Abstract
High-risk mutations in several genes predispose to both colorectal cancer (CRC) and endometrial cancer (EC). We therefore hypothesised that some lower-risk genetic variants might also predispose to both CRC and EC. Using CRC and EC genome-wide association series, totalling 13,265 cancer cases and 40,245 controls, we found that the protective allele [G] at one previously-identified CRC polymorphism, rs2736100 near TERT, was associated with EC risk (odds ratio (OR) = 1.08, P = 0.000167); this polymorphism influences the risk of several other cancers. A further CRC polymorphism near TERC also showed evidence of association with EC (OR = 0.92; P = 0.03). Overall, however, there was no good evidence that the set of CRC polymorphisms was associated with EC risk and neither of two previously-reported EC polymorphisms was associated with CRC risk. A combined analysis revealed one genome-wide significant polymorphism, rs3184504, on chromosome 12q24 (OR = 1.10, P = 7.23 × 10−9) with shared effects on CRC and EC risk. This polymorphism, a missense variant in the gene SH2B3, is also associated with haematological and autoimmune disorders, suggesting that it influences cancer risk through the immune response. Another polymorphism, rs12970291 near gene TSHZ1, was associated with both CRC and EC (OR = 1.26, P = 4.82 × 10−8), with the alleles showing opposite effects on the risks of the two cancers.
Similar content being viewed by others
Introduction
Colorectal carcinoma (CRC) is the fourth commonest cancer in the western world and cancer of the uterine corpus, or endometrial carcinoma (EC), is the fourth commonest cancer among women. Both cause significant morbidity and mortality worldwide. There is evidence from rare, Mendelian cancer predisposition syndromes that CRC and EC can have a common aetiology. Specifically, germline mutations in mismatch repair (MMR) genes MLH1, MSH2, MSH6 and PMS21 and in DNA polymerases POLD1 and POLE2 predispose to a high incidence (lifetime risk 30–71%2,3,4,5) of both CRC and EC. The MMR system maintains genomic stability by correcting mismatched nucleotide pairs that arise during DNA replication and MMR mutations cause a microsatellite instability (MSI+) phenotype in CRCs and ECs6. Bi-allelic MLH1 promoter methylation7,8 and a few somatic mutations in MLH1 and MSH29 are seen in sporadic CRCs and ECs, causing the same MSI+ and hypermutator phenotype. Histologically, MMR-deficient CRCs and ECs are characterised by poor differentiation and the presence of mucinous and signet-cell features and tumour-infiltrating lymphocytes10,11. POLE and POLD1 encode polymerases that synthesise respectively the leading and lagging strand of the DNA replication fork. The exonuclease (proofreading) domains of these polymerases increase replication fidelity by recognising and excising mispaired bases12,13. Germline missense mutations in the exonuclease domains of POLD1 and POLE predispose to both CRC and EC and somatic POLE mutations occur in sporadic CRCs and ECs2,14,15,16. Polymerase exonuclease domain mutations (EDMs) do not cause MSI, but lead to an ultramutator phenotype, with over one million base substitutions in some cancers.
Genome-wide association studies (GWAS) have successfully identified tens of common single nucleotide polymorphisms (SNPs) associated with a modestly increased risk (typically 10–25%) of CRC. In addition, one EC SNP, near HNF1B, has been reported at stringent levels of statistical significance. To date, the lists of CRC and EC SNPs are non-overlapping. Since CRC and EC may share mechanisms of pathogenesis, as evidenced by the high-penetrance germline mutations and the somatic (epi)mutations discussed above, we hypothesised (i) that some CRC SNPs may predispose to EC and vice versa and (ii) that there exist unidentified SNPs that predispose to both CRC and EC. In this study, we tested these hypotheses using 16 different CRC and EC GWAS data sets, totalling 13,265 cancer cases and 40,245 cancer-free or population controls.
Methods
GWAS data sets
Five CRC GWAS data sets genotyped on various Illumina tag-SNP arrays were available, comprising: (i) CORGI (UK1), (ii) Scotland 1, (iii) VICTOR/QUASAR2/BC58, (iv) CFR1 and (v) CFR2/CGEMS (total 5,725 cases and 6,671 controls)17,18,19,20,21. The VQ58, CORGI and Scotland 1 series were genotyped using Illumina Hap300, Hap240S, Hap370, Hap550 or Omni2.5M arrays. BC58 genotyping was performed as part of the WTCCC2 study on Hap1.2M-Duo Custom arrays. The CCFR samples were genotyped using Illumina Hap1M, Hap1M-Duo or Omni-express arrays. CGEMS samples (all controls) were genotyped using Illumina Hap300 and Hap240 or Hap550 arrays. Standard quality -control measures were applied as reported17. Moreover, any duplicate or cryptically related samples were excluded by pairwise identity by descent (IBD) analysis.
