Abstract
EPH kinases and their ligands, ephrins (EFNs), have vital and diverse biological functions. We recently reported that Efnb3 gene deletion results in hypertension in female but not male mice. These data suggest that EFNB3 regulates blood pressure in a sex- and sex hormone-dependent way. In the present study, we conducted a human genetic study to assess the association of EFNB3 single nucleotide polymorphisms with human hypertension risks, using 3,448 patients with type 2 diabetes from the ADVANCE study (Action in Diabetes and Vascular Disease: Peterax and Diamicron MR Controlled Evaluation). We have observed significant association between 2 SNPs in the 3′ untranslated region or within the adjacent region just 3′ of the EFNB3 gene with hypertension, corroborating our findings from the mouse model. Thus, our investigation has shown that EFNB3 is a hypertension risk gene in certain individuals.
Similar content being viewed by others
Introduction
EPH erythropoietin-producing human hepatocellular carcinoma receptor kinases are the largest family of receptor tyrosine kinases. They are divided into A and B subfamilies according to sequence homology1. Ephrins (EFNs), which are also cell surface molecules, are ligands of EPHs. EFNs are classified into A and B subfamilies. Members of the A subfamily attach to the cell surface through glycosylphosphatidylinositol anchoring, whereas members of the B subfamily attach through transmembrane tails2,3,4. Interactions among EPHs and EFNs are promiscuous but, in general, EPH A members interface preferentially with EFN A family members, and EPH B members with EFN B family members2,3,4. Such redundancy suggests that these kinases are crucial in various biological contexts. EFNs can stimulate EPH receptors, and this is called forward signaling. Interestingly, EPHs are also capable of stimulating EFNs which then transmit signaling reversely into cells, a phenomenon known as reverse signaling.
EPHs and EFNs are expressed in many tissues and organs. They play important roles in the central nervous system2,4, immune system5,6,7,8,9,10,11,12,13,14, digestive system15, bone metabolism16,17, angiogenesis18 and other processes19,20,21,22,23,24,25.
We recently reported that members in the EPH/EFN families, i.e., EPHB6, EFNB1, EFNB3, EFNB2 and EPHB4 are crucial in blood pressure (BP) regulation in mouse models26,27,28,29,30,31. While the default function of the first 3 is to reduce BP, the default function of the last 2 is opposite, i.e., to increase BP: gene deletion of Ephb6, Efnb1 and Efnb3 results in increased BP, but the deletion of Efnb2 and Ephb4, decreased BP. These genes are expressed in vascular smooth muscle cells (VSMCs), providing a molecular framework for their function in these cells. Through smooth muscle-specific gene deletion and in vitro mechanistic dissection, we have proved that VSMCs are the major targets through which EPHs/EFNs exert their effect on BP modulation26,27,28,29,30,31. The BP phenotype in the knock out (KO) mice is often sex- and sex hormone-level dependent. For example, hypertension in EPHB6 KO is only observed in castrated mice26; hypertension after Efnb3 KO is present in females but not males; the BP phenotype in Efnb3 KO mice is reversed after gonadectomy: castrated KO males become hypertensive, while ovariectomized KO females, normotensive29; on the other hand, male but not female Efnb2 and Ephb4 KO mice are hypotensive27,30.
The findings from the above-mentioned mouse studies prompted us to investigate the relevance of EPHB and EFNB BP-regulating function to human hypertension. Indeed, we have previously revealed that 5 single nucleotide polymorphisms (SNPs) in the EFNB2 gene are associated with hypertension in male patients in the ADVANCE (Action in Diabetes and Vascular Disease: Peterax and Diamicron MR Controlled Evaluation) study30, a clinical trial in type 2 diabetes (T2D)32. Also, a SNP in the GRIP1 gene, whose product is in the EFNB3 signalling pathway, is approaching the Boferroni-corrected p-value in International Blood Pressure Consortium cohorts31.
