Abstract
Diverse (sub)cellular materials are secreted by cells into the systemic circulation at different stages of disease progression. These circulating biomarkers include whole cells, such as circulating tumour cells, subcellular extracellular vesicles and cell-free factors such as DNA, RNA and proteins. The biophysical and biomolecular state of circulating biomarkers carry a rich repertoire of molecular information that can be captured in the form of liquid biopsies for disease detection and monitoring. In this Review, we discuss miniaturized platforms that allow the minimally invasive and rapid detection and analysis of circulating biomarkers, accounting for their differences in size, concentration and molecular composition. We examine differently scaled materials and devices that can enrich, measure and analyse specific circulating biomarkers, outlining their distinct detection challenges. Finally, we highlight emerging opportunities in biomarker and device integration and provide key future milestones for their clinical translation.
Key points
-
Circulating biomarkers (whole cells, extracellular vesicles and cell-free factors) exist in various biophysical and biomolecular forms and carry a rich repertoire of molecular information.
-
The diversity of circulating biomarkers, that is, differences in size, concentration and molecular composition, makes their specific detection and clinical application challenging.
-
Miniaturized device designs, nanomaterials and information-revealing assay strategies can be integrated to detect circulating biomarkers in the form of liquid biopsies to reveal disease-reflective information.
-
Capturing the dynamic spectrum of circulating biomarkers will empower personalized medicine, particularly disease monitoring and drug selection.
-
To promote the clinical translation of liquid biopsies, platforms and devices should be developed considering specific clinical needs, regulatory guidelines and reimbursement models.
Similar content being viewed by others
Introduction
The monitoring of molecular markers is key to stratifying patients, guiding treatment decisions and monitoring drug efficacy1,2, particularly in precision medicine. However, common diagnostic approaches, such as imaging and tissue biopsy, are insensitive and often performed at a late disease stage, thereby delaying medical decision-making. For example, imaging lacks specificity to molecular mechanisms and cannot detect early, subtle changes. Although tissue biopsy remains the gold standard for cancer molecular diagnostics, it is highly invasive and has limited sampling capacity to capture dynamic disease biology such as spatial heterogeneity and temporal evolution3,4,5. Alternatively, liquid biopsies allow minimally invasive disease characterization6,7,8 and monitoring9,10. Compared to tissue-based detection, liquid biopsy measures circulating biomarkers in a variety of bodily fluids (for example, blood, urine, saliva and sweat) to enable accessible molecular characterization and repeated monitoring.
At different stages of disease progression, cells release (sub)cellular materials into the systemic circulation11,12,13. These circulating biomarkers are secreted in an orchestrated manner, exist in diverse forms, and can be broadly classified according to their biogenesis, biophysical structure and biomolecular composition. They include whole living cells (for example, circulating tumour cells (CTCs)14), nanoscale membrane-bound extracellular vesicles (EVs)15 and cell-free factors (for example, nucleic acids, proteins, lipids and metabolites)16,17. These structurally and molecularly diverse circulating biomarkers carry a rich repertoire of molecular information and play active roles in mediating pathophysiological processes in a variety of diseases (for example, cancer, neurodegenerative disorders and infectious diseases)17,18,19. Capturing this dynamic network of circulating information could thus present a transformative approach for precision diagnostics and personalized treatment.
Despite their clinical potential, the highly heterogeneous nature of circulating biomarkers makes their specific detection and clinical translation challenging20,21,22. In particular, in biomarker discovery, current approaches, such as mass spectrometry and gene expression microarrays, lack performance in characterizing and distinguishing circulating biomarkers. Owing to mismatches between their mode of measurement and the requirements of the different biomarkers (for example, in relation to their respective size and concentration), key information, such as molecular types and organizational states, is often missed, especially against a complex biological background of native biofluids. In addition, these approaches require large sample volumes and extensive sample processing, are laborious and time-consuming, and thus impede clinical application and adoption.
Size-matching device designs and information-revealing assay strategies may allow the precise detection of features of circulating biomarkers by exploiting distinct signal transduction mechanisms for direct detection, even amidst a complex biological background. Moreover, modular technology units can be readily integrated into miniaturized devices to achieve collaborative functions, enabling the monitoring of a multitude of biomarker information at high resolution and new clinical opportunities in a variety of diseases (for example, different cancers, cardiovascular diseases, neurodegenerative disorders and infectious diseases)8,23,24.
In this Review, we discuss key characteristics of different types of circulating biomarkers in various diseases, their respective detection challenges and important strategies to overcome these challenges. We highlight miniaturized technologies and assay designs for circulating biomarkers, with a particular focus on how this field is contributing to the development of transformative and informative liquid biopsy tests. Finally, we provide an outlook on emerging opportunities in biomarker and device integration for pre-clinical development and clinical translation.
Circulating biomarkers
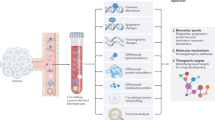
Cells secrete diverse (sub)cellular materials into the systemic circulation (Fig. 1a), including CTCs, EVs and cell-free factors. These biomarkers carry molecular information in their various biophysical and biomolecular features, representing multifaceted surrogates for disease evaluation.
a, Cells secrete diverse (sub)cellular materials into the systemic circulation. Circulating tumour cells (CTCs) are whole cancer cells disseminated from the primary tumour. Extracellular vesicles (EVs) are small (10–2,000 nm) vesicles released by a variety of cells. Living cells, particularly rapidly dividing cells, secrete exosomes and microvesicles. Free-floating molecules (circulating nucleic acids and proteins) are released into the bloodstream by different mechanisms such as necrosis and apoptosis. b, CTCs can circulate as single cells or clusters, both of which have metastatic and invasive potential. CTCs can be distinguished from blood cells through their biophysical and biomolecular properties. CTCs are bigger and stiffer than blood cells and they contain epithelial markers as well as tumour markers. c, EVs carry diverse molecular content (proteins, nucleic acids, lipids and their various modifications) inherited from their parent cells. EVs also possess acquired markers through surface interactions with different molecules (proteins and nucleic acids). As vehicles of intercellular communication, EVs deliver molecular cargoes to recipient cells to modulate biological processes. d, Changes in the abundance of cell-free factors can serve as a biomarker in distinguishing healthy and diseased states. Mutational status, epigenetic and post-translational modifications, and interactions with other factors can further provide multifaceted information to inform specific disease profiles.
Circulating tumour cells
CTCs were first discovered in the peripheral blood of a patient with metastatic cancer in 1869 (ref. 25). CTCs, which are whole cancer cells disseminated from the primary tumour into the systemic circulation (Fig. 1a), bear hallmarks of living cells, are relatively large (typical diameter >8 μm) compared to cells of blood origin and are intricately involved in the metastatic cascade26. During the progression of epithelial cancers, local tumour cells acquire migratory and invasive ability through epithelial–mesenchymal transition27, which allows them to intravasate into the bloodstream and/or the lymphatic system as CTCs, before eventually extravasating at distant organs to seed metastatic tumours. Notably, CTCs can circulate as single cells or clusters, both of which possess metastatic and invasive potential28,29 (Fig. 1b). Owing to the complex blood environment and selection pressure at different stages of metastasis, only a small fraction of CTCs (or CTC clusters) progress, making viable CTCs extremely rare in the peripheral blood, with an estimated concentration of ~1–10 cells per 109 haematological cells20.
Despite their low concentration, CTCs can be distinguished from blood cells through their biophysical and/or biomolecular properties. Single CTCs are typically bigger and stiffer than blood cells20 and commonly harbour epithelial markers that are not of blood origin30 (Fig. 1b). Importantly, CTCs are heterogeneous in individual patients and across patients3,31. Comprehensive characterization of CTC quantity and biophysical and biomolecular changes can thus provide insight into tumour classification, disease progression and treatment evaluation.
Extracellular vesicles
EVs are nanoscale, heterogeneous membrane vesicles secreted by a variety of cell types15,32. EVs play important roles in mediating intercellular communication and regulating diverse physiological processes, for example, homeostasis, vascular biology and the immune system33,34. According to their size and biogenesis pathways, EVs can be further classified into exosomes (10–200 nm), microvesicles (200–2,000 nm) and apoptotic bodies (500–2,000 nm)35,36. Exosomes and microvesicles are secreted by living cells, particularly by rapidly dividing cells (Fig. 1a), whereas apoptotic bodies are released by dying cells. Microvesicles are formed through direct outward budding of cellular plasma membrane, whereas exosomes are created by repeated inward budding of the endosomal limiting membrane, leading to the generation of multivesicular bodies and exocytosis of exosomes. EVs are found in many bodily fluids, including blood, urine, ascites, milk and cerebrospinal fluid37,38, with a typical concentration of 1010–1012 vesicles per millilitre of blood39,40.
EVs contain diverse molecular content, for example, proteins41,42, nucleic acids43,44 and lipids45,46, and various sugar modifications47,48 reflective of their parent cells and extracellular environment (Fig. 1c). The molecular cargo of EVs is either inherited from parental cells49 or acquired from extracellular space50 (Fig. 1c) and can thus offer multifaceted biological insights. Inherited constituents are sorted and packaged into vesicles during EV biogenesis and can therefore serve as circulating surrogates of the parent cells to provide cell origin-specific biomarkers. Alternatively, different molecules, such as proteins and nucleic acids, can associate with the EV membrane through various surface interactions (for example, electrostatic interactions, hydrophobic interactions and hydrogen bonding), which can give insight into the biophysical properties of the bound molecules and/or the EVs to unmask biomarker subpopulations51. As mediators of intercellular communication47,52, EVs can readily cross anatomical and physiological barriers, including the highly selective blood–brain barrier53, and deliver molecular cargoes to mediate biological processes of recipient cells and regulate microenvironments at distant sites. Therefore, EVs can also serve as functional biomarkers54 (Fig. 1c).
Cell-free factors
Various molecules, including cell-free DNA (cfDNA), RNA and proteins, are also found in blood. These molecules are released through multiple mechanisms, including necrosis, apoptosis and secretion6 (Fig. 1a), and depending on their biogenesis, they not only vary in abundance but also bear heterogeneous modifications (Fig. 1d); for example, cfDNA are extracellular fragments of DNA with a typical concentration of 1–10 ng ml–1 of plasma55,56, which increases in cancer and other physiological conditions such as acute trauma57, cerebral infarction58, exercise59, transplantation60 and infection61. In addition to differences in abundance, genetic and epigenetic alterations of cfDNA reflect the cells of origin62,63,64; for example, circulating tumour DNA (ctDNA) is released by multiple types of tumours, from different tumour regions and during disease progression. Changes in ctDNA (abundance and molecular properties) may thus reflect spatiotemporal tumour heterogeneity and have been associated with tumour burden and malignant progression65.