EC GWAS comprised: (i) NSECG, (ii) ANECS and (iii) SEARCH (total 2,212 cases and 6,725 controls)22. All samples were of European ancestry with the majority of samples from the UK and others from USA and Australia. Standard quality control measures were performed for each GWAS, as described in the referenced publications and details about each dataset are shown in Table 1. Some of the control datasets, including the Wellcome Trust Case Control Consortium 2 (WTCCC2)23, have previously been used in both CRC and EC GWAS. We ensured that such controls were assigned proportionately to case data sets and were not used more than once (Table 1).
Principal component analysis (PCA) was conducted for all samples together, to ensure that all individuals were of European ancestry and we excluded all individuals who clustered outside the main centroid in pairwise plots of the first 4 PCs. The adequacy of case-control matching and possibility of differential genotyping of cases and controls was assessed using Q-Q plots of test statistics. λGC values for the CORGI, Scotland1, VQ58, CCFR1 and CCFR2 studies were 1.02, 1.01, 1.01, 1.02 and 1.03 respectively and those for NSECG, ANECS and SEARCH were 1.02, 1.02 and 1.00 respectively.
EC targeted genotyping data sets
A further 4,330 EC cases and 26,849 female controls were genotyped as part of the Endometrial Cancer Association Consortium (ECAC), with samples from seven countries: UK, USA, Belgium, Germany, Norway, Sweden and Australia. The controls were selected from healthy females participating in the Breast Cancer Association Consortium (BCAC) and Ovarian Cancer Association Consortium (OCAC) part of the iCOGS project and matched and analysed with cases in eight groups by geographical location (see Table 1). These samples were genotyped using a custom Illumina Infinium iSelect array with 211,155 SNPs designed by the COGS (Collaborative Oncological Gene-environment Study) initiative24,25,26,27. The SNPs on this array were chosen based on regions of interest from previous breast, prostate, ovarian and endometrial cancer studies, rather than on genome-wide coverage. We did not impute genotypes from the COGS studies, but included directly-genotyped SNPs in the discovery meta-analysis. These SNPs were not used for locus fine mapping.
Association study and meta-analysis
Whole-genome imputation using two reference panels (1000 Genomes 2012 release28 and 196 high-coverage whole genome-sequenced UK individuals) was performed with IMPUTE229, yielding up to 6 million SNPs either typed or imputed with high quality (info score >0.9). Case-control analysis for each GWAS data set was performed using frequentist tests with a logistic regression model using SNPTEST (v2.4)30. There was no evidence of systematic over-dispersion of the test statistic for any of the 16 studies (lambdaGC = 1.01–1.04 based on weakly correlated SNPs, r2 < 0.2). Fixed-effects, inverse variance weighted meta-analysis was conducted for the 6 million well-imputed SNPs in the eight CRC and EC GWAS (8,935 cases, 13,396 controls) across the genome using GWAMA (v2.1)31. For the ~200,00 SNPs genotyped on the COGS array, the additional 4,330 EC cases and 26,849 controls from ECAC were included in a meta-analysis of 16 studies yielding a total of 13,265 cases and 40,245 controls for these loci. SNPs with globally significant CRC/EC associations (Pmeta < 5 × 10−8) were identified and the regions examined using standard fine mapping and annotation methods.
Previously reported CRC and EC SNPs
The effects of 25 previously published tag-SNPs that have been formally associated with CRC risk in GWAS were investigated in EC (Table 2). We additionally assessed two SNPs (near TERT32 and MTHFR33,34) with convincing CRC associations from focussed studies. We estimated that our EC sample set provided 72% power to detect the effect of a typical CRC SNP (allele frequency = 0.25, per allele odds ratio = 1.1) at P = 0.05 and 23% power to detect a similar allele at P = 0.001, corresponding to a false discovery rate of q = 0.05 in our sample. Two EC SNPs from GWAS22 were similarly investigated in CRC. All of these SNPs were either discovered or replicated in European populations and were genotyped directly or had near-perfect proxies on the Illumina GWAS arrays used; 13 of the SNPs were also present on the iCOGS arrays. Three EC SNPs in the TERT-CLPTM1L region35 were not included in this analysis, owing to poor tagging on the GWAS arrays and hence sub-optimal imputation.