As a continued effort to establish the relevance of our mouse findings to human hypertension, we queried the association of SNPs in the EFNB3 gene to hypertension in ADVANCE. We restricted our query specifically to EFNB3, excluding other EPH/EFN genes to a penalty from excessive multiple testing. The results reveal that 2 SNPs in the EFNB3 gene were significant for their association with hypertension in T2D patients.
Methods
Patient population
The patient cohort consisted of 3,448 patients with T2D of European descent and at high risk for macrovascular or microvascular diabetes complications who were recruited in the ADVANCE clinical study, a factorial, multicentre, randomised controlled clinical trial of 11,140 participants recruited from 215 centers in 20 countries32,33. All individuals were T2D subjects 65 years old or older, or they were T2D subjects 55 years old or older who were diagnosed at age of 30 years or older, and had one of the following: a history of major macrovascular disease; a history of major microvascular disease; diagnosis of T2D over 10 years prior to entry into study; presence of another major risk factor for vascular disease including: smoking, dyslipidemia, microalbuminuria.
The 3,448 patients were classified as normotensive or hypertensive. Hypertension was defined as treated for hypertension or having a measurement of systolic pressure >140 mm Hg or diastolic pressure >90 mmHg at entry into the study. As detailed in Table 1, 70.5% and 78.8% of male and female cases, respectively, who were admitted into the study were under anti-hypertensive treatment, This 140/90 mmHg threshold for hypertension diagnosis for the remaining cases and controls was adopted according to the U.S. National Institute of Health guidelines proposed for hypertension diagnosis34, and this threshold is used in most clinical trials including the ADVANCE study32.
There were 2219 males in the cohort of whom 1794 were hypertensive and 425 were normotensive. There were 1229 females of whom 1016 were hypertensive and 213 were normotensive. There were 4.4 times as many hypertensive as normotensive diabetic subjects and 1.8 times as many males as females.
Ethics statement
This human genetic study was carried out in accordance with relevant guidelines of the participating institutions. The research protocol was approved by ADVANCE Study Ethics Committees. Informed consent was obtained from all the subjects used in this study.
Genotyping and data analysis
The 3,448 individuals were genotyped for 724,847 SNPs with Affymetrix Genome-Wide Human SNP Arrays 5.0 or 6.0 at the genomic platform of the CRCHUM. A further 8,117,344 SNP genotypes were then imputed with the IMPUTE2 program35,36. A subset (147,088) of the genotyped SNPs that were in Linkage equilibrium (at r2 ≤ 0.8) was selected to perform a principal component (PC) analysis. using the Eigenstrat software37 in order to test for population stratification in the European wide sample. The first two independent principal components from this analysis (PC1 and PC2) which account for the majority of the covariance among genotypes due to population structure were able to clearly separate samples according to geographic origin within Europe and were subsequently used as covariates in the association analysis to correct for any population stratification effects. Association analysis was performed for 39 SNPs, 2 genotyped and 37 imputed, that fell within the 25,066-bp region of the EFNB3 gene plus additional 10-kb stretches 5′ and 3′ of the gene, between positions 7,599,260 and 7,624,326 (Build 37/hg19) on chromosome 17. Association analysis was performed for males-only, females-only and combined male and female samples of hypertensive patients versus normotensive controls using SNPTEST, the companion program to IMPUTE235,36. A logistic regression model with additive genotype coding and with PC1, PC2, age, body mass index (BMI), and genotyping batch and genotyping array (5.0 Vs. 6.0) as covariates was used to perform the association tests. Sex was also included as a covariate for the combined analyses of males and females. The number of tagging SNPs among the 39 analyzed SNPs located within the gene region was determined for an r2 value > 0.8 and a minor allele frequency cut-off of 0.05 using the LDSELECT program38. Seven tag SNPs were identified across the EFNB3 region for the CEU population (Utah residents with ancestry from northern and western Europe), indicating the presence of 7 independent LD blocks and 7 independent tests across these genotypes. We therefore corrected for 7 independent tests for 3 sets of analysis (males only, females only and combined) resulting in 21 independent tests and thus used a Bonferroni significance threshold of 0.05/21 giving a critical p-value of 0.0024, or a critical −log10 p-value of 2.62. We note that this correction is conservative. The male only and female only samples are subsets of the combined sample, so that the combined sample does not represent a set of completely new independent tests. Nevertheless the patterns of association of SNPs are such that this over-correction is of little concern (see Locus Zoom plots below).