Proteins in blood are often the end products of biological processes and can thus reflect the physiological and/or pathological state of cells to reveal additional disease pathogenicity. Protein abundance in bodily fluids and post-translational modifications, which can alter protein function66, can serve as biomarkers in many diseases, including cancer17 and cardiovascular67 and neurodegenerative diseases18; for example, measuring antibody levels in blood allows monitoring of the dynamic humoral response to vaccination and pathogen infection68. Similarly, the detection of viral proteins in bodily fluids reveals the presence of actively replicating viruses19.
Challenges in detecting circulating biomarkers
The diversity of circulating biomarkers, including differences in size, concentration and molecular composition, makes their specific detection and clinical application challenging; for example, micron-sized CTCs are rare and exist against a high background of blood cells. By contrast, EVs are more abundant in body fluids, but their nanoscale dimension makes it challenging to characterize and distinguish them from small non-vesicular components (for example, similar-sized lipoproteins and identical proteins in free-floating forms)69. Moreover, cell-free factors carry specific epigenetic and post-translational modifications and thus require distinct measurement strategies70.
Commonly used purification and analytical approaches face limitations in probing the diversity of circulating biomarkers; for example, pre-analytical purification of CTCs typically requires CTC enrichment through lysis and centrifugation to deplete blood cells, which may lead to loss of viable tumour cells8. Similarly, EV isolation relies on ultracentrifugation or size-exclusion chromatography, and purification of free-floating molecules is based on immunoprecipitation, which requires a large sample amount, may lead to loss of molecules, and is time-consuming and laborious. In addition, analytical protein assays commonly rely on enzyme-linked immunosorbent assay (ELISA), western blotting and/or mass spectrometry to measure purified biomarkers. These approaches require extensive processing and advanced instrumentation, have limited sensitivity for scarce targets, and lack capabilities to differentiate biomarker features such as biophysical and biomolecular specificity. Similarly, nucleic acid detection (for example, PCR) requires complex procedures and is almost exclusively performed in large, centralized clinical laboratories.
Furthermore, these techniques lack the resolution to distinguish features within individual classes of biomarkers (for example, biophysical and biomolecular subpopulations) and cannot be adapted for multiparametric profiling across biomarker classes, which hinders biomarker discovery. Importantly, laborious and time-consuming approaches that require large sample volumes may be difficult to translate to the clinic.
Miniaturized detection technologies
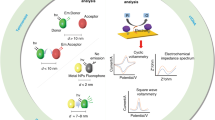
Manipulation and detection technologies with dimensions and modes of action in the size range of circulating biomarkers can account for the heterogeneity in size and concentration (Fig. 2a); for example, micron-level manipulation can be achieved using micropatterned systems (such as microfluidics71, microelectronics72 and micromechanics systems73) or prepared by fabrication approaches (such as photolithography74) or by assembly technologies (such as 3D printing75) (Fig. 2a). Sub-micron manipulation requires devices and systems at the nanoscale; for example, top-down fabricated nanodevices, such as by electron-beam lithography76 and nanoimprint lithography77, can be tuned to exhibit and exploit size-complementary near-field effects, including optical78,79,80, piezoelectric81 and magnetoelastic82 effects, to achieve precise spatial manipulation (Fig. 2a). In addition, bottom-up synthesized nanomaterials of different composition, size and shape83,84 can act as modular units with multimodal capabilities; for example, metal–organic frameworks, which can be used for biomarker enrichment, detection and fingerprinting85, or core–shell nanoparticles, which have been used in drug delivery, bioimaging, catalysis and optical spectroscopy86 (Fig. 2). Finally, direct molecular manipulation, that is, nano and sub-nano control, can be achieved using biological building blocks such as nucleic acids and proteins; for example, programmable DNA origami87,88, hybrid molecular switches89,90 and integrated circuitries91,92 can be assembled exploiting predictable interactions to achieve complex architectures and precise spatial and biochemical control (Fig. 2a). Moreover, through hybrid assembly with active ingredients, for example, with catalytic enzymes, responsive molecular systems can be designed that perform specific reactions against a complex biological background (Fig. 2a). These size-matching technologies can serve as modular components in devices intended for the detection and manipulation of circulating biomarkers.
a, Size-matching technologies can cater to the various sizes of circulating biomarkers. Micron-scale manipulation can be achieved by micropatterned systems such as microfluidics and microelectronics. Sub-micron assessment requires nanodevices that exhibit size-complementary near-field effects. In addition, nanomaterials with different compositions and hierarchical organizations can be synthesized to construct different architectures with multimodal capabilities. Molecular control can be achieved using biological building blocks such as nucleic acids and proteins. b, Biomarkers can be selectively enriched by exploiting their biophysical and/or biomolecular properties; for example, a deterministic lateral displacement array allows size-based enrichment, cationized polymers enable charge-based enrichment, and functionalized nanoparticles and molecular nanostructures allow biochemical enrichment. Integrated enrichment strategies are often applied to achieve high-coverage biomarker enrichment. c, An analytical platform that integrates enrichment, culturing, processing and detection modules can extract multiparametric information from diverse circulating biomarkers. The acquired information can be processed and modelled using large-scale machine learning algorithms114 to develop composite signatures and accurate classifications. CTC, circulating tumour cell; t-SNE, t-distributed stochastic neighbour embedding.
Information-revealing assays
Integrated enrichment
Efficient enrichment is crucial to detecting rare circulating biomarkers, particularly amongst a heterogeneous and complex biological background of bodily fluids. Biomarkers can be positively or negatively enriched. In positive enrichment, targeted biomarkers are isolated from heterogeneous background materials; in negative enrichment, background components are depleted. Various biophysical (for example, size and charge) and/or biomolecular (for example, proteins and nucleic acids) properties distinguish biomarkers from background components, which can be exploited for enrichment (Fig. 2b); for example, microfluidics with well-designed flow fields allow size-based biophysical separation of CTCs from smaller blood cells93 (Fig. 2b). Similarly, integrated microfluidics with matching field effects (for example, acoustic94,95, thermal96, electric97 and dielectric98) enable the size-based and/or charge-based detection of small circulating biomarkers, including EVs, proteins and cfDNAs (Fig. 2b).
Complementary to biophysical enrichment, biomolecular enrichment provides high-resolution separation (that is, subpopulations within the same biomarker type). Biomolecular enrichment systems are typically hybrid, that is, at the interface of physical and biological technologies, to achieve specific biochemical recognition and precise physical manipulation (Fig. 2b); for example, tumour-derived EVs with high expression of surface antigens (for example, epidermal growth factor receptor (EGFR) and epithelial cell adhesion molecule (EpCAM)) can be selectively captured with magnetic nanomaterials that are functionalized with the complementary biological recognition element99,100. Protein enrichment is crucial for the isolation of low-abundance proteins and to reduce sample complexity, particularly against a complex biological background. Free-floating proteins in the circulation can be enriched using surface-anchored antibodies101, nanobodies102 or aptamers103.
These different strategies are often combined to improve biomarker enrichment (Fig. 2b); for example, the sequential application of negative selection (such as depletion of leukocytes through CD45) and positive selection (such as size-selection of large CTCs) reduces fluidic clogging and improves CTC purity104. Similarly, selection by post-translational modifications of circulating proteins, interactions with both metal ions and antibodies can be exploited105,106.
Complementary measurement
To extract multiparametric information from diverse circulating biomarkers, sensors, assays and analytics can be integrated within one platform (Fig. 2c). Sensors can be complemented with size-matched transduction mechanisms; for example, CTCs can be quantified using microfluidic platforms equipped with electrochemical detectors that function as cytosensors for single-cell analysis107. By varying the geometric design of nanodevices, the probing depth of plasmonic sensors can be tuned to detect nanoscale EVs108. Responsive molecular nanostructures allow the detection of sub-nanoscale features such as DNA methylation109. Biomarker activity can be measured in situ and in real time using assay mechanisms inspired by microtechnology and nanotechnology110,111,112 that exploit the interplay of molecular and physical technologies; for example, biological amplifiers, such as molecular probes and enzymes, can be integrated with responsive physical transducers such as electrodes and optical resonators113.
The integration of multiple functional modules in one platform (Fig. 2c) is particularly useful to detect rare biomarkers that require substantial sample processing and to extract information from a spectrum of circulating biomarkers (that is, biomarker combinations). Such all-in-one miniaturized systems that incorporate enrichment, sample preparation, culture, molecular characterization and functional studies in one chip simplify sample handling and provide rich biomarker information71. In addition, multiparametric information (for example, abundance, activities and interactions)114,115,116 can be collected from different types of circulating biomarkers at different time points in disease progression (Fig. 2c). Through clinical evaluation, such complementary information may not only refine established clinical biomarkers but also reveal new ones.
Circulating tumour cell detection
Molecular characterization
CTCs can be genotypically and phenotypically characterized using enrichment-based and enrichment-free approaches. Multiple isolation techniques have been developed for enrichment-based detection, including biophysical separation, through size and geometry, and biomolecular isolation, through CTC surface antigens; for example, label-free size-based separation of viable CTCs can be achieved by applying hydrodynamic forces in a spiral microfluidic device117 (Fig. 3a). The controlled laminar flow within the channels can be manipulated to generate size-dependent cell trajectories for high-throughput CTC enrichment. Alternatively, acoustofluidics allows contact-free, size-based enrichment of CTCs in complex biofluids using a combination of acoustic waves and fluid flows118. As the enrichment is label-free, purified CTCs are compatible with downstream biomolecular characterization. In addition, high-throughput isolation of CTCs preserves their structural and biological integrity119. Geometry-based enrichment exploits the distinct geometries of cellular aggregates; for example, a microchip with three triangular pillars that act as bifurcating traps enables the selective, label-free capture of CTC clusters (down to two-cell clusters) under low-shear stress, preserving CTC integrity and thus allowing downstream proteomic and transcriptomic profiling120 (Fig. 3b).
a, Size-based enrichment of circulating tumour cells (CTCs) using spiral microfluidics. Larger CTCs and smaller blood cells are separated under the combined effects of an inertial lift force and Dean drag force. b, Geometry-based enrichment of CTC clusters using microfluidic bifurcating traps. CTC clusters are captured at the bifurcation formed by a set of three triangular micropillars. Scale bars, 60 μm. c, Positive biomolecular enrichment using epithelial cell adhesion molecule (EpCAM)-coated silicon micropillars. Scale bar, 50 μm. d, Negative selection through immunomagnetic removal of blood cells. e, Direct quantification of CTCs using microfabricated Hall sensors. The sensors measure the magnetic moment of magnetically labelled CTCs in unprocessed blood. f, In vivo detection of CTCs based on photoacoustic (PA) flow cytometry. Laser heating of light-absorbing melanin expressed in melanoma CTCs causes acoustic waves to travel to the transducers, enabling label-free and in situ quantification of CTCs. g, Evaluation of migratory and proliferative potential of glioblastoma CTCs using a Y-shape microfluidic channel. The microfluidic device contains a series of Y-shaped microchannels. The time-lapse images show migrating CTCs with different motilities. h, Quantification of CTC intercellular adhesion based on harmonic acoustic manipulation. Single cells are reversibly paired or separated by tuning the acoustic wells. i, Single-CTC immunoblotting using a miniaturized projection electrophoresis device. The integrated system enables cell loading, lysis, protein separation, immunoprobing and data readout. AFU, arbitrary fluorescence unit; BSA, bovine serum albumin; CBC, circulating blood clot; IgG, immunoglobulin G; OVA, ovalbumin. Part a reprinted from ref. 117, Springer Nature Limited. Part b adapted from ref. 120, Springer Nature Limited. Part c reprinted from ref. 123, Springer Nature Limited. Panel d reprinted with permission from ref. 130, Proceedings of the National Academy of Sciences. Part e adapted/reprinted from ref. 131. ©The Authors, some rights reserved; exclusive licensee AAAS. Part f reprinted from ref. 133 with permission from AAAS. Part g adapted from ref. 135, Springer Nature Limited. Panel h adapted from ref. 137, Springer Nature Limited. Part i adapted from ref. 140, Springer Nature Limited.