Genome-wide enrichment of susceptibility SNPs between CRC and EC
Beyond the 29 previously published associations, we investigated the presence of genome-wide enrichment for CRC and EC. After removing previous associations, we pruned the set of 6 million typed or well-imputed SNPs (r2 < 0.1) to 246,896. Using several P value thresholds, we determined whether there was a tendency for the same SNPs to co-occur in the lists of putative CRC and EC SNPs, irrespective of direction of effect.
Results
We initially investigated the 29 previously-identified CRC and EC polymorphisms (Table 2). One SNP, rs2736100, originally reported in CRC32, was significantly associated with EC risk (OR: 0.93, 95% confidence interval (95% CI): 0.89-0.96, P = 0.000167) after correcting for multiple testing (P<0.001). The risk allele for CRC [A] was protective in EC. rs2736100 lies in the intronic region of the telomerase reverse transcriptase TERT. It or highly correlated SNPs have previously been associated with the risk of multiple different cancer types and we ourselves have previously found evidence that these TERT SNPs are associated with EC risk35. Two other CRC SNPs (rs6691170 and rs10936599) were nominally associated with EC risk (P < 0.05). Interestingly, the latter of these lies close to the telomerase RNA component TERC locus; it is a multi-cancer risk SNP36,37,38 and has been associated with longer telomeres. Overall, 15 of the 29 SNPs showed the same direction of effect in both cancer types (that is, same nominal risk allele, irrespective of effect size) and this evidently was not a significant deviation from randomness (P = 1, binomial sign test).
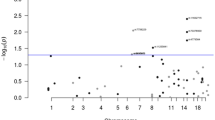
Meta-analysis of all CRC and EC data sets revealed a single genome-wide significant SNP, rs3184504, on chromosome 12q24 (OR: 1.10, 95% CI 1.07–1.13, Pmeta: 7.23 × 10−9, heterogeneity I2 = 0; Fig. 1, Supplementary Table 1). This SNP is a missense variant (p.Trp262Arg) in exon 4 of SH2B3. It has not previously been associated with either CRC or EC. The major [C] allele was consistently the risk allele in all datasets, including those analysed using the iCOGS array, on which the SNP was included due to promising, but unproven, associations below genome-wide significance in previous breast cancer and EC GWAS. An additional 3 SNPs (Fig. 2) in strong pairwise linkage disequilibrium (LD) with rs3184504 (r2 > 0.9) showed strong evidence of CRC-EC association (Pfine mapping < 10−5). These 4 SNPs lie in a 68kb region, that includes the genes SH2B3 and ATXN2 and their functional annotation is shown in Supplementary Table 2. None of the 4 SNPs was associated with the mRNA level of SH2B3, ATXN2 or other nearby genes in public eQTL databases (details not shown).
Forest plot showing association between cancer risk and rs3184504 genotype in each data set.
Studies are shown in order of EC GWAS, EC iCOGS and CRC GWAS (Table 1). Black squares represent the point estimate of the odds ratio and have areas proportional to study size. Lines represent 95% confidence intervals. The diamond shows the summary statistic. The overall heterogeneity statistic is shown. There is also no evidence of heterogeneity between the pooled CRC and pooled EC studies (details not shown).
Regional association plot for region around rs3184504.
Plots are produced in LocusZoom and show the most strongly associated SNP, rs3184504 (purple diamond). rs7137828, intron of ATXN2, is the SNP with the second lowest P value. The primary aim of this analysis is to compare association signals among SNPs in the region. Therefore, the data are derived from a meta-analysis of genotyped or high-quality imputed SNPs in the GWAS data sets and because imputation quality was more variable in iCOGS than in the GWAS data, the iCOGS samples are not included.
There are SNPs that have previously been independently identified in GWAS of different phenotypes where the risk allele for one phenotype is the protective allele for another39,40. In order to search for SNPs for which the same allele has differing directions of effect in CRC and EC, we conducted a fixed-effect meta-analysis with the odds ratios of all the CRC SNPs GWAS inverted (Supplementary Table 3). In this analysis, we discovered rs12970291 on chromosome 18q22, where the major G allele is protective in CRC (OR:0.78, 95%CI:0.69-0.90, 3.42 × 10−4) and confers risk in EC (OR:1.24, 95%CI: 1.11–1.38, p:1.11 × 10−4). In meta-analysis, the rs12970291 association reached genome-wide significance (OR:1.26, 95%CI:1.16–1.38, Pmeta:4.82 × 10−8; Fig. 3). Fine mapping analysis identified a large number of SNPs in high pairwise LD with rs12970291 (r2 > 0.85), in a 70 kb region that includes the gene TSHZ1, which is ~15 kb proximal to rs12970291 (Fig. 4). Seventeen SNPs had a stronger disease association than rs12970291 in fine mapping, with the lowest P value at rs35185115 (Pfine mapping = 1.08 × 10−6). Fine mapping of CRC and EC GWAS separately (Supplementary Figure 1) showed an association peak occurring in the same LD block between 10.5–51.8 kb downstream of TSHZ1, while an additional suggestive association signal near rs17263435 (PEC = 4.35 × 10−5) was not present in CRC (PCRC = 0.1). Several SNPs in the region have potential functional importance (Supplementary Table 4) and of particular note is the missense SNP rs3390274 (p.Ala468Thr) in the last exon of TSHZ1. SNPs with a pairwise LD of >0.4 with rs12970291 in the region were not significantly associated with mRNA level of TSHZ1 or other nearby genes in public eQTL databases (details not shown).