Results
The levels of two cardiovascular disease-related biomarkers, troponin T and brain natriuretic peptide (BNP), in the sera of the hypertension case and control subjects were registered. The levels of these two biomarkers were significantly elevated in both male and female cases, compared to those of the controls (Fig. 1). High serum troponin T levels reflect myocardium damage39, which is often a complication of hypertension, while BNP levels have been reported to be progressively elevated with increasing severity of hypertension, particularly when left ventricular hypertrophy is present40. Thus, the elevation of both these biomarkers in the hypertension cases serves as a validation for the presence of hypertension and/or its complications in these subjects.
The data were converted to natural log (Ln) to achieve normal distribution for Student’s t test. Means + SD are presented. Sample sizes (n) are indicated. *p < 0.05. (A) Serum troponin T levels in male (left panel) and female (right panel) subjects (B) Serum BNP levels in male (left panel) and female (right panel) subjects.
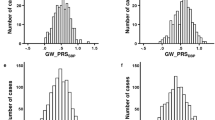
The results of association analysis of the 39 SNPs present within the EFNB3 gene and surrounding regions and hypertension are presented in Table 2 and Fig. 2. Two SNPs with reference SNP cluster ID 3744263 (rs3744263) (genotyped) and rs7141 (imputed) located within the 3′ untranslated region (3′UTR) of the EFNB3 gene were associated with hypertension at p-values below the Bonferroni critical p-value for the combined sample (0.00034 and 0.00026, respectively) and for the males-only sample (0.00148 and 0.00195, respectively). The p-values for the females-only sample for these SNPs were nominally significant but not significant when corrected for multiple testing (0.047 and 0.038, respectively). These two SNPs were in high linkage disequilibrium (LD) (r2 = 0.896, CEU) (Fig. 3), so that most likely they are both serving as proxy for a functional polymorphism, probably located within the 3′UTR of EFNB3.
The symbols (circles and diamonds) and the left-hand vertical axis illustrate –log10 p-values of the SNPs. Blue lines and the right-hand vertical axis represent rates of recombination in the chromosomal regions concerned. Red lines represent the Bonforroni-corrected p-value. The color of the symbols represents r2 values. The horizontal axis indicates the position in the chromosomes. The position of the EFNB3 gene is shown in the bottom of the figures.
Table 3 shows the allele frequencies for the two SNPs, rs7141 (A and G) and rs3744263 (C and T) along with odds ratios and their confidence limits for the combined sample, and for the male-only and the female-only samples. For both SNPs, the major allele (A or C) was more frequent among cases than among controls, in combined sample, males-only sample and females-only sample. The minor allele (G or T) was more frequent among controls in all the samples. Both SNPs had essentially the same major allele odds ratios in all the samples, which were higher among males than females (1.364 versus 1.249 for rs7141, 1.364 versus 1.242 for rs3744263), although the difference was not significant.
Table 4 presents the results of a test of independence of allele frequencies between males and females for cases and controls. Although the associations between hypertension and the 2 SNPs were significant in males and combined samples but not in females as shown above in Table 1, the χ2 tests of independence in both sexes were not significant for either cases or controls, suggesting that allele frequencies are not significantly different between cases and controls in both sexes. The major allele frequencies for both SNPs (A and C) were slightly higher among the male cases, but the difference was not significant. The major allele frequencies among male and female controls were essentially the same. These tests suggest that the association of the 2 SNPs to hypertension is similar between male and female subjects.