Positive biomolecular enrichment of CTCs in blood often relies on tumour-enriched molecular markers; for example, cancers of epithelial origin are characterized by specific protein markers, such as EpCAM, which are used for the characterization of solid tumours (for example, breast121 and prostate cancer122). A silicon microfluidic platform that contains micropillars coated with antibodies against EpCAM and a herringbone structure can capture CTCs without requiring further processing or labelling of blood samples123 (Fig. 3c). Its herringbone structure enhances interactions between CTCs and the antibody-coated chip by creating microvortices that enhance the enrichment of target cells through antibody capture124. In addition, nanomaterials have been explored to increase CTC capture efficiency ex vivo and in vivo; for example, immunomagnetic enrichment of CTCs ex vivo can be achieved using on-chip, near-infrared, fluorescence-enhanced detection in blood samples125. Alternatively, antibody-functionalized graphene oxide nanosheets on a patterned gold surface enable single-digit capture of CTCs per millilitre of blood126. Similarly, a thiolated ligand-exchange reaction with gold nanoparticles on a herringbone chip enables the release of antibody-captured CTCs without compromising their viability127. The metal–thiol interactions can readily be disrupted in the presence of excess thiol molecules, replacing the immobilized antibodies with biocompatible thiol molecules128. CTCs can also be captured in vivo; for example, an intravascular magnetic nanowire allows the collection of CTCs from the systemic circulation. Inserted through a standard intravenous catheter into the auricular vein of a porcine model, the wire captures CTCs previously labelled with injected magnetic particles129.
Although EpCAM expression allows positive selection of CTCs in several cancer types, it cannot be applied for cancers with low or no EpCAM expression (for example, neuroendocrine cancers, blood cancers). Alternatively, negative selection (such as targeted removal of leukocytes) can be used in conjunction with other enrichment strategies; for example, an inertial separation array and a magnetic sorter can be integrated into a single chip for the high-throughput depletion of blood cells and separation of CTCs against a complex background (for example, leukapheresis product)130 (Fig. 3d).
CTCs can also be directly detected without prior enrichment; for example, a miniaturized magnetic sensor leverages the micro-Hall effect to quantify CTCs131 (Fig. 3e); here, magnetically labelled CTCs subjected to an external magnetic field are distinguished from unbound magnetic nanoparticles based on their magnetic moment associated with biomarker expression, enabling direct CTC detection without purification. An automated microfluidic platform with an integrated field-effect transistor also allows direct quantification of CTCs in blood samples. The platform is equipped with a cell-trapping device based on hydrodynamic microstructures for continuous CTC trapping and an array of aptamer-bound field-effect transistor sensors for specific, molecular detection of CTCs132. Alternatively, CTCs can be quantified in vivo using photoacoustic flow cytometry with transcutaneous delivery of a high-pulse-rate laser and focused ultrasound transducers in patients with melanoma133 (Fig. 3f).
Functional characterization
Functional characterization of CTCs, which is fundamental to the investigation of the biology of metastatic cancers and to devising therapeutic strategies, requires isolation of viable CTCs (that is, cells that can be expanded) and in-depth characterization (for example, of cellular behaviour and drug susceptibility). Understanding CTC function at single-cell resolution could give quantitative insights into tumour heterogeneity. Based on distinct features of invasive CTCs (for example, high motility and weak cell–cell contacts), microfluidic platforms can be applied to sort and isolate specific CTCs for downstream analyses; for example, a microfluidic assay comprising two parallel seeding and collection channels connected by Y-shaped microchannels (Fig. 3g) allows quantification of CTC migration and proliferation134. The Y-shaped channels are designed to have a 20 μm-wide feeder channel bifurcating to either 10 μm-wide or 3 μm-wide branches depending on the migratory potential of cells. This relative abundance of migratory cells and proliferative index (using the protein biomarker Ki-67) enable assessment of the metastatic potential of breast cancer cells with high accuracy134 and prediction of patient-specific outcomes in glioblastoma135. CTC migration dynamics at single-cell resolution can be measured on a microchip that contains an array of fibronectin-coated micropillars designed to confine cells, promoting individual cell scattering from a collectively advancing front. Based on the cellular dynamics, the cells can be profiled into distinct migration phenotypes; this profile is in good agreement with epithelial or mesenchymal biomarker expression of cells136. Alternatively, an acoustic tweezer enables the quantification of CTC–CTC adhesion with high throughput137 (Fig. 3h) by measuring intercellular adhesion forces.
CTC proteome studies have revealed distinct surface and secreted proteins138. Proteomic analysis at single-CTC resolution has been demonstrated using rare-cell western blotting on a microfluidic device to quantify multiple surface and intracellular signalling proteins in individual CTCs139. Here, the microfluidic device comprises an array of microwells stippled into a polyacrylamide gel on a glass slide. Integrated with a label-free CTC collection tool that selects based on size and deformability, the system can detect variations in protein expression and biophysical phenotypes among CTCs. This platform has also been optimized for synchronized analyses of general and/or specific cytosolic and nuclear proteins from hundreds of single CTCs140,141 (Fig. 3i).
Treatment response can also be evaluated by functionally characterizing CTCs. Many drug-screening platforms have been developed using cancer cell lines, requiring large quantities of samples and long durations142,143. To enable in situ drug screening with minimal processing and small sample volumes, a microfluidic culture platform can be applied144 that allows co-culturing of CTCs and immune cells from the same patients within specialized tapered microwells, enabling rapid and efficient establishment of CTC clusters. With its cost-effectiveness and rapid readout, the platform could facilitate clinical prediction of the response of a specific patient to various therapeutic strategies, thereby bringing precision medicine closer to reality.
Extracellular vesicle detection
Constituent markers
EVs are characterized by their biophysical size and biomolecular composition. Through different biogenesis pathways, EVs inherit molecular cargoes from their parent cells, including characteristic vesicle markers, such as tetraspanins and ALG2-interacting protein X (ALIX), as well as other proteins and nucleic acids145,146, which can provide valuable information about the parent cells (for example, cells of origin and pathophysiological states). Enrichment-based and enrichment-free approaches enable the detection of both biophysical and biomolecular properties of EVs to distinguish EV constituent markers from non-vesicular components.
Enrichment-based detection relies on biophysical and biomolecular separation approaches; for example, in acoustofluidics, the frequency of acoustic waves can be tuned to match the nanoscale dimensions of EVs, allowing contact-free EV enrichment from cell culture media in a device with standing surface acoustic waves147. This device has further been optimized to isolate EVs from saliva and to profile small RNAs in purified EVs94. Ultrafast EV purification (in less than 10 min) can be achieved by negative pressure oscillation and double-coupled harmonic oscillator-enabled membrane vibration; here, the transverse waves generated by the oscillators reduce fouling effects and particle aggregation, resulting in an improved processing speed, yield and purity148 (Fig. 4a). In this automated system, EVs could be isolated from urine samples of patients with cancer and validated through transcriptomic profiling.
a, Biophysical enrichment of extracellular vesicles (EVs) by coupled membrane vibration. The system uses negative pressure oscillation and double-coupled harmonic oscillator-enabled membrane vibration to enable automated label-free purification of exosomes from varied biofluids. b, Biomolecular enrichment using a functionalized, self-assembled 3D nanoporous herringbone (HB) structure. The nanoHB structures promote mass transfer, increase surface area and enhance probe–EV interactions to achieve efficient and rapid EV binding. c, Integrated EV analysis technology with solution-phase magnetic enrichment and electrochemical detection. A 96-well chip enables high-throughput evaluation of colorectal cancer EV markers. d, Direct and dual-specific EV detection enables simultaneous evaluation of biophysical and biomolecular properties of the same EVs. EVs are labelled by immunofluorescent probes and gold nanoparticles that promote the formation of EV-templated plasmonic nanoshells; the nanoshells have distinct dimensions (and thus optical properties) and locally quench fluorescent probes only if they are target-bound on the same EV. e, Activity-based assessment of EV–drug target interactions. Bio-orthogonal probe amplification and spatial patterning of molecular reactions within EV size-matched plasmonic nanoring resonators enable in situ analysis of EV–drug dynamics. f, 3D tracking of EV activity in live cells. A holographic fluorescence imaging technology can recover the full electric field of fluorescent light, thereby achieving 3D tracking of individual EVs. AP, air pressure; HF, high frequency; HiMEX, high-throughput integrated magneto-electrochemical extracellular vesicle; LF, low frequency; NP, negative pressure; PCB, printed circuit board; PDMS, polydimethylsiloxane. Part a reprinted from ref. 148, Springer Nature Limited. Part b adapted from ref. 149, Springer Nature Limited. Part c adapted from ref. 151, Springer Nature limited. Part d adapted/reprinted from ref. 154. ©The Authors, some rights reserved; exclusive licensee AAAS. Part e adapted from ref. 158, Springer Nature Limited. Part f Reprinted with permission of AAAS from ref. 159. © The Authors, some right reserved; exclusive licensee AAAS. Distributed under a CC BY-NC 4.0 License (http://creativecommons.org/licenses/by-nc/4.0/).