Forest plot showing association between cancer risk and rs12970291 genotype in each data set.
Legend is as for Fig. 1.
Regional association plot for region around rs12970291.
Legend is as for Fig. 2, except as follows. The most strongly associated SNP from the full discovery meta-analysis (rs12970291, purple diamond) is not the most strongly associated in the GWAS data sets. The most strongly associated SNP, rs35185115, lies about 30kb downstream of TSHZ1, but this SNP imputed poorly in iCOGS and was therefore assessed in fewer samples in the discovery meta-analysis than rs12970291, which was directly genotyped in iCOGS.
Finally, we performed genome-wide enrichment analysis for nearly 250,000 independent SNPs (r2 < 0.1) below genome-wide significance levels to investigate whether there was a set of cryptic shared CRC and EC risk loci (Supplementary Table 5). Using P value thresholds of 10−3, 10−2 and 0.05, we found no evidence of a significant sharing of CRC and EC SNPs using this method.
Discussion
Using a combined CRC and EC GWAS meta-analysis, we have identified a region on chromosome 12q24.1 spanning two genes, SH2B3 and ATXN2, which contains a SNP that is formally associated at GWAS thresholds of significance with cancer risk. Of the variants in this region, rs3184504 is of particular interest, because it is a non-synonymous change (TGG → CGG; p.Trp262Arg) in the pleckstrin homology domain of SH2B3, which is a priori a much stronger candidate than the spinocerebellar ataxia gene ATXN2. SH2B3 is a member of the SH2B adaptor family of proteins and is involved in a range of signalling activities by growth factor and cytokine receptors. It is a key negative regulator in cytokine signalling in haematopoiesis and is expressed at a high level in the bone marrow and white blood cells, but at a low level in the normal bowel and endometrium (EMBL-EBI expression atlas). Comparative genomics shows that the rs3184504 risk allele (C, Arg residue) is conserved in all primates and some vertebrates (Supplementary Figure 1) and has a much lower allele frequency (~0.5) in Europeans than in African, Asian and admixed American populations (~1.0). Amino acids Trp (tryptophan) and Arg (arginine) present in the two forms of the polymorphic SH2B3 protein possess a hydrophobic (uncharged) and positively charged side chain respectively. Different programs that predict the effect of this variation on protein function vary in their assessment (Grantham score = 121 (range 0–215)41, Polyphen2 = 0.1242, SIFT = 1.043, CADD score PHRED-scaled = 5.53244); overall, the possibility remains that the amino acid change has a modest or greater effect on protein function. The NHGRI GWAS Catalog shows that SNPs in the SH2B3/ATNX2 region including rs3184504 and rs653178 have been previously associated with immune-mediated conditions: coeliac disease45, rheumatoid arthritis43, type 1 diabetes46, autoimmune hepatitis47 and also cardiovascular traits including coronary artery disease48 and blood pressure49. The genotype at rs653178 has been linked to levels of SH2B3 mRNA expression in peripheral blood cell eQTL analysis (p = 9.24 × 10−12), although this association is not present in public eQTL data sets. Interestingly, rs3184504 T is generally the risk allele in autoimmune traits, suggesting opposing effects of the functional polymorphism on cancer and other traits, perhaps via shared effects on immune activation. A similar phenomenon has been found for the HNF1B SNP rs4430796 which has opposing effects on EC and type 2 diabetes risk50.
The TERT-CLPTM1L locus has been identified in multiple cancer susceptibility GWAS51,52,53,54,55,56,57,58 and it is of interest that the CRC SNP rs2736100 also shows signs of significance in EC in our analysis (OR:1.08, 95%CI:1.04-1.12, P = 1.67 × 10−4). In parallel with this study and using overlapping data sets, we have recently performed a detailed analysis of the TERT-CLPTM1L locus in EC which provided evidence that rs7705526 is associated with EC risk (Passoc = 7.7 × 10−5), albeit at locus-specific rather than genome-wide significance thresholds35. rs7705526 is moderately correlated with rs2736100 (r2 ~ 0.5) but is poorly tagged in most Illumina GWAS arrays. Supplementary Figure 2 shows the complex LD structure between these two SNPs and 4 other SNPs previously associated with CRC and EC at varying levels of significance (P = 8.4 × 10−3 to 4.9 × 10−6) at this locus.
