 2× SDS loading buffer, and boiled for 5 min. Denatured proteins were resolved by SDS-polyacrylamide gel electrophoresis and transferred to a polyvinylidene difluoride membrane for western blotting detection by KIF20A (L13) antibody (1 : 500) and horseradish-conjugated donkey anti-goat secondary antibody (1 : 2500) with chemiluminescent substrate (ThermoFisher Scientific, Cat: 34095).
Histology, immunocytochemistry, and immunohistochemistry
Animal were perfused with 1× PBS followed by 4% paraformaldehyde (PFA). The whole brains were removed, fixed in 4% PFA overnight, cryoprotected in 30% sucrose, and embedded in Tissue Tek OCT Compound (Fisher HealthCare). For histological analysis, whole brain sagittal sections (6 μm) were stained with hematoxylin and eosin (Sigma). For immunohistochemistry, whole brain sagittal sections (16 μm) or fixed tumor cells were blocked and permeabilized for 2 h at room temperature in blocking solution (1× PBS, 0.1% Triton X-100, 10% Donkey serum, 5% AffiniPure Fab fragment Donkey anti-Mouse IgG, 0.2% Sodium Azide), followed by incubation with primary antibody in blocking solution at 4 °C overnight. The sections or cells were rinsed three times with 1× PBS and incubated with secondary antibody in 1× PBS for 1 h at room temperature. Sections or cells were counterstained with Hoechst 33342 dye and rinsed three times with 1× PBS followed by once in water, then the sections or cells were mounted with Fluoromount-G (SouthernBiotech) for imaging with microscope.
Analyses of inducible knockout mice
The pups of Atoh1-CreER; Ptcfl/fl and Atoh1-CreER; Ptcfl/fl; Kif20afl/fl mice were treated with tamoxifen at P4 by oral gavage (0.6 mg/30 μl) using 24 G gavaging needles. Tamoxifen (T5648, Sigma) was prepared as a 20 mg/ml stock solution in corn oil (Sigma). EdU was then given by intraperitoneal injection at P5 with the dosage of 0.1 mg per gram of the pups. After 24 or 48 h post EdU treatment, the brain samples were collected at P6 and P7 for analyses. Cryosections of P6 or P7 brains (16 μm) were analyzed for immunostaining on EdU, Ki67 (1 : 200), NeuN (1 : 100), or activated caspase 3 (1 : 200). Stained sections were mounted with Fluoromount-G for imaging. For MB formation analysis, after pups were treated with tamoxifen at P4 by gavage, they were monitored until symptoms of brain tumor growth occurred. Brain tumor samples were removed and cryosections were made for histological documentation.
For fluorescence-activated cell sorting (FACS)-based cell cycle analysis, pieces of P6 cerebellums were gently dissociated in cold Hank’s balanced salt solution (HBSS) (Mediatech) with 5 mM EDTA. After centrifugation with 300 × g at 4 °C for 10 min, cells were washed twice with cold HBSS buffer. Subsequently, large cell/aggregates were removed with a cell strainer (40 µm). After fixation with 70% ethanol drop by drop and then being stored for overnight at 4 °C, cells were washed in cold PBS twice. For propidium iodide staining, cells were resuspended in 0.5 ml staining buffer (PBS + 10 μg/ml PI + 0.1% Triton X-100 + 0.2 mg/ml DNAse-free RNAse A) and were incubated at 37 °C for 30 min. Cells were filtered before FACS analysis. Measurements were done using a CyAn ADP instrument (DakoCytomation) in combination with summit software.
Expansion of mouse MB cells in culture
Brains with MB were dissected out and transferred into PBS on ice. Blood and fat tissues were removed from the tumor mass under a dissecting microscope and the tumor tissue pieces were washed again with PBS. Tumor tissues were minced with a scalpel, passed through syringes with 18- and 22-gauge needles, and then incubated in a 1 : 1 mixture of Accutase (Sigma) and TrypLE (Invitrogen) for 15 min at 37 °C with occasionally vortex. Undigested tissues were removed by filtering with 40 μm cell strainer (Falcon, 352340). Dissociated cells were washed twice with Dulbecco’s modified Eagle medium (DMEM)/F12 medium followed by centrifugation at 2000 r.p.m. for 5 min. Cells were then resuspended and plated onto uncoated dishes in Neurobasal and DMEM/F12 media (1 : 1 mix) containing N2 and B27 supplements (Invitrogen), and human recombinant FGF2 and EGF (20 ng/ml, PEPROTECH). Five to 7 days later, cell spheres were dissociated in Accutase (Sigma-Aldrich) and plated onto Primaria dishes (BD Biosciences) coated with Poly-l-ornithine solution (Sigma-Aldrich) and mouse laminin (Sigma-Aldrich) to allow adherent growth.