Biomolecular enrichment can be implemented at various scales; for example, a microfluidic chip can be fabricated with a 3D nanoporous herringbone structure by guided colloidal self-assembly of silica nanoparticles. The herringbone structure promotes microscale mass transfer of bioparticles and increases the surface area and probe density to improve the efficiency and speed of exosome binding, permitting drainage of the boundary fluid to reduce near-surface hydrodynamic resistance, thereby promoting particle–surface interactions for exosome binding and immunosensing of tumour-derived EVs149 (Fig. 4b). Magnetic nanoparticles can be applied for solution-phase enrichment; for example, by functionalizing magnetic nanomaterials with biological recognition elements, such as antibodies and aptamers, they can be used to capture and profile EVs. This enrichment approach can be integrated, together with electrochemical EV detection, in a compact sensor150. In the format of a 96-well setup, this sensor allows high-throughput evaluation of colorectal cancer EV markers151 (Fig. 4c). Polycore magnetic particles in the size range of EVs can transduce EV glycans through EV-induced nanoparticle aggregation, using a built-in giant magnetoresistance sensor48 to measure the resultant magnetic signals. This assay enables dual-specific measurement of EV size and glycan composition to reveal disease signatures.
EVs can also be directly detected without prior enrichment, for example, using surface plasmon resonance (SPR). SPR refers to the collective oscillation of conduction electrons at the metal–dielectric interface when illuminated by incident light152. Plasmonic nanostructures can be designed with a probing depth matching EV dimensions to detect local biomolecular binding. Applying this sensing principle, a label-free SPR platform with periodic arrays of gold nanoholes153, each functionalized with antibodies, has been designed to detect the specific binding of EVs with complementary protein biomarkers, resulting in sensitive optical signals for ovarian cancer detection. A solution-phase, dual-specific plasmonic assay can simultaneously evaluate biophysical and biomolecular properties of EVs154 (Fig. 4d); here, EV protein biomarkers are first labelled with fluorescent probes, followed by in situ growth of gold nanoshells on the EVs. The EV-templated plasmonic nanoshells have distinct spectral properties and quench co-localized fluorescent probes on the same EVs, whereas non-EV targets remain unresponsive. This platform has been clinically evaluated in patient ascites, demonstrating that, compared to free-floating proteins, EV constituent markers show better accuracy in differentiating patient prognosis154.
Acquired markers
EVs interact and associate with various types of biomolecules in the circulation, including proteins and nucleic acids. This acquisition of biomarkers, that is, EV-bound biomarkers, can reflect additional biophysical and biochemical properties of EVs and/or bound molecules, allowing the identification of EV subpopulations and other information (for example, protein aggregation)155. EV-bound biomarker detection has been particularly explored in the context of neurodegenerative diseases because EVs associate with pathological amyloid proteins, which are enriched in human brain amyloid plaques, suggesting that EV–amyloid complexes could contribute to plaque formation in the brain156. For example, an analytical platform that contains a size-matching double-photonic structure (sensor probing depth is less than 200 nm and is matched to EV size) enables quantitative detection of different organizational states of amyloid-β (Aβ) proteins, that is, both EV-bound Aβ and circulating free Aβ, directly in blood samples of patients with Alzheimer disease51. The platform is further coupled with enzymatic amplification of localized optical deposits (3,3′-diaminobenzidine tetrahydrochloride) to enhance the plasmonic signal. Here, multiplex quantification is achieved because the enzymatic deposits are formed only when multiple biomarkers co-localize in the same EVs. Using the platform, different subpopulations of Aβ could be identified that differ in their organizational states, propensity to form fibrillar structures and affinities to associate with EVs. Specifically, a prefibrillar Aβ population, which shows a high tendency to form fibrillar structures, preferentially binds to EVs, particularly to neuronal EVs. Therefore, EV association can serve as a measure of Aβ biophysical aggregation state, and circulating EV-bound Aβ protein can accurately reflect brain amyloid deposition to categorize patients.
Activity markers
EVs can also serve as activity markers; for example, a microfluidic 3D nanochip, fabricated by high-resolution colloidal inkjet printing, can perform integrative molecular analysis and functional phenotyping of EV-bound matrix metalloproteinase 14 (MMP14)157. Using the technology, biofunctional signatures of EVs could be established that correlate with tumour invasion and metastasis in mouse models and plasma samples of patients with breast cancer. An integrated nanotechnology platform allows the evaluation of EVs as a functional marker of drug dynamics158 (Fig. 4e). Here, EVs are first competitively labelled with bio-orthogonal drug probes and enzymatically amplified for direct detection in a nanoring plasmonic resonator. EVs with low drug occupancy are labelled with more drug probes, triggering a stronger plasmonic signal compared to EVs with high drug occupancy. These molecular interactions (EV protein typing and probe amplification) are spatially patterned within the most sensitive in-ring region to enable a real-time, multiparametric analysis of EV dynamics. This platform can accurately reflect disease status and distinguish drug responses within 24 h of treatment initiation in blood samples from patients with lung cancer. A holographic fluorescence imaging approach can track individual EVs inside live cells in 3D159 (Fig. 4f) using a wide-field fluorescence microscope with good spatial resolution and high sensitivity. This technology allows monitoring of EV uptake, transport and functional response in recipient cells in real time.
Cell-free biomarker detection
Cell-free biomarkers, including nucleic acids and proteins with distinct modifications, such as methylation and phosphorylation, can be detected at high resolution using nanodevices and molecular assays.
Direct quantification profiling
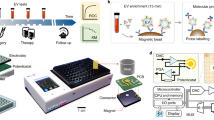
Nanomaterials can be applied to directly and sensitively detect cell-free biomarkers in clinical samples; for example, DNA-functionalized magnetic nanoparticles can function as dispersible electrodes for the highly sensitive detection of nucleic acids in whole blood160 (Fig. 5a). The nanoparticles bind to analytes through nucleic acid hybridization, including microRNA160 and ctDNA161, in unprocessed blood samples and act as electrochemical transducers, enabling a rapid response time. Ultrasensitive protein analysis can be achieved using a digital ELISA platform that contains beads functionalized with antibodies to capture one or zero target protein molecules per bead162. The immunocomplexes are then labelled with enzymatic fluorescent reporters, which allows the isolation of single proteins in individual reaction chambers for fluorescence imaging. This ELISA platform can detect concentrations of 0.4 fM prostate-specific antigen (PSA) in patient sera. Proteins in complex biofluids can also be quantitatively analysed using antibody-modified magnetic nanoparticles that block an array of solid-state nanopores163. This sensor achieves a detection limit of 0.8 fM for PSA in whole blood at high throughput and has the potential to be miniaturized for point-of-care and mobile use. A sandwich assay based on a hybrid mechanical and optoplasmonic nanosensor can detect carcinoembryonic antigen and PSA in serum with a detection limit of 1 × 10−16 g ml−1 (ref. 101). A CRISPR-based assay can measure anti-SARS-CoV-2 antibodies in serum samples164, with a detection limit of 10 aM, enabling the detection of previously undetectable antibodies in vaccinated kidney transplant recipients. The assay converts the detection of anti-SARS-CoV-2 antibodies into the production of a double-stranded DNA barcode through proximity-based primer extension and recombinase polymerase amplification to amplify the signal. The double-stranded DNA barcode then activates CRISPR–Cas12a to cleave fluorescently quenched DNA probes.
a, Nanoparticle-based detection of microRNA. Gold-coated magnetic nanoparticles are functionalized with DNA probes that act as dispersible electrodes. After target hybridization and magnetic collection on the surface of a gold microelectrode, stable peak currents are measured to reflect the target analyte amount. b, Direct detection of nucleic acids through catalytic molecular nanostructures. A DNA–protein hybrid nanocomplex comprising an inactivating DNA duplex and a DNA polymerase allows direct and sensitive detection of nucleic acid targets in clinical samples. Upon specific binding of target nucleic acid, the complex destabilizes and releases active polymerase. The signalling element translates polymerase activity into quantifiable optical or electrochemical signals. c, Single-molecule measurement of circulating proteins using an integrated microdroplet platform. Miniaturized digital droplet assays are integrated into a mobile device, allowing sensitive, rapid and low-cost measurements. d, Single-nucleotide discrimination of DNA targets using a portable device incorporating toroidal convection PCR and a microarray of fluorescently quenched probes. By setting a temperature gradient across the device, the reaction solution moves circularly through convection, achieving thermal cycling. Specific detection of targets is achieved through strand displacement reactions of microarrayed toehold probes. e, Real-time analysis of protein–protein interactions using a genetically encoded protein nanopore sensor. The sensor comprises a truncated ferric hydroxamate uptake component A (t-FhuA) protein pore scaffold, a flexible hexapeptide glycine-glycine-serine-glycine-glycine-serine ((GGS)2) tether, a barnase (Bn) protein receptor and a peptide adaptor (O). Reversible protein interaction events are captured electrochemically as current transitions. Oon represents the Bn-released open substate; Ooff represents the Bn-captured open substate; τon represents the interevent duration; τoff represents the dwell time. Bs, barstar; dNTP, deoxynucleoside triphosphate; HRP, horseradish peroxidase; ssDNA, single-stranded DNA. Part a reprinted from ref. 160, Springer Nature Limited. Part b reprinted from ref. 89, Springer Nature Limited. Part c adapted with permission from ref. 168, Proceedings of the National Academy of Sciences. Part d adapted from ref. 171, Springer Nature Limited. Part e adapted from ref. 173, Springer Nature Limited.
Functional nanostructures can be precisely constructed in a programmable manner165, for example, to design responsive catalytic nanocomplexes. Such molecular switches can be integrated into miniaturized portable assays to sensitively detect nucleic acids in native biofluids89 (Fig. 5b). Here, each switch contains a DNA–enzyme hybrid assembly and an inhibitory DNA complex that binds and inactivates an enzyme. Upon binding of specific nucleic acids (DNA or RNA), the DNA complex dissociates to activate strong enzymatic activity. Through ratiometric tuning of molecular components within individual switches and collaborative coupling among multiple catalytic networks, this technology can sensitively detect SARS-CoV-2 RNA in clinical swab lysates, bypassing the technical requirements and processing steps of PCR detection (for example, sample purification, reverse transcription and thermal cycling), while achieving comparable sensitivity to PCR detection (detection limits as low as single copies)166,167.
Digital assays enable absolute quantification of molecules with high sensitivity; however, current technologies (for example, digital PCR and digital ELISA) require bulk instrumentation, hindering portable use. To achieve portable applications, digital droplet assays can be miniaturized168 (Fig. 5c) by parallelizing droplet generation, incubation and detection of single proteins onto a mobile platform, thereby achieving higher throughput than conventional droplet detection, while maintaining high sensitivity and a dynamic range. Magnetic nanoparticles coupled to a photonic crystal also allow digital-resolution biomolecule detection169. The magneto-plasmonic nanoparticles accelerate single-molecule sensing, enabling quantification of microRNA directly from unprocessed human serum with a 1-min response time and single-base mismatch selectivity.