The rs2736100 A allele is the risk allele for CRC and testicular germ cell tumour, while the same allele is protective for EC, glioma and lung cancer, suggesting that this variant has its effects in a tissue-specific manner. Interestingly, we have found evidence in this study for a SNP (rs12970291, chromosome 18q22) that has opposing allelic effects on CRC and EC risk. The top candidate gene in this region is TSHZ1 which encodes zinc finger homeodomain factor teashirt zinc finger family member 1, a protein involved in skin, skeletal, brain and gut development59 that is functionally related to the CRC gene BMP460. One of several candidate SNPs near and within TSHZ1 is the uncommon missense variant rs33930274 (p.Ala468Thr) in the last exon of TSHZ1, although the predicted functional consequences of this change are inconsistent (Grantham score = 58, SIFT = 0.0, Polyphen2 = 0.0, CADD score PHRED-scaled: 0.001).
Apart from the SH2B3 and TERT SNPs, only two of 27 previously-reported CRC SNPs, including one near TERC, showed any good evidence of association with EC and neither of the known EC SNPs was associated with CRC risk. Otherwise, there was no convincing evidence for a shared EC and CRC predisposition based on common polymorphisms, although it will be important to keep repeating multi-cancer GWAS as more risk SNPs are identified and sub-set analyses – for example of MSI+ ECs and CRCs – might also be fruitful. It remains a little puzzling that, like breast and ovarian cancer, CRC and EC share high-penetrance risk alleles, yet relatively few common risk alleles of modest effect.
Additional Information
How to cite this article: Cheng, T. H.T. et al. Meta-analysis of genome-wide association studies identifies common susceptibility polymorphisms for colorectal and endometrial cancer near SH2B3 and TSHZ1. Sci. Rep. 5, 17369; doi: 10.1038/srep17369 (2015).
References
Lynch, H. T. et al. Review of the Lynch syndrome: history, molecular genetics, screening, differential diagnosis and medicolegal ramifications. Clin Genet 76, 1–18, 10.1111/j.1399-0004.2009.01230.x (2009).
Palles, C. et al. Germline mutations affecting the proofreading domains of POLE and POLD1 predispose to colorectal adenomas and carcinomas. Nat Genet 45, 136–144, 10.1038/ng.2503 (2013).
Dunlop, M. G. et al. Cancer risk associated with germline DNA mismatch repair gene mutations. Hum Mol Genet 6, 105–110 (1997).
Hendriks, Y. M. et al. Cancer risk in hereditary nonpolyposis colorectal cancer due to MSH6 mutations: impact on counseling and surveillance. Gastroenterology 127, 17–25 (2004).
Senter, L. et al. The clinical phenotype of Lynch syndrome due to germ-line PMS2 mutations. Gastroenterology 135, 419–428, 10.1053/j.gastro.2008.04.026 (2008).
Jass, J. R. et al. Diagnostic use of microsatellite instability in hereditary non-polyposis colorectal cancer. Lancet 346, 1200–1201 (1995).
Herman, J. G. et al. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc Natl Acad Sci USA 95, 6870–6875 (1998).
Simpkins, S. B. et al. MLH1 promoter methylation and gene silencing is the primary cause of microsatellite instability in sporadic endometrial cancers. Hum Mol Genet 8, 661–666 (1999).
Haraldsdottir, S. et al. Colon and endometrial cancers with mismatch repair deficiency can arise from somatic, rather than germline, mutations. Gastroenterology 147, 1308–1316 e1301, 10.1053/j.gastro.2014.08.041 (2014).
Hampel, H. et al. Screening for Lynch syndrome (hereditary nonpolyposis colorectal cancer) among endometrial cancer patients. Cancer Res 66, 7810–7817, 10.1158/0008-5472.can-06-1114 (2006).
Smyrk, T. C., Watson, P., Kaul, K. & Lynch, H. T. Tumor-infiltrating lymphocytes are a marker for microsatellite instability in colorectal carcinoma. Cancer 91, 2417–2422 (2001).
Goldsby, R. E. et al. Defective DNA polymerase-delta proofreading causes cancer susceptibility in mice. Nat Med 7, 638–639, 10.1038/88963 (2001).