Discussion
Our genetic investigation in patients recruited from the ADVANCE study revealed significant association of 2 SNPs within the EFNB3 gene with hypertension in T2D patients. This finding corroborates our results obtained from Efnb3 KO mice based on in vivo, ex vivo and in vitro experiments, which showed that EFNB3 deletion results in hypertension29.
In this human genetic study, we have two inclusion criteria for hypertension cases: 1) patients under active anti-hypertensive treatment; the majority (73.5%) of the cases are in this category; or 2) patients with >140 mmHg for systolic or >90 mmHg for diastolic pressure. The 140/90 mmHg threshold is adopted for this study based on the U.S. National Institute of Health guidelines34, and this threshold is also used in most clinical trials including the ADVANCE study32. There could exist some “white-coat hypertension” when the 140/90 mmHg threshold is used. However, the reported incidence of “white-coat hypertension” is about 15–30% in subjects diagnosed with elevated office blood pressure measurements41,42. In our study, since only 26.5% of the hypertension cases were recruited based on office blood pressure measurement at the time of recruitment, the possible contamination of “white-coat hypertension” in this group will only be 15–30% of 26.5%, i.e., a negligible 4–8% of the total hypertension cases.
If we adopt a higher BP cut-off threshold for the inclusion criteria, such as >160/100 mmHg, to reduce the potential contamination from “white-coat hypertension”, we will not be able to use all those patients recruited based on prior diagnosis and under active anti-hypertensive treatment as cases. Therefore, we would lose 70.5% and 78.8% of our total male and female cases, respectively, who are certainly not experiencing “white-coat hypertension”. This will greatly reduce our statistical power. Based on these considerations, for better statistical power, we adopted the classical 140/90 mmHg threshold for the definition of hypertension for those recruited based on their BP measurements.
No genes in the EPHB/EFNB family were identified as hypertension risk genes in several large-scale GWAS43,44,45,46,47,48,49,50,51, including that of the International Blood Pressure Consortium (IBPC)43. It is possible that the contribution of genes in the EPHB/EFNB family to the BP phenotype is relatively small, and the possible association is rendered undetectable due to heavy multiple testing statistical penalties in GWAS. We then specifically queried the IBPC dataset for association of EFNB3 and a group of related genes with blood pressure. The results have been published recently31. The SNPs in the region of EFNB3 gene are highly under-represented in the ICBP meta-analysis genotype map, and no significant association of available EFNB3 SNPs with blood pressure is revealed. However, we noticed that the p-value of a SNP in GRIP1, a signalling molecule in the EFNB3 signalling pathway, is 0.000389, approaching the Bonferroni-corrected critical p-value of 0.000302. Considering the very conservative nature of Bonferroni correction, it is indicative of the implication of EFNB3 in BP control.
Why can we detect the association of 2 EFNB3 SNPs with hypertension in the ADVANCE study but none in the IBPC dataset? One possible reason is that these 2 studies have different inclusion criteria of samples. The ICPB meta-analysis avoids diabetic patients and attempts to reflect the range of blood pressures of the general population, while the ADVANCE samples are all diabetic patients. Then why can we detect a significant GRIP1 SNP but not EFNB3 SNPs in the IBPC study? GRIP1 is in the signalling pathways of several other EFN/EPH molecules (e.g., EPHB6 and EFNB1), and these molecules are also involved in BP regulation26,28; consequently, GRIP1 might carry higher statistical weight than EFNB3 in BP control, hence the detection of its SNP association with BP.
The 2 SNPs for which significant association was found (Table 2) are all in the 3′ untranslated region or within the adjacent region just 3′ of the EFNB3 gene (Fig. 2), and are in LD (Fig. 3). None of the 2 SNPs were found to alter directly sequences involved in well-defined functions such as messenger stability (UA-rich sequences), binding sites for microRNA species which may induce increased mRNA turnover, or alter the poly (A) tail which is also involved in mRNA stability. These SNPs are in LD and may be serving as proxy for an unidentified polymorphism that is affecting these functions. However we are not able, at this time, to pin-point the exact functional polymorphism.