Multiparametric profiling
Cells and EVs circulate as packaged units, whereas cell-free factors are disparately secreted and individually suspended in the circulation. This lack of organization makes tracing their origins difficult. Multiparametric measurements could assess the abundance of a particular cell-free factor and its distinct features, such as mutation status and epigenetic modification, and synergize complementary information such as dynamics monitoring and multi-omics integration.
Mutations, including single-nucleotide differences, play important roles in various diseases (for example, cancer, cardiovascular diseases and infectious diseases)170. Mutations in circulating nucleic acids can be detected using diagnostic technologies with single-nucleotide specificity; for example, a portable device based on a microarray system that is functionalized with fluorescently quenched toehold probes, capable of multiplex single-nucleotide discrimination, enables single-nucleotide polymorphism (SNP) typing171 (Fig. 5d). This battery-powered device can be further integrated with a toroidal convection chamber for PCR, allowing device miniaturization and point-of-care applications. SNPs can also be detected using an electrochemical sensor platform172 that contains DNA-nanostructured probes on the sensor surface with high selectivity for single-base mismatches, increasing the accessibility of target molecules to surface-immobilized probes.
Protein dynamics can provide important insights into pathophysiological processes but are difficult to measure in body fluids. A protein nanopore-based sensor that enables real-time measurement of protein–protein interactions can be integrated into nanofluidic devices173 (Fig. 5e). The sensor comprises a truncated outer membrane protein pore, a flexible tether, a protein receptor and a peptide adaptor. When a target protein ligand binds to the receptor, reversible capture and release events of the receptor can be measured as current transitions. Similarly, a microfluidic device-based single-molecule imaging technique can reveal conformational properties of proteins in blood174. Here, proteins of interest, for example, p53 complexes, are pulled down as clusters in an oscillating flow chamber, followed by targeting with fluorescently labelled antibodies, enabling the investigation of protein aggregation and selection of conformational p53 mutants.
Multimodal information about cfDNA characteristics, including DNA methylation, tissue of origin and DNA fragmentation, can assist in early cancer detection in clinical plasma samples175. These epigenetic and genetic features can be extracted by modified ten-eleven translocation-assisted pyridine borane sequencing and analysed in an integrated manner for cancer diagnosis. The blood test CancerSEEK combines assays for genetic alterations and protein biomarkers176. By integrating information from different omics technologies, CancerSEEK can identify early-stage cancers and localize the cancer organs of origin.
Outlook
The development of new diagnostic technologies should consider the requirements and needs of key stakeholders, that is, clinicians and patients2,7,18. From a clinical perspective, timely and actionable clinical information is crucial for stratifying patients, guiding treatment decisions and monitoring therapeutic response. Therefore, diagnostic platforms should demonstrate robust clinical performance, minimal sample processing, automated handling and high-throughput measurement to improve their integration with existing clinical workflows. From the perspective of a patient, minimally invasive procedures that provide rapid outputs at an affordable price are preferred. However, although routinely used in clinical practice, imaging modalities (for example, CT, MRI and PET) are costly, have limited sensitivity to molecular mechanisms and can delay detection, in particular, when gross abnormalities are present. Alternatively, tissue biopsy can provide some molecular information; however, the procedure is invasive, may cause complications and cannot be readily repeated. By contrast, liquid biopsy of circulating biomarkers offers a minimally invasive diagnostic strategy that can provide molecular information at high throughput for precise disease characterization, complementing conventional diagnostic approaches.
Single biomarkers are typically tested in diagnostic assays; however, they often lack sufficient discriminatory power18,177. Alternatively, assessment of multiple circulating biomarkers may allow early diagnosis (for example, stage I cancer detection) and high-coverage characterization of various diseases (for example, CancerSEEK detects breast, colorectal, gastric, liver, lung, oesophageal, ovarian and pancreatic cancer in a single blood test)176,178. Miniaturized devices in combination with information-revealing assays enable the integrated assessment of multiple biomarkers in small sample volumes, requiring only microlitres of bodily fluids158,175,176 (Box 1).
As more circulating biomarkers are being discovered, a better understanding of their release mechanisms and dynamics will guide improved biomarker signatures. In particular, different combinations of biomarkers are released into body fluids at different stages of disease progression6,26,32. Capturing this dynamic spectrum of circulating information will empower personalized medicine, including disease monitoring and drug selection. Technology platforms at the interface of biological systems, physical engineering and data analytics can acquire multiplex information at high resolution and throughput; for example, simultaneous interrogation of multidimensional properties, such as multi-omics and spatiotemporal features, could enable comprehensive and synergistic measurements. In addition, high-resolution profiling at the single-cell179,180 or single-EV level181,182 could reveal complex heterogeneity and define biomarker subpopulations. Moreover, big data analytics with artificial intelligence systems could enable rapid modelling, reveal refined signatures and improve classification accuracy178,183.
In addition to these advances in biomarker understanding, various technological hurdles need to be overcome for these analytical devices to progress to the clinic. At different stages of technology development, multiple considerations should be made with respect to its intended clinical use and platform performance. For example, when prototyping the technology, the system should be designed with functional modules, such as target enrichment, biomarker detection and data analysis, to promote its eventual integration with the clinical workflow. As compatibility with existing workflows and system automation are key considerations to promote clinical adoption, such integration will not only enhance analytical performance but also improve clinical uptake. Likewise, during the scaling-up of development to enable high-throughput manufacturing, reproducible and consistent analytical performance needs to be ensured, while achieving scalable production at low cost.
To promote the clinical translation of liquid biopsy tests, the specific clinical needs, regulatory guidelines and reimbursement models, among others184,185, should also be considered. In particular, translating pre-clinical prototypes into clinical platforms requires careful product development strategies. Depending on the development complexity of liquid biopsy technologies as well as relevant market demands, these technologies can be either deployed as laboratory-developed tests, for example, FoundationOne Liquid CDx, which are run in dedicated Clinically Laboratory Improvement Amendment (CLIA)-certified or equivalent clinical laboratories, or as in vitro diagnostic tests, for example, CELLSEARCH Circulating Tumour Cell Kit186, which represent higher-volume tests and have broader market potential. This decision has important regulatory implications187. For example, laboratory-developed tests are designed based on in-house protocols, can only be used in a single CLIA-certified laboratory and thus are exempted from extensive regulatory oversight. By contrast, in vitro diagnostic tests are marketed as standalone assay kits (and/or devices), can be used in multiple laboratories and are subjected to stringent regulatory clearance. Specific regulatory clearance requires the technology platforms to ensure analytical validity (analytical performance in terms of assay sensitivity, specificity, reliability and robustness), clinical validity (accuracy and reliability of the assay in achieving a defined clinical outcome) and clinical utility (clinical benefit by the assay as compared to standard methods in disease management). Importantly, clinical utility is a key consideration by reimbursement authorities in determining cost-effectiveness188,189. Reimbursement and clinical adoption of liquid biopsy tests require a value proposition weighing the incremental test costs against the clinical benefit.
Encouragingly, several liquid biopsy tests have already received FDA clearance and are commercially available (Table 1). These assays will serve as predicates and motivate the development of new liquid biopsy tests across multiple indications. To continue the clinical deployment of liquid biopsy tests, collaborations across multiple key stakeholders, including the scientific and medical community, patient organizations, commercial developers, policy-makers and funders, are important. Such efforts will not only nurture the translation of liquid biopsy technologies but also transform the way we practice medicine.
Citation diversity statement
We acknowledge that papers authored by scholars from historically excluded groups are systematically under-cited. Here, we have made every attempt to reference relevant papers in a manner that is equitable in terms of racial, ethnic, gender and geographical representation.
References
Ashley, E. A. Towards precision medicine. Nat. Rev. Genet. 17, 507–522 (2016).
Collins, F. S. & Varmus, H. A new initiative on precision medicine. N. Engl. J. Med. 372, 793–795 (2015).
Gerlinger, M. et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 366, 883–892 (2012).
McGranahan, N. & Swanton, C. Clonal heterogeneity and tumor evolution: past, present, and the future. Cell 168, 613–628 (2017).
Vitale, I., Shema, E., Loi, S. & Galluzzi, L. Intratumoral heterogeneity in cancer progression and response to immunotherapy. Nat. Med. 27, 212–224 (2021).
Wan, J. C. M. et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat. Rev. Cancer 17, 223–238 (2017). This review provides an overview of ctDNA biology and its translational potential for prognostication, molecular profiling and disease monitoring.
Ignatiadis, M., Sledge, G. W. & Jeffrey, S. S. Liquid biopsy enters the clinic-implementation issues and future challenges. Nat. Rev. Clin. Oncol. 18, 297–312 (2021).
Altug, H., Oh, S. H., Maier, S. A. & Homola, J. Advances and applications of nanophotonic biosensors. Nat. Nanotechnol. 17, 5–16 (2022).
Kim, J., Campbell, A. S., de Ávila, B. E. & Wang, J. Wearable biosensors for healthcare monitoring. Nat. Biotechnol. 37, 389–406 (2019).
Bariya, M., Nyein, H. Y. Y. & Javey, A. Wearable sweat sensors. Nat. Electron. 1, 160–171 (2018).
Heitzer, E., Haque, I. S., Roberts, C. E. S. & Speicher, M. R. Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat. Rev. Genet. 20, 71–88 (2019).
Ramalingam, N. & Jeffrey, S. S. Future of liquid biopsies with growing technological and bioinformatics studies: opportunities and challenges in discovering tumor heterogeneity with single-cell level analysis. Cancer J. 24, 104–108 (2018).
Huang, C. et al. Extracellular vesicles in cardiovascular disease. Adv. Clin. Chem. 103, 47–95 (2021).
Krebs, M. G. et al. Molecular analysis of circulating tumour cells-biology and biomarkers. Nat. Rev. Clin. Oncol. 11, 129–144 (2014).
Raposo, G. & Stoorvogel, W. Extracellular vesicles: exosomes, microvesicles, and friends. J. Cell Biol. 200, 373–383 (2013).
Schwarzenbach, H., Hoon, D. S. & Pantel, K. Cell-free nucleic acids as biomarkers in cancer patients. Nat. Rev. Cancer 11, 426–437 (2011).
Hanash, S. M., Pitteri, S. J. & Faca, V. M. Mining the plasma proteome for cancer biomarkers. Nature 452, 571–579 (2008).
Teunissen, C. E. et al. Blood-based biomarkers for Alzheimer’s disease: towards clinical implementation. Lancet Neurol. 21, 66–77 (2022).