Simon, M., Giot, L. & Faye, G. The 3′ to 5′ exonuclease activity located in the DNA polymerase delta subunit of Saccharomyces cerevisiae is required for accurate replication. EMBO J 10, 2165–2170 (1991).
Network, T. C. G. A. Comprehensive molecular characterization of human colon and rectal cancer. Nature 487, 330–337, 10.1038/nature11252 (2012).
Cerami, E. et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2, 401–404, 10.1158/2159-8290.cd-12-0095 (2012).
Church, D. N. et al. DNA polymerase epsilon and delta exonuclease domain mutations in endometrial cancer. Hum Mol Genet 22, 2820–2828, 10.1093/hmg/ddt131 (2013).
Dunlop, M. G. et al. Common variation near CDKN1A, POLD3 and SHROOM2 influences colorectal cancer risk. Nat Genet 44, 770–776, 10.1038/ng.2293 (2012).
Houlston, R. S. et al. Meta-analysis of three genome-wide association studies identifies susceptibility loci for colorectal cancer at 1q41, 3q26.2, 12q13.13 and 20q13.33. Nat Genet 42, 973–977, 10.1038/ng.670 (2010).
Newcomb, P. A. et al. Colon Cancer Family Registry: an international resource for studies of the genetic epidemiology of colon cancer. Cancer Epidemiol Biomarkers Prev 16, 2331–2343, 10.1158/1055-9965.epi-07-0648 (2007).
Tomlinson, I. P. et al. A genome-wide association study identifies colorectal cancer susceptibility loci on chromosomes 10p14 and 8q23.3. Nat Genet 40, 623–630, 10.1038/ng.111 (2008).
Yeager, M. et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet 39, 645–649, 10.1038/ng2022 (2007).
Spurdle, A. B. et al. Genome-wide association study identifies a common variant associated with risk of endometrial cancer. Nat Genet 43, 451–454, 10.1038/ng.812 (2011).
2, T. W. T. C.-C. C. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447, 661–678, 10.1038/nature05911 (2007).
Eeles, R. A. et al. Identification of 23 new prostate cancer susceptibility loci using the iCOGS custom genotyping array. Nat Genet 45, 385–391, 391e381-382, 10.1038/ng.2560 (2013).
Michailidou, K. et al. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat Genet 45, 353–361, 361e351-352, 10.1038/ng.2563 (2013).
Pharoah, P. D. et al. GWAS meta-analysis and replication identifies three new susceptibility loci for ovarian cancer. Nat Genet 45, 362–370, 370e361-362, 10.1038/ng.2564 (2013).
Sakoda, L. C., Jorgenson, E. & Witte, J. S. Turning of COGS moves forward findings for hormonally mediated cancers. Nat Genet 45, 345–348, 10.1038/ng.2587 (2013).
Abecasis, G. R. et al. An integrated map of genetic variation from 1,092 human genomes. Nature 491, 56–65, 10.1038/nature11632 (2012).
Howie, B., Marchini, J. & Stephens, M. Genotype imputation with thousands of genomes. G3 (Bethesda) 1, 457–470, 10.1534/g3.111.001198 (2011).
Marchini, J., Howie, B., Myers, S., McVean, G. & Donnelly, P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet 39, 906–913, 10.1038/ng2088 (2007).
Magi, R. & Morris, A. P. GWAMA: software for genome-wide association meta-analysis. BMC Bioinformatics 11, 288, 10.1186/1471-2105-11-288 (2010).
Kinnersley, B. et al. The TERT variant rs2736100 is associated with colorectal cancer risk. Br J Cancer 107, 1001–1008, 10.1038/bjc.2012.329 (2012).
Hubner, R. A. & Houlston, R. S. MTHFR C677T and colorectal cancer risk: A meta-analysis of 25 populations. Int J Cancer 120, 1027–1035, 10.1002/ijc.22440 (2007).
Hubner, R. A., Lubbe, S., Chandler, I. & Houlston, R. S. MTHFR C677T has differential influence on risk of MSI and MSS colorectal cancer. Hum Mol Genet 16, 1072–1077, 10.1093/hmg/ddm055 (2007).
Carvajal-Carmona, L. G. et al. Candidate locus analysis of the TERT-CLPTM1L cancer risk region on chromosome 5p15 identifies multiple independent variants associated with endometrial cancer risk. Hum Genet 134, 231–245, 10.1007/s00439-014-1515-4 (2015).
Chubb, D. et al. Common variation at 3q26.2, 6p21.33, 17p11.2 and 22q13.1 influences multiple myeloma risk. Nat Genet 45, 1221–1225, 10.1038/ng.2733 (2013).