In mice, the female Efnb3 KO manifested BP increase, but in female T2D patients, the 2 SNPs in the EFNB3 gene were not significantly associated with hypertension. There was no significant difference between males and females, for both cases and controls, with regard to major allele frequency. A lack of detection of the association of 2 SNPs to hypertension in female was possibly the result of insufficient statistical power due to a small female T2D patient sample size, as only 1229 females (1016 hypertensive and 213 normotensive) were genotyped. A second reason could be that the T2D females in the ADVANCE study are all above 55 year old, and likely to have reduced estrogen levels. We demonstrated that EFNB3′s role in BP was estrogen-dependent, as female KO mice after ovariectomy no longer had increased BP29. It is therefore conceivable that with a larger female patient sample size and a younger population, these 2 SNPs might become significantly associated with hypertension in females.
In mice, Efnb3 KO males were normotensive and they only became hypertensive after castration, indicating that testosterone in the absence of EFNB3 is protective against hypertension. However, in male T2D patients, 2 EFNB3 SNPs (rs7141 and rs3744263) were significantly and positively associated with hypertension. There could be several possible explanations. 1) The minimal age of male patients in the ADVANCE study was 55-years old. Plasma total testosterone levels range from 270 to 1,070 ng/dL in “normal” adult males52. Considering 346 ng/dL as a cut-off for the diagnosis of hypogonadism, as recommended by the International Society for the Study of the Aging Male, about 30% of men older than 40 years are hypogonadic53. Further, male patients with the metabolic syndrome are prone to hypogonadism54. Therefore, it is conceivable that more than 30% of the male patients in the ADVANCE study suffer from hypogonadism, reminiscent of the reduced testosterone levels in the castrated Efnb3 KO males. Therefore, for the ADVANCE patients with EFNB3 mutations, they might have lost the protective effect of testosterone. Indeed, some reports indicate that hypogonadism in men, whether evoked by ageing, diabetes, chemical castration or other unidentified factors, correlates with hypertension53,54,55,56,57,58,59. It is possible that some of these hypogonadic hypertension patients suffer from EFNB3 mutations, and the concerted effect of lower testosterone levels and EFNB3 mutation renders them prone to hypertension, as is the case in castrated Efnb3 KO mice. 2) Another contributing factor could be that these T2D males are generally overweight or obese. Adipose tissues are rich in aromatase, a rate-limiting enzyme converting testosterone to estrogen60. Therefore, obese males tend to have relatively higher estrogen levels, which in the absence of EFNB3, could increase BP, as we have demonstrated in our Efnb3 KO mouse model29. Hence, the ADVANCE cohort provides us with a unique situation to reveal the association of the EFNB3 SNPs with hypertension in these male T2D patients. 3) In this genetic study, we demonstrated that 2 SNPs in the EFNB3 genes were associated with hypertension in males, but the association could be either positive or negative. If the association is negative in male humans, it is not incompatible with our findings that male KO mice show no increased BP.
In the discussion of the two previous paragraphs regarding the discrepant results from the Efnb3 KO mouse model versus those from the current ADVANCE study regarding the sex differences, we assumed that the mutation in the EFNB3 gene in this study is a loss-of-function one. However, there is no evidence that this is the case. The responsible mutations in LD with the 2 significant SNPs could actually result in alteration-of-function or gain-of-function. If that is the case, there would be concordance between our findings that EFNB3 deletion in mice causes increased BP in females, while in humans, the 2 EFNB3 SNPs are associated with hypertension in males, as alteration-of-function and gain-of-function mutations could have quite different consequences from loss-of-function mutations. This possibility discussed in this paragraph would override the explanations in the previous two paragraphs.
In summary, in this study, we discovered a significant association of 2 SNPs in the EFNB3 gene with hypertension in a human genetic study. This has opened a new area of investigation into the pathogenesis, diagnosis and personalized therapy of hypertension.