Kevadiya, B. D. et al. Diagnostics for SARS-CoV-2 infections. Nat. Mater. 20, 593–605 (2021).
Alix-Panabières, C. & Pantel, K. Challenges in circulating tumour cell research. Nat. Rev. Cancer 14, 623–631 (2014). This article discusses challenges and strategies in enriching, detecting and characterizing CTCs in translational cancer research.
Shao, H. et al. New technologies for analysis of extracellular vesicles. Chem. Rev. 118, 1917–1950 (2018). This article presents a comprehensive and critical overview of emerging analytical technologies for EV detection and their clinical applications.
Song, P. et al. Limitations and opportunities of technologies for the analysis of cell-free DNA in cancer diagnostics. Nat. Biomed. Eng. 6, 232–245 (2022).
Huang, Q. et al. Critical review: digital resolution biomolecular sensing for diagnostics and life science research. Lab Chip 20, 2816–2840 (2020).
Rufo, J., Cai, F., Friend, J., Wiklund, M. & Huang, T. J. Acoustofluidics for biomedical applications. Nat. Rev. Methods Primers 2, 30 (2022).
Ashworth, T. R. A case of cancer in which cells similar to those in the tumours were seen in the blood after death. Aust. Med. J. 14, 146 (1869).
Vanharanta, S. & Massagué, J. Origins of metastatic traits. Cancer Cell 24, 410–421 (2013).
Chaffer, C. L. & Weinberg, R. A. A perspective on cancer cell metastasis. Science 331, 1559–1564 (2011).
Psaila, B. & Lyden, D. The metastatic niche: adapting the foreign soil. Nat. Rev. Cancer 9, 285–293 (2009).
Aceto, N. et al. Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell 158, 1110–1122 (2014).
Pantel, K. & Alix-Panabières, C. Real-time liquid biopsy in cancer patients: fact or fiction. Cancer Res. 73, 6384–6388 (2013).
Keller, L. & Pantel, K. Unravelling tumour heterogeneity by single-cell profiling of circulating tumour cells. Nat. Rev. Cancer 19, 553–567 (2019).
Théry, C., Zitvogel, L. & Amigorena, S. Exosomes: composition, biogenesis and function. Nat. Rev. Immunol. 2, 569–579 (2002).
Raposo, G. et al. B lymphocytes secrete antigen-presenting vesicles. J. Exp. Med. 183, 1161–1172 (1996).
Zitvogel, L. et al. Eradication of established murine tumors using a novel cell-free vaccine: dendritic cell-derived exosomes. Nat. Med. 4, 594–600 (1998).
EL Andaloussi, S., Mäger, I., Breakefield, X. O. & Wood, M. J. Extracellular vesicles: biology and emerging therapeutic opportunities. Nat. Rev. Drug Discov. 12, 347–357 (2013).
Minciacchi, V. R., Freeman, M. R. & Di Vizio, D. Extracellular vesicles in cancer: exosomes, microvesicles and the emerging role of large oncosomes. Semin. Cell Dev. Biol. 40, 41–51 (2015).
Simpson, R. J., Lim, J. W., Moritz, R. L. & Mathivanan, S. Exosomes: proteomic insights and diagnostic potential. Expert Rev. Proteom. 6, 267–283 (2009).
Tong, L. et al. Milk-derived extracellular vesicles alleviate ulcerative colitis by regulating the gut immunity and reshaping the gut microbiota. Theranostics 11, 8570–8586 (2021).
Caradec, J. et al. Reproducibility and efficiency of serum-derived exosome extraction methods. Clin. Biochem. 47, 1286–1292 (2014).
Melo, S. A. et al. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell 26, 707–721 (2014).
Choi, D. S., Kim, D. K., Kim, Y. K. & Gho, Y. S. Proteomics of extracellular vesicles: Exosomes and ectosomes. Mass. Spectrom. Rev. 34, 474–490 (2015).
Shao, H. et al. Protein typing of circulating microvesicles allows real-time monitoring of glioblastoma therapy. Nat. Med. 18, 1835–1840 (2012).
Valadi, H. et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 9, 654–659 (2007).
Skog, J. et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 10, 1470–1476 (2008).
Mathivanan, S., Fahner, C. J., Reid, G. E. & Simpson, R. J. ExoCarta 2012: database of exosomal proteins, RNA and lipids. Nucleic Acids Res. 40, D1241–D1244 (2012).
Kalra, H. et al. Vesiclepedia: a compendium for extracellular vesicles with continuous community annotation. PLoS Biol. 10, e1001450 (2012).
Maas, S. L. N., Breakefield, X. O. & Weaver, A. M. Extracellular vesicles: unique intercellular delivery vehicles. Trends Cell Biol. 27, 172–188 (2017).
Wang, Z. et al. Dual-selective magnetic analysis of extracellular vesicle glycans. Matter 2, 150–166 (2020).
van Niel, G., D’Angelo, G. & Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 19, 213–228 (2018).
Buzás, E. I., Tóth, E. Á., Sódar, B. W. & Szabó-Taylor, K. É. Molecular interactions at the surface of extracellular vesicles. Semin. Immunopathol. 40, 453–464 (2018).
Lim, C. Z. J. et al. Subtyping of circulating exosome-bound amyloid β reflects brain plaque deposition. Nat. Commun. 10, 1144 (2019).
Mathieu, M., Martin-Jaular, L., Lavieu, G. & Théry, C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat. Cell Biol. 21, 9–17 (2019).
Gustafson, D., Veitch, S. & Fish, J. E. Extracellular vesicles as protagonists of diabetic cardiovascular pathology. Front. Cardiovasc. Med. 4, 71 (2017).
Théry, C., Ostrowski, M. & Segura, E. Membrane vesicles as conveyors of immune responses. Nat. Rev. Immunol. 9, 581–593 (2009).
Mouliere, F. et al. High fragmentation characterizes tumour-derived circulating DNA. PLoS One 6, e23418 (2011).
Mouliere, F., El Messaoudi, S., Pang, D., Dritschilo, A. & Thierry, A. R. Multi-marker analysis of circulating cell-free DNA toward personalized medicine for colorectal cancer. Mol. Oncol. 8, 927–941 (2014).
Rodrigues Filho, E. M. et al. Elevated cell-free plasma DNA level as an independent predictor of mortality in patients with severe traumatic brain injury. J. Neurotrauma 31, 1639–1646 (2014).
Tsai, N. W. et al. The value of serial plasma nuclear and mitochondrial DNA levels in patients with acute ischemic stroke. Clin. Chim. Acta 412, 476–479 (2011).
Breitbach, S., Sterzing, B., Magallanes, C., Tug, S. & Simon, P. Direct measurement of cell-free DNA from serially collected capillary plasma during incremental exercise. J. Appl. Physiol. 117, 119–130 (2014).
De Vlaminck, I. et al. Circulating cell-free DNA enables noninvasive diagnosis of heart transplant rejection. Sci. Transl. Med. 6, 241ra77 (2014).
De Vlaminck, I. et al. Noninvasive monitoring of infection and rejection after lung transplantation. Proc. Natl Acad. Sci. USA 112, 13336–13341 (2015).
Snyder, M. W., Kircher, M., Hill, A. J., Daza, R. M. & Shendure, J. Cell-free DNA comprises an in vivo nucleosome footprint that informs its tissues-of-origin. Cell 164, 57–68 (2016).
Lehmann-Werman, R. et al. Identification of tissue-specific cell death using methylation patterns of circulating DNA. Proc. Natl Acad. Sci. USA 113, E1826–E1834 (2016).
Sun, K. et al. Plasma DNA tissue mapping by genome-wide methylation sequencing for noninvasive prenatal, cancer, and transplantation assessments. Proc. Natl Acad. Sci. USA 112, E5503–E5512 (2015).
Cescon, D. W., Bratman, S. V., Chan, S. M. & Siu, L. L. Circulating tumor DNA and liquid biopsy in oncology. Nat. Cancer 1, 276–290 (2020).
Mann, M. & Jensen, O. N. Proteomic analysis of post-translational modifications. Nat. Biotechnol. 21, 255–261 (2003).
Zethelius, B. et al. Use of multiple biomarkers to improve the prediction of death from cardiovascular causes. N. Engl. J. Med. 358, 2107–2116 (2008).
Krammer, F. & Simon, V. Serology assays to manage COVID-19. Science 368, 1060–1061 (2020).
Théry, C. et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 7, 1535750 (2018).
Aebersold, R. et al. How many human proteoforms are there. Nat. Chem. Biol. 14, 206–214 (2018).
Nge, P. N., Rogers, C. I. & Woolley, A. T. Advances in microfluidic materials, functions, integration, and applications. Chem. Rev. 113, 2550–2583 (2013).
Xu, Y. et al. A review of impedance measurements of whole cells. Biosens. Bioelectron. 77, 824–836 (2016).
Arlett, J. L., Myers, E. B. & Roukes, M. L. Comparative advantages of mechanical biosensors. Nat. Nanotechnol. 6, 203–215 (2011).
Jha, S. K., Ahmed, Z., Agio, M., Ekinci, Y. & Löffler, J. F. Deep-UV surface-enhanced resonance Raman scattering of adenine on aluminum nanoparticle arrays. J. Am. Chem. Soc. 134, 1966–1969 (2012).
Ouyang, L. et al. Expanding and optimizing 3D bioprinting capabilities using complementary network bioinks. Sci. Adv. 6, eabc5529 (2020).
Chen, Y. Nanofabrication by electron beam lithography and its applications: a review. Microelectron. Eng. 135, 57–72 (2015).
Truskett, V. N. & Watts, M. P. Trends in imprint lithography for biological applications. Trends Biotechnol. 24, 312–317 (2006).
Willets, K. A. & Van Duyne, R. P. Localized surface plasmon resonance spectroscopy and sensing. Annu. Rev. Phys. Chem. 58, 267–297 (2007).
Mayer, K. M. & Hafner, J. H. Localized surface plasmon resonance sensors. Chem. Rev. 111, 3828–3857 (2011).
Jahani, Y. et al. Imaging-based spectrometer-less optofluidic biosensors based on dielectric metasurfaces for detecting extracellular vesicles. Nat. Commun. 12, 3246 (2021).
Dufrêne, Y. F. et al. Imaging modes of atomic force microscopy for application in molecular and cell biology. Nat. Nanotechnol. 12, 295–307 (2017).
Lapine, M., Shadrivov, I. V., Powell, D. A. & Kivshar, Y. S. Magnetoelastic metamaterials. Nat. Mater. 11, 30–33 (2011).
Yang, X., Yang, M., Pang, B., Vara, M. & Xia, Y. Gold nanomaterials at work in biomedicine. Chem. Rev. 115, 10410–10488 (2015).