Codd, V. et al. Identification of seven loci affecting mean telomere length and their association with disease. Nat Genet 45, 422–427, 427e421-422, 10.1038/ng.2528 (2013).
Figueroa, J. D. et al. Genome-wide association study identifies multiple loci associated with bladder cancer risk. Hum Mol Genet 23, 1387–1398, 10.1093/hmg/ddt519 (2014).
Chung, D., Yang, C., Li, C., Gelernter, J. & Zhao, H. GPA: a statistical approach to prioritizing GWAS results by integrating pleiotropy and annotation. PLoS Genet 10, e1004787, 10.1371/journal.pgen.1004787 (2014).
Sivakumaran, S. et al. Abundant pleiotropy in human complex diseases and traits. Am J Hum Genet 89, 607–618, 10.1016/j.ajhg.2011.10.004 (2011).
Grantham, R. Amino acid difference formula to help explain protein evolution. Science 185, 862–864 (1974).
Adzhubei, I. A. et al. A method and server for predicting damaging missense mutations. Nat Methods 7, 248–249, 10.1038/nmeth0410-248 (2010).
Stahl, E. A. et al. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat Genet 42, 508–514, 10.1038/ng.582 (2010).
Kircher, M. et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet 46, 310–315, 10.1038/ng.2892 (2014).
Hunt, K. A. et al. Newly identified genetic risk variants for celiac disease related to the immune response. Nat Genet 40, 395–402, 10.1038/ng.102 (2008).
Barrett, J. C. et al. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet 41, 703–707, 10.1038/ng.381 (2009).
de Boer, Y. S. et al. Genome-wide association study identifies variants associated with autoimmune hepatitis type 1. Gastroenterology 147, 443–452 e445, 10.1053/j.gastro.2014.04.022 (2014).
Dichgans, M. et al. Shared genetic susceptibility to ischemic stroke and coronary artery disease: a genome-wide analysis of common variants. Stroke 45, 24–36, 10.1161/strokeaha.113.002707 (2014).
Wain, L. V. et al. Genome-wide association study identifies six new loci influencing pulse pressure and mean arterial pressure. Nat Genet 43, 1005–1011, 10.1038/ng.922 (2011).
Painter, J. N. et al. Fine-mapping of the HNF1B multicancer locus identifies candidate variants that mediate endometrial cancer risk. Hum Mol Genet 24, 1478–1492, 10.1093/hmg/ddu552 (2015).
Haiman, C. A. et al. A common variant at the TERT-CLPTM1L locus is associated with estrogen receptor-negative breast cancer. Nat Genet 43, 1210–1214, 10.1038/ng.985 (2011).
Kote-Jarai, Z. et al. Seven prostate cancer susceptibility loci identified by a multi-stage genome-wide association study. Nat Genet 43, 785–791, 10.1038/ng.882 (2011).
McKay, J. D. et al. Lung cancer susceptibility locus at 5p15.33. Nat Genet 40, 1404–1406, 10.1038/ng.254 (2008).
Petersen, G. M. et al. A genome-wide association study identifies pancreatic cancer susceptibility loci on chromosomes 13q22.1, 1q32.1 and 5p15.33. Nat Genet 42, 224–228, 10.1038/ng.522 (2010).
Shete, S. et al. Genome-wide association study identifies five susceptibility loci for glioma. Nat Genet 41, 899–904, 10.1038/ng.407 (2009).
Stacey, S. N. et al. New common variants affecting susceptibility to basal cell carcinoma. Nat Genet 41, 909–914, 10.1038/ng.412 (2009).
Turnbull, C. et al. Variants near DMRT1, TERT and ATF7IP are associated with testicular germ cell cancer. Nat Genet 42, 604–607, 10.1038/ng.607 (2010).
Wang, Y. et al. Common 5p15.33 and 6p21.33 variants influence lung cancer risk. Nat Genet 40, 1407–1409, 10.1038/ng.273 (2008).
Manfroid, I., Caubit, X., Kerridge, S. & Fasano, L. Three putative murine Teashirt orthologues specify trunk structures in Drosophila in the same way as the Drosophila teashirt gene. Development 131, 1065–1073, 10.1242/dev.00977 (2004).
Tomlinson, I. P. et al. Multiple common susceptibility variants near BMP pathway loci GREM1, BMP4 and BMP2 explain part of the missing heritability of colorectal cancer. PLoS Genet 7, e1002105, 10.1371/journal.pgen.1002105 (2011).