Additional Information
How to cite this article: Tremblay, J. et al. Evidence from single nucleotide polymorphism analyses of ADVANCE study demonstrates EFNB3 as a hypertension risk gene. Sci. Rep. 7, 44114; doi: 10.1038/srep44114 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Unified nomenclature for Eph family receptors and their ligands, the ephrins. Eph Nomenclature Committee. Cell. 90, 403–404 (1997).
Flanagan, J. G. & Vanderhaeghen, P. The ephrins and Eph receptors in neural development. Annu Rev Neurosci. 21, 309–345 (1998).
Pasquale, E. B. Eph-ephrin bidirectional signaling in physiology and disease. Cell. 133, 38–52 (2008).
Wilkinson, D. G. Eph receptors and ephrins: regulators of guidance and assembly. Int Rev Cytol. 196, 177–244 (2000).
Luo, H. et al. Efnb1 and Efnb2 proteins regulate thymocyte development, peripheral T cell differentiation, and antiviral immune responses and are essential for interleukin-6 (IL-6) signaling. J Biol Chem. 286, 41135–41152 (2011).
Luo, H., Wan, X., Wu, Y. & Wu, J. Cross-linking of EphB6 resulting in signal transduction and apoptosis in Jurkat cells. J Immunol. 167, 1362–1370 (2001).
Luo, H. et al. Ephrinb1 and Ephrinb2 are associated with interleukin-7 receptor alpha and retard its internalization from the cell surface. J Biol Chem. 286, 44976–44987 (2011).
Luo, H., Yu, G., Tremblay, J. & Wu, J. EphB6-null mutation results in compromised T cell function. J Clin Invest. 114, 1762–1773 (2004).
Luo, H., Yu, G., Wu, Y. & Wu, J. EphB6 crosslinking results in costimulation of T cells. J Clin Invest. 110, 1141–1150 (2002).
Wu, J. & Luo, H. Recent advances on T-cell regulation by receptor tyrosine kinases. Curr Opin Hematol. 12, 292–297 (2005).
Yu, G., Luo, H., Wu, Y. & Wu, J. Ephrin B2 induces T cell costimulation. J Immunol. 171, 106–114 (2003).
Yu, G., Luo, H., Wu, Y. & Wu, J. Mouse ephrinB3 augments T-cell signaling and responses to T-cell receptor ligation. J Biol Chem. 278, 47209–47216 (2003).
Yu, G., Luo, H., Wu, Y. & Wu, J. EphrinB1 is essential in T-cell-T-cell co-operation during T-cell activation. J Biol Chem. 279, 55531–55539 (2004).
Yu, G., Mao, J., Wu, Y., Luo, H. & Wu, J. Ephrin-B1 is critical in T-cell development. J Biol Chem. 281, 10222–10229 (2006).
Batlle, E. et al. Beta-catenin and TCF mediate cell positioning in the intestinal epithelium by controlling the expression of EphB/ephrinB. Cell. 111, 251–263 (2002).
Davy, A., Bush, J. O. & Soriano, P. Inhibition of gap junction communication at ectopic Eph/ephrin boundaries underlies craniofrontonasal syndrome. PLoS Biol. 4, e315 (2006).
Zhao, C. et al. Bidirectional ephrinB2-EphB4 signaling controls bone homeostasis. Cell Metab. 4, 111–121 (2006).
Wang, H. U., Chen, Z. F. & Anderson, D. J. Molecular distinction and angiogenic interaction between embryonic arteries and veins revealed by ephrin-B2 and its receptor Eph-B4. Cell. 93, 741–753 (1998).
Dravis, C. et al. EphB2 and ephrin-B2 regulate the ionic homeostasis of vestibular endolymph. Hear Res. 223, 93–104 (2007).
Hashimoto, T. et al. Ephrin-B1 localizes at the slit diaphragm of the glomerular podocyte. Kidney Int. 72, 954–964 (2007).
Konstantinova, I. et al. EphA-Ephrin-A-mediated beta cell communication regulates insulin secretion from pancreatic islets. Cell. 