Howes, P. D., Chandrawati, R. & Stevens, M. M. Bionanotechnology. Colloidal nanoparticles as advanced biological sensors. Science 346, 1247390 (2014).
Furukawa, H., Cordova, K. E., O’Keeffe, M. & Yaghi, O. M. The chemistry and applications of metal-organic frameworks. Science 341, 1230444 (2013).
Li, J. F., Zhang, Y. J., Ding, S. Y., Panneerselvam, R. & Tian, Z. Q. Core-Shell nanoparticle-enhanced Raman spectroscopy. Chem. Rev. 117, 5002–5069 (2017).
Lin, C. et al. Submicrometre geometrically encoded fluorescent barcodes self-assembled from DNA. Nat. Chem. 4, 832–839 (2012).
Li, Y. et al. Controlled assembly of dendrimer-like DNA. Nat. Mater. 3, 38–42 (2004).
Ho, N. R. Y. et al. Visual and modular detection of pathogen nucleic acids with enzyme-DNA molecular complexes. Nat. Commun. 9, 3238 (2018).
Gootenberg, J. S. et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 356, 438–442 (2017).
Wang, C., Yue, L. & Willner, I. Controlling biocatalytic cascades with enzyme–DNA dynamic networks. Nat. Catal. 3, 941–950 (2020).
Schaffter, S. W. & Schulman, R. Building in vitro transcriptional regulatory networks by successively integrating multiple functional circuit modules. Nat. Chem. 11, 829–838 (2019).
Karabacak, N. M. et al. Microfluidic, marker-free isolation of circulating tumor cells from blood samples. Nat. Protoc. 9, 694–710 (2014).
Wang, Z. et al. Acoustofluidic salivary exosome isolation: a liquid biopsy compatible approach for human papillomavirus-associated oropharyngeal cancer detection. J. Mol. Diagn. 22, 50–59 (2020).
Hao, N. et al. Acoustofluidics-assisted engineering of multifunctional three-dimensional zinc oxide nanoarrays. ACS Nano 14, 6150–6163 (2020).
Liu, C. et al. Low-cost thermophoretic profiling of extracellular-vesicle surface proteins for the early detection and classification of cancers. Nat. Biomed. Eng. 3, 183–193 (2019).
Clegg, J. R. & Peppas, N. A. Molecular recognition with soft biomaterials. Soft Matter 16, 856–869 (2020).
Ayala-Mar, S., Perez-Gonzalez, V. H., Mata-Gómez, M. A., Gallo-Villanueva, R. C. & González-Valdez, J. Electrokinetically driven exosome separation and concentration using dielectrophoretic-enhanced PDMS-based microfluidics. Anal. Chem. 91, 14975–14982 (2019).
Shao, H. et al. Chip-based analysis of exosomal mRNA mediating drug resistance in glioblastoma. Nat. Commun. 6, 6999 (2015).
Jiang, Y. et al. Aptamer/AuNP biosensor for colorimetric profiling of exosomal proteins. Angew. Chem. Int. Ed. Engl. 56, 11916–11920 (2017).
Kosaka, P. M. et al. Detection of cancer biomarkers in serum using a hybrid mechanical and optoplasmonic nanosensor. Nat. Nanotechnol. 9, 1047–1053 (2014).
Guo, K. et al. Rapid single-molecule detection of COVID-19 and MERS antigens via nanobody-functionalized organic electrochemical transistors. Nat. Biomed. Eng. 5, 666–677 (2021).
Cho, H. et al. Single-step nanoplasmonic VEGF165 aptasensor for early cancer diagnosis. ACS Nano 6, 7607–7614 (2012).
Sun, N., Li, X., Wang, Z., Li, Y. & Pei, R. High-purity capture of CTCs based on micro-beads enhanced isolation by size of epithelial tumor cells (ISET) method. Biosens. Bioelectron. 102, 157–163 (2018).
Mertins, P. et al. Integrated proteomic analysis of post-translational modifications by serial enrichment. Nat. Methods 10, 634–637 (2013).
Zecha, J. et al. Linking post-translational modifications and protein turnover by site-resolved protein turnover profiling. Nat. Commun. 13, 165 (2022).
Wu, L. et al. Beyond capture: circulating tumor cell release and single‐cell analysis. Small Methods 3, 1800544 (2019).
Liang, K. et al. Nanoplasmonic quantification of tumor-derived extracellular vesicles in plasma microsamples for diagnosis and treatment monitoring. Nat. Biomed. Eng. 1, 0021 (2017).
Li, W. et al. Label-free colorimetric assay for methyltransferase activity based on a novel methylation-responsive DNAzyme strategy. Anal. Chem. 82, 1935–1941 (2010).
Zhang, L. et al. Plasmonic Al nanopyramid array sensor for monitoring the attaching and spreading of cells. Sens. Actuators B Chem. 279, 503–508 (2019).
Cheong, R., Wang, C. J. & Levchenko, A. Using a microfluidic device for high-content analysis of cell signaling. Sci. Signal. 2, pl2 (2009).
Boneschansker, L., Yan, J., Wong, E., Briscoe, D. M. & Irimia, D. Microfluidic platform for the quantitative analysis of leukocyte migration signatures. Nat. Commun. 5, 4787 (2014).
Culver, H. R., Wechsler, M. E. & Peppas, N. A. Label-free detection of tear biomarkers using hydrogel-coated gold nanoshells in a localized surface plasmon resonance-based biosensor. ACS Nano 12, 9342–9354 (2018).
John-Herpin, A., Kavungal, D., von Mücke, L. & Altug, H. Infrared metasurface augmented by deep learning for monitoring dynamics between all major classes of biomolecules. Adv. Mater. 33, e2006054 (2021).
Camacho, D. M., Collins, K. M., Powers, R. K., Costello, J. C. & Collins, J. J. Next-generation machine learning for biological networks. Cell 173, 1581–1592 (2018).
Topol, E. J. High-performance medicine: the convergence of human and artificial intelligence. Nat. Med. 25, 44–56 (2019).
Warkiani, M. E. et al. Ultra-fast, label-free isolation of circulating tumor cells from blood using spiral microfluidics. Nat. Protoc. 11, 134–148 (2016).
Li, P. et al. Acoustic separation of circulating tumor cells. Proc. Natl Acad. Sci. USA 112, 4970–4975 (2015).
Wu, M. et al. Circulating tumor cell phenotyping via high-throughput acoustic separation. Small 14, e1801131 (2018).
Sarioglu, A. F. et al. A microfluidic device for label-free, physical capture of circulating tumor cell clusters. Nat. Methods 12, 685–691 (2015).
Riethdorf, S. et al. Detection of circulating tumor cells in peripheral blood of patients with metastatic breast cancer: a validation study of the CellSearch system. Clin. Cancer Res. 13, 920–928 (2007).
Gorin, M. A. et al. Circulating tumour cells as biomarkers of prostate, bladder, and kidney cancer. Nat. Rev. Urol. 14, 90–97 (2017).
Nagrath, S. et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature 450, 1235–1239 (2007).
Stott, S. L. et al. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proc. Natl Acad. Sci. USA 107, 18392–18397 (2010).
Zhang, R. et al. Magnetic “squashing” of circulating tumor cells on plasmonic substrates for ultrasensitive NIR fluorescence detection. Small Methods 3, 1800474 (2019).
Yoon, H. J. et al. Sensitive capture of circulating tumour cells by functionalized graphene oxide nanosheets. Nat. Nanotechnol. 8, 735–741 (2013).
Park, M. H. et al. Enhanced isolation and release of circulating tumor cells using nanoparticle binding and ligand exchange in a microfluidic chip. J. Am. Chem. Soc. 139, 2741–2749 (2017).
Hong, R. et al. Glutathione-mediated delivery and release using monolayer protected nanoparticle carriers. J. Am. Chem. Soc. 128, 1078–1079 (2006).
Vermesh, O. et al. An intravascular magnetic wire for the high-throughput retrieval of circulating tumour cells in vivo. Nat. Biomed. Eng. 2, 696–705 (2018).
Mishra, A. et al. Ultrahigh-throughput magnetic sorting of large blood volumes for epitope-agnostic isolation of circulating tumor cells. Proc. Natl Acad. Sci. USA 117, 16839–16847 (2020).
Issadore, D. et al. Ultrasensitive clinical enumeration of rare cells ex vivo using a micro-hall detector. Sci. Transl. Med. 4, 141ra92 (2012).
Chen, Y. H., Pulikkathodi, A. K., Ma, Y. D., Wang, Y. L. & Lee, G. B. A microfluidic platform integrated with field-effect transistors for enumeration of circulating tumor cells. Lab Chip 19, 618–625 (2019).
Galanzha, E. I. et al. In vivo liquid biopsy using Cytophone platform for photoacoustic detection of circulating tumor cells in patients with melanoma. Sci. Transl. Med. 11, eaat5857 (2019).
Yankaskas, C. L. et al. A microfluidic assay for the quantification of the metastatic propensity of breast cancer specimens. Nat. Biomed. Eng. 3, 452–465 (2019).
Wong, B. S. et al. A microfluidic cell-migration assay for the prediction of progression-free survival and recurrence time of patients with glioblastoma. Nat. Biomed. Eng. 5, 26–40 (2021).
Wong, I. Y. et al. Collective and individual migration following the epithelial-mesenchymal transition. Nat. Mater. 13, 1063–1071 (2014).
Yang, S. et al. Harmonic acoustics for dynamic and selective particle manipulation. Nat. Mater. 21, 540–546 (2022).
Alix-Panabières, C. EPISPOT assay: detection of viable DTCs/CTCs in solid tumor patients. Recent Results Cancer Res. 195, 69–76 (2012).
Sinkala, E. et al. Profiling protein expression in circulating tumour cells using microfluidic western blotting. Nat. Commun. 8, 14622 (2017).
Grist, S. M., Mourdoukoutas, A. P. & Herr, A. E. 3D projection electrophoresis for single-cell immunoblotting. Nat. Commun. 11, 6237 (2020).
Vlassakis, J. et al. Measuring expression heterogeneity of single-cell cytoskeletal protein complexes. Nat. Commun. 12, 4969 (2021).
Horvath, P. et al. Screening out irrelevant cell-based models of disease. Nat. Rev. Drug Discov. 15, 751–769 (2016).
Wilding, J. L. & Bodmer, W. F. Cancer cell lines for drug discovery and development. Cancer Res. 74, 2377–2384 (2014).
Khoo, B. L. et al. Expansion of patient-derived circulating tumor cells from liquid biopsies using a CTC microfluidic culture device. Nat. Protoc. 13, 34–58 (2018).
Colombo, M., Raposo, G. & Théry, C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 30, 255–289 (2014).