Acknowledgements
We are grateful for funding to the Oxford NIHR Comprehensive Biomedical Research Centre. Core funding to the Wellcome Trust Centre for Human Genetics was provided by the Wellcome Trust (090532/Z/09/Z). The European colorectal cancer data were supported by COST Action BM1206. We thank Breakthrough Breast Cancer and the Institute of Cancer Research for support and funding of the Breakthrough Generations Study and the study participants, study staff and the doctors, nurses and other health care providers and health information sources who have contributed to the study. We acknowledge NHS funding to the Royal Marsden/ICR NIHR Biomedical Research Centre. The Colon CFR was supported by grant UM1 CA167551 from the National Cancer Institute and through cooperative agreements with the following CCFR centres: Australasian Colorectal Cancer Family Registry, Mayo Clinic Cooperative Family Registry for Colon Cancer Studies, Ontario Familial Colorectal Cancer Registry, Seattle Colorectal Cancer Family Registry, USC Consortium Colorectal Cancer Family Registry. The Colon CFR GWAS was supported by funding from the National Cancer Institute, National Institutes of Health (U01 CA122839 and R01 CA143237 to Graham Casey). The content of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating centres in the Colon Cancer Family Registry (CCFR), nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government or the CCFR. Jeremy P. Cheadle was funded by the Bobby Moore Fund from Cancer Research UK, Tenovus, the Kidani Trust, Cancer Research Wales and the National Institute for Social Care and Health Research Cancer Genetics Biomedical Research Unit. Nada A. Al-Tassan and Brian F. Meyer were funded and supported by KFSHRC. Luis Carvajal-Carmona receives founding from The V Foundation for Cancer Research.The authors would also like to acknowledge The Australian Ovarian Cancer Study (AOCS) group. A full list of the participants and their affiliations appears in the suppplementary information file that is appended to this article.
Author information
Authors and Affiliations
Contributions
Recruited study participants, obtained samples and provided data. T.H.T.C., M.G., L.M., C.P., A.J., D.D.B., A.K.W., J.H., M.J., N.M.L., P.A.N., S.G., D.C., F.S., G.C., G.G.G., P.P., J.P., A.n.C., A.n.S., F.C., J.M.C., D.L., P.F., B.B., H.e.B., H.i.B., J.C.C., H.B.S., V.K., H.D., J.L., T.L., A.L., P.e.r.H., M.E.P., M.o.S., A.C.a., S.C.B., A.R.M., S.a.A., M.R.T., W.M.D., J.o.D., G.O., T.P., E.H., J.A., K.A., R.J.S., M.M.c.E., E.L.G., S.C.D., S.J.W., B.L.F., H.M.J.W., J.T., T.S.N., E.T., M.M., I.R., P.e.t.H., T.D., F.A., S.S., A.H., M.W.B., A.E., K.C., A.M., M.K.B., K.M., J.P.T., Q.W., S.h.A., C.S.H., M.i.S., D.A., J.e.D., A.O.C.S., N.A.l.T., R.e.H., B.F.M., N.W., F.J.H., B.K., S.M.F., M.T., A.T., H.C., R.W.H., S.H., L.C.C., J.P.C., D.E., M.D., R.i.H., A.m.S. and I.T. Performed additional molecular analyses: T.C., J.P.a., T.O.’M., A.J., A.D., M.S. and L.C.-C. Analysed data: T.C., D.T., C.P. and I.T. Planned and supervised study: D.E., M.D.u.n., R.H.o., A.S.p. and I.T. Obtained funding: M.E.P., M.S., A.C.a., A.C., S.C.-B., A.R.M., S.A., M.T., P.P., P.H., H.S., H.W., J.T., T.N., D.L., P.F., E.T., M.M., I.R., M.D.u.r., T.D., F.A., L.C., S.S., A.H., M.B., S.R., K.C., A.M., D.E., A.S.p. and I.T. Wrote manuscript: T.C., D.T., R.H.o., A.S.p. and I.T.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Cheng, T., Thompson, D., Painter, J. et al. Meta-analysis of genome-wide association studies identifies common susceptibility polymorphisms for colorectal and endometrial cancer near SH2B3 and TSHZ1. Sci Rep 5, 17369 (2015). https://doi.org/10.1038/srep17369
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep17369
This article is cited by
-
Linkage analysis revealed risk loci on 6p21 and 18p11.2-q11.2 in familial colon and rectal cancer, respectively
European Journal of Human Genetics (2019)
-
Linear isoforms of the long noncoding RNA CDKN2B-AS1 regulate the c-myc-enhancer binding factor RBMS1
European Journal of Human Genetics (2019)
-
Endometrial cancer gene panels: clinical diagnostic vs research germline DNA testing
Modern Pathology (2017)
-
Five endometrial cancer risk loci identified through genome-wide association analysis
Nature Genetics (2016)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.