Generation of Daoy cell lines with stably integrated Tet-on-inducible shRNA expression system
Daoy cells were infected with lentivirus of Tet-on-inducible human KIF20A shRNA-726 and GFP-Luciferase prepared from Tet-pLKO-shKIF20A726 plasmid and Hiv7CMV-GFP-IRES-Luciferase plasmid, respectively. The transduced Daoy cells were selected for stable single clones with Puromycin (1 μg/ml, Sigma-Aldrich) and GFP fluorescence signal in DMEM (Invitrogen) with 10% fetal bovine serum. Single clones were picked up by glass cloning cylinders (Sigma-Aldrich) in 10 cm plate. Three clones were obtained and tested for growth ability and doxycycline (0.5 μg/ml) inducibility using growth curve assay, western blottings, and luciferase reporter assay system. Clone #2 was selected for further use.
Growth assay of tumor cells
Mouse MB cell lines or Daoy clones stably integrated with Tet-on-inducible human KIF20A shRNA-726, or wild-type Daoy cells were seeded into multiple wells of 24-well plates (2 × 104 or 1 × 104 cells per well). Cells in quadruplicate were then dissociated and counted using a hemocytometer at consecutive days after initial plating.
Analyses of KIF20A expression using transcriptome databases of MB patient samples
Raw microarray CEL files were obtained from the two published MB cohorts, GSE8521738 and EGAS0000100195339. Expression data were quantified using custom chip definition files corresponding to Ensembl genes (v23.0.0) from brain array40 and then each dataset was normalized using robust multichip averaging from the oligo package v1.54.141. Duplicate samples were removed and then datasets were integrated for batch correction using ComBat from the sva package v3.36.042.
Intracranial tumor transplantation and bioluminescence imaging
NSG mice (8–10 weeks old) were anesthetized with isoflurane and oxygen, and were placed under a microscope. After exposing the skull with a scalpel, a cell suspension (1 × 105 cells in 4 μl PBS) of Daoy clone #2 stably integrated with Tet-on-inducible human KIF20A shRNA (shRNA-726) and Luciferase gene was slowly injected into the cerebellum at a depth of 2.5 mm using a 10 μl Hamilton syringe with a 26 G needle, using a plastic blocker to control the injection depth. After injection, the incision was closed using wound clips. The transplanted mice were then monitored weekly with luciferase-based bioluminescence imaging for tumor growth. When tumor growth was evident (4 weeks after cell implantation), the mice were separated into two groups—control group and doxycycline treatment group. For the treatment group, mice were first gavaged with 0.2 ml of Doxycycline (10 mg/ml) for 2 consecutive days. Then the mice were fed continuously with Doxycycline-containing food (TestDiet, 625 p.p.m.), with the food being replaced every other day. Tumor growths in two mice groups were monitored weekly by bioluminescence imaging. Survivals of mice after cell transplantation were recorded and analyzed.
Image acquisition and processing
Fluorescent images were taken with Zeiss Observer II or confocal microscope of Zeiss LSM 700 or Zeiss LSM 880.
Statistics and reproducibility
The numbers of tumor-forming knockout or NSG mice (animal survival experiment), brain samples used in each genetic analysis, or replicates of cell samples (wells) in cultured cell experiments, were indicated in the histograms of the figures. All data were presented as mean ± SD. Student’s t-test or two-way analysis of variance were performed and indicated in histograms of the figures. Log-rank test was performed for the Kaplan–Meier curve. In all descriptions, *p < 0.05, **p < 0.01, and ***p < 0.001; n.s. represented not significant (p > 0.05). Cultured cell-related experiments were repeated twice and similar results were obtained.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
All data and materials are available from the authors upon reasonable request. The sources of most materials were indicated in the “Methods” section. All source data underlying the graphs and charts presented in the main figures are available as Supplementary Data.
References
Caussinus, E. & Gonzalez, C. Induction of tumor growth by altered stem-cell asymmetric division in Drosophila melanogaster. Nat. Genet. 37, 1125–1129 (2005).
Betschinger, J., Mechtler, K. & Knoblich, J. A. Asymmetric segregation of the tumor suppressor brat regulates self-renewal in Drosophila neural stem cells. Cell 124, 1241–1253 (2006).
Lee, C. Y., Wilkinson, B. D., Siegrist, S. E., Wharton, R. P. & Doe, C. Q. Brat is a Miranda cargo protein that promotes neuronal differentiation and inhibits neuroblast self-renewal. Dev. Cell 10, 441–449 (2006).
Lee, C. Y. et al. Drosophila Aurora-A kinase inhibits neuroblast self-renewal by regulating aPKC/Numb cortical polarity and spindle orientation. Genes Dev. 20, 3464–3474 (2006).
Wang, H. et al. Aurora-A acts as a tumor suppressor and regulates self-renewal of Drosophila neuroblasts. Genes Dev. 20, 3453–3463 (2006).
Wang, H. Y., Ouyang, Y. S., Somers, W. G., Chia, W. & Lu, B. W. Polo inhibits progenitor self-renewal and regulates Numb asymmetry by phosphorylating Pon. Nature 449, 96–U70 (2007).
Gonzalez, C. Spindle orientation, asymmetric division and tumour suppression in Drosophila stem cells. Nat. Rev. Genet. 8, 462–472 (2007).
Neumuller, R. A. & Knoblich, J. A. Dividing cellular asymmetry: asymmetric cell division and its implications for stem cells and cancer. Genes Dev. 23, 2675–2699 (2009).
Geng, A. et al. KIF20A/MKLP2 regulates the division modes of neural progenitor cells during cortical development. Nat. Commun. 9, 2707 (2018).
Qiu, R. X. et al. SEPT7 interacts with KIF20A and regulates the proliferative state of neural progenitor cells during cortical development. Cereb. Cortex 30, 3030–3043 (2020).
Dirks, P. B. Brain tumor stem cells: the cancer stem cell hypothesis writ large. Mol. Oncol. 4, 420–430 (2010).
Chen, J., Mckay, R. M. & Parada, L. F. Malignant glioma: lessons from genomics, mouse models, and stem cells. Cell 149, 36–47 (2012).
Wang, J. & Wechsler-Reya, R. J. The role of stem cells and progenitors in the genesis of medulloblastoma. Exp. Neurol. 260, 69–73 (2014).
Fine, H. A. New strategies in glioblastoma: exploiting the new biology. Clin. Cancer Res 21, 1984–1988 (2015).
Gimple, R. C., Bhargava, S., Dixit, D. & Rich, J. N. Glioblastoma stem cells: lessons from the tumor hierarchy in a lethal cancer. Genes Dev. 33, 591–609 (2019).
Kawai, Y. et al. KIF20A expression as a prognostic indicator and its possible involvement in the proliferation of ovarian clearcell carcinoma cells. Oncol. Rep. 40, 195–205 (2018).
Sheng, Y. et al. Upregulation of KIF20A correlates with poor prognosis in gastric cancer. Cancer Manag. Res. 10, 6205–6216 (2018).
Zhao, X. et al. Overexpression of KIF20A confers malignant phenotype of lung adenocarcinoma by promoting cell proliferation and inhibiting apoptosis. Cancer Med. 7, 4678–4689 (2018).
Zhou Y. M., et al. Identification of potential biomarkers in glioblastoma through bioinformatic analysis and evaluating their prognostic value. BioMed Res. Int. 2019, 6581576 (2019).
Yang, Z. J. et al. Medulloblastoma can be initiated by deletion of Patched in lineage-restricted progenitors or stem cells. Cancer cell 14, 135–145 (2008).
Wiederschain, D. et al. Single-vector inducible lentiviral RNAi system for oncology target validation. Cell Cycle 8, 498–504 (2009).
Ozawa T. & James C. D. Establishing intracranial brain tumor xenografts with subsequent analysis of tumor growth and response to therapy using bioluminescence imaging. J. Vis. Exp. 1986 (2010).
Schuller, U. et al. Acquisition of granule neuron precursor identity is a critical determinant of progenitor cell competence to form Shh-induced medulloblastoma. Cancer Cell 14, 123–134 (2008).
Grammel, D. et al. Sonic hedgehog-associated medulloblastoma arising from the cochlear nuclei of the brainstem. Acta Neuropathol. 123, 601–614 (2012).
Li, P. et al. A population of Nestin-expressing progenitors in the cerebellum exhibits increased tumorigenicity. Nat. Neurosci. 16, 1737–1744 (2013).
Holland, E. C. et al. Combined activation of Ras and Akt in neural progenitors induces glioblastoma formation in mice. Nat. Genet. 25, 55–57 (2000).
Bachoo, R. M. et al. Epidermal growth factor receptor and Ink4a/Arf: convergent mechanisms governing terminal differentiation and transformation along the neural stem cell to astrocyte axis. Cancer Cell 1, 269–277 (2002).
Uhrbom, L., Kastemar, M., Johansson, F. K., Westermark, B. & Holland, E. C. Cell type-specific tumor suppression by Ink4a and Arf in kras-induced mouse gliomagenesis. Cancer Res. 65, 2065–2069 (2005).
Zhu, Y. et al. Early inactivation of p53 tumor suppressor gene cooperating with NF1 loss induces malignant astrocytoma. Cancer Cell 8, 119–130 (2005).
Alcantara Llaguno, S. et al. Malignant astrocytomas originate from neural stem/progenitor cells in a somatic tumor suppressor mouse model. Cancer Cell 15, 45–56 (2009).
Patel, A. P. et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 344, 1396–1401 (2014).
Lee, J. H. et al. Human glioblastoma arises from subventricular zone cells with low-level driver mutations. Nature 560, 243 (2018).
Neftel, C. et al. An integrative model of cellular states, plasticity, and genetics for glioblastoma. Cell 178, 835 (2019).
Vladoiu, M. C. et al. Childhood cerebellar tumours mirror conserved fetal transcriptional programs. Nature 572, 67 (2019).
Hovestadt, V. et al. Resolving medulloblastoma cellular architecture by single-cell genomics. Nature 572, 74–79 (2019).
Jessa, S. et al. Stalled developmental programs at the root of pediatric brain tumors. Nat. Genet. 51, 1702 (2019).
Zhang, L. G. et al. Single-cell transcriptomics in medulloblastoma reveals tumor-initiating progenitors and oncogenic cascades during tumorigenesis and relapse. Cancer Cell 36, 302 (2019).
Cavalli, F. M. G. et al. Intertumoral heterogeneity within medulloblastoma subgroups. Cancer Cell 31, 737–754 (2017). e736.
Northcott, P. A. et al. The whole-genome landscape of medulloblastoma subtypes. Nature 547, 311–317 (2017).
Dai, M. et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 33, e175 (2005).
Carvalho, B. S. & Irizarry, R. A. A framework for oligonucleotide microarray preprocessing. Bioinformatics 26, 2363–2367 (2010).
Leek, J. T., Johnson, W. E., Parker, H. S., Jaffe, A. E. & Storey, J. D. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 28, 882–883 (2012).
Acknowledgements
We thank Jeremy LaDou and animal center staff for assistance with animal breeding and care, and Brian Armstrong and staff for the use of imaging facility. This work is supported by NIH grant NS096130 to Q.L.
Author information
Authors and Affiliations
Contributions
R.X.Q. and Q.L. conceived and designed the study. R.X.Q. and J.W. conducted the experiments. B.G. and P.A.N. provided analyses of KIF20A expression in medulloblastoma patient transcriptome databases. R.W.R. provided Atoh1-CreER; Ptcfl/fl mice and insightful discussions on medulloblastoma biology. R.X.Q. and Q.L. analyzed data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Qiu, R., Wu, J., Gudenas, B. et al. Depletion of kinesin motor KIF20A to target cell fate control suppresses medulloblastoma tumour growth. Commun Biol 4, 552 (2021). https://doi.org/10.1038/s42003-021-02075-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-021-02075-4
This article is cited by
-
A Four-Gene Panel for the Prediction of Prognosis and Immune Cell Enrichment in Gliomas
Molecular Biotechnology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.