Yáñez-Mó, M. et al. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 4, 27066 (2015).
Lee, K., Shao, H., Weissleder, R. & Lee, H. Acoustic purification of extracellular microvesicles. ACS Nano 9, 2321–2327 (2015).
Chen, Y. et al. Exosome detection via the ultrafast-isolation system: EXODUS. Nat. Methods 18, 212–218 (2021).
Zhang, P. et al. Ultrasensitive detection of circulating exosomes with a 3D-nanopatterned microfluidic chip. Nat. Biomed. Eng. 3, 438–451 (2019).
Jeong, S. et al. Integrated magneto-electrochemical sensor for exosome analysis. ACS Nano 10, 1802–1809 (2016).
Park, J. et al. An integrated magneto-electrochemical device for the rapid profiling of tumour extracellular vesicles from blood plasma. Nat. Biomed. Eng. 5, 678–689 (2021).
Xin, H., Namgung, B. & Lee, L. P. Nanoplasmonic optical antennas for life sciences and medicine. Nat. Rev. Mater. 3, 228–243 (2018).
Im, H. et al. Label-free detection and molecular profiling of exosomes with a nano-plasmonic sensor. Nat. Biotechnol. 32, 490–495 (2014).
Wu, X. et al. Exosome-templated nanoplasmonics for multiparametric molecular profiling. Sci. Adv. 6, eaba2556 (2020).
Lim, C. Z. J., Zhang, L., Zhang, Y., Sundah, N. R. & Shao, H. New sensors for extracellular vesicles: insights on constituent and associated biomarkers. ACS Sens. 5, 4–12 (2020).
Lim, C. Z. J., Natalia, A., Sundah, N. R. & Shao, H. Biomarker organization in circulating extracellular vesicles: new applications in detecting neurodegenerative diseases. Adv. Biosyst. 4, e1900309 (2020).
Zhang, P. et al. Molecular and functional extracellular vesicle analysis using nanopatterned microchips monitors tumor progression and metastasis. Sci. Transl. Med. 12, eaaz2878 (2020).
Pan, S. et al. Extracellular vesicle drug occupancy enables real-time monitoring of targeted cancer therapy. Nat. Nanotechnol. 16, 734–742 (2021).
Liebel, M. et al. 3D tracking of extracellular vesicles by holographic fluorescence imaging. Sci. Adv. 6, eabc2508 (2020).
Tavallaie, R. et al. Nucleic acid hybridization on an electrically reconfigurable network of gold-coated magnetic nanoparticles enables microRNA detection in blood. Nat. Nanotechnol. 13, 1066–1071 (2018).
Chen, D., Wu, Y., Hoque, S., Tilley, R. D. & Gooding, J. J. Rapid and ultrasensitive electrochemical detection of circulating tumor DNA by hybridization on the network of gold-coated magnetic nanoparticles. Chem. Sci. 12, 5196–5201 (2021).
Rissin, D. M. et al. Single-molecule enzyme-linked immunosorbent assay detects serum proteins at subfemtomolar concentrations. Nat. Biotechnol. 28, 595–599 (2010).
Chuah, K. et al. Nanopore blockade sensors for ultrasensitive detection of proteins in complex biological samples. Nat. Commun. 10, 2109 (2019).
Tang, Y. et al. A CRISPR-based ultrasensitive assay detects attomolar concentrations of SARS-CoV-2 antibodies in clinical samples. Nat. Commun. 13, 4667 (2022).
Seeman, N. C. & Sleiman, H. F. DNA nanotechnology. Nat. Rev. Mat. 3, 17068 (2017).
Sundah, N. R. et al. Catalytic amplification by transition-state molecular switches for direct and sensitive detection of SARS-CoV-2. Sci. Adv. 7, eabe5940 (2021).
Chen, Y. et al. Collaborative equilibrium coupling of catalytic DNA nanostructures enables programmable detection of SARS-CoV-2. Adv. Sci. 8, e2101155 (2021).
Yelleswarapu, V. et al. Mobile platform for rapid sub-picogram-per-milliliter, multiplexed, digital droplet detection of proteins. Proc. Natl Acad. Sci. USA 116, 4489–4495 (2019).
Che, C. et al. Accelerated digital biodetection using magneto-plasmonic nanoparticle-coupled photonic resonator absorption microscopy. ACS Nano 16, 2345–2354 (2022).
1000 Genomes Project Consortium et al. A global reference for human genetic variation. Nature 526, 68–74 (2015).
Khodakov, D., Li, J., Zhang, J. X. & Zhang, D. Y. Highly multiplexed rapid DNA detection with single-nucleotide specificity via convective PCR in a portable device. Nat. Biomed. Eng. 5, 702–712 (2021).
Pei, H. et al. A DNA nanostructure-based biomolecular probe carrier platform for electrochemical biosensing. Adv. Mater. 22, 4754–4758 (2010).
Thakur, A. K. & Movileanu, L. Real-time measurement of protein-protein interactions at single-molecule resolution using a biological nanopore. Nat. Biotechnol. 37, 96–101 (2019).
Mao, C. P. et al. Protein detection in blood with single-molecule imaging. Sci. Adv. 7, eabg6522 (2021).
Siejka-Zielińska, P. et al. Cell-free DNA TAPS provides multimodal information for early cancer detection. Sci. Adv. 7, eabh0534 (2021).
Cohen, J. D. et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 359, 926–930 (2018).
Borrebaeck, C. A. Precision diagnostics: moving towards protein biomarker signatures of clinical utility in cancer. Nat. Rev. Cancer 17, 199–204 (2017).
Cristiano, S. et al. Genome-wide cell-free DNA fragmentation in patients with cancer. Nature 570, 385–389 (2019).
Cheow, L. F. et al. Single-cell multimodal profiling reveals cellular epigenetic heterogeneity. Nat. Methods 13, 833–836 (2016).
Wang, N. et al. Single-cell microRNA-mRNA co-sequencing reveals non-genetic heterogeneity and mechanisms of microRNA regulation. Nat. Commun. 10, 95 (2019).
Lee, K. et al. Multiplexed profiling of single extracellular vesicles. ACS Nano 12, 494–503 (2018).
Penders, J. et al. Single particle automated raman trapping analysis of breast cancer cell-derived extracellular vesicles as cancer biomarkers. ACS Nano 15, 18192–18205 (2021).
Zeune, L. L. et al. Deep learning of circulating tumour cells. Nat. Mach. Intell. 2, 124–133 (2020).
IJzerman, M. J. et al. Towards routine implementation of liquid biopsies in cancer management: it is always too early, until suddenly it is too late. Diagnostics 11, 103 (2021).
Horgan, D. et al. Accelerating the development and validation of liquid biopsy for early cancer screening and treatment tailoring. Healthcare 10, 1714 (2022).
Goodsaid, F. M. The labyrinth of product development and regulatory approvals in liquid biopsy diagnostics. Clin. Transl. Sci. 12, 431–439 (2019).
Gatter, K. FDA oversight of laboratory-developed tests: where are we now. Arch. Pathol. Lab. Med. 141, 746–748 (2017).
IJzerman, M. J., Berghuis, A. M. S., de Bono, J. S. & Terstappen, L. W. M. M. Health economic impact of liquid biopsies in cancer management. Expert Rev. Pharmacoecon. Outcomes Res. 18, 593–599 (2018).
Douglas, M. P., Gray, S. W. & Phillips, K. A. Private payer and medicare coverage for circulating tumor DNA testing: a historical analysis of coverage policies from 2015 to 2019. J. Natl Compr. Canc Netw. 18, 866–872 (2020).
Cohen, S. J. et al. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J. Clin. Oncol. 26, 3213–3221 (2008).
Cristofanilli, M. et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N. Engl. J. Med. 351, 781–791 (2004).
Danila, D. C. et al. Circulating tumor cell number and prognosis in progressive castration-resistant prostate cancer. Clin. Cancer Res. 13, 7053–7058 (2007).
Riethdorf, S., O’Flaherty, L., Hille, C. & Pantel, K. Clinical applications of the CellSearch platform in cancer patients. Adv. Drug Deliv. Rev. 125, 102–121 (2018).
McKiernan, J. et al. A novel urine exosome gene expression assay to predict high-grade prostate cancer at initial biopsy. JAMA Oncol. 2, 882–889 (2016).
McKiernan, J. et al. A prospective adaptive utility trial to validate performance of a novel urine exosome gene expression assay to predict high-grade prostate cancer in patients with prostate-specific antigen 2-10ng/ml at initial biopsy. Eur. Urol. 74, 731–738 (2018).
Margolis, E. et al. Predicting high-grade prostate cancer at initial biopsy: clinical performance of the ExoDx (EPI) Prostate Intelliscore test in three independent prospective studies. Prostate Cancer Prostatic Dis. 25, 296–301 (2022).
Lamb, Y. N. & Dhillon, S. Epi proColon® 2.0 CE: a blood-based screening test for colorectal cancer. Mol. Diagn. Ther. 21, 225–232 (2017).
Malapelle, U. et al. Profile of the Roche cobas® EGFR mutation test v2 for non-small cell lung cancer. Expert Rev. Mol. Diagn. 17, 209–215 (2017).
Martínez-Sáez, O. et al. Frequency and spectrum of PIK3CA somatic mutations in breast cancer. Breast Cancer Res. 22, 45 (2020).
Kim, S. T. et al. Prospective feasibility study for using cell-free circulating tumor DNA-guided therapy in refractory metastatic solid cancers: an interim analysis. JCO Precis. Oncol. 1, PO.16.00059 (2017).
Aggarwal, C. et al. Clinical implications of plasma-based genotyping with the delivery of personalized therapy in metastatic non-small cell lung cancer. JAMA Oncol. 5, 173–180 (2019).
Woodhouse, R. et al. Clinical and analytical validation of FoundationOne Liquid CDx, a novel 324-gene cfDNA-based comprehensive genomic profiling assay for cancers of solid tumor origin. PLoS One 15, e0237802 (2020).
Acknowledgements
The authors received support from the National University of Singapore (NUS), NUS Research Scholarship, Ministry of Education and National Medical Research Council.
Author information
Authors and Affiliations
Contributions
All authors contributed to the preparation of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Bioengineering thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Natalia, A., Zhang, L., Sundah, N.R. et al. Analytical device miniaturization for the detection of circulating biomarkers. Nat Rev Bioeng 1, 481–498 (2023). https://doi.org/10.1038/s44222-023-00050-8
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s44222-023-00050-8
This article is cited by
-
Identification of exosome protein panels as predictive biomarkers for non-small cell lung cancer
Biological Procedures Online (2023)
-
Multiplexed RNA profiling by regenerative catalysis enables blood-based subtyping of brain tumors
Nature Communications (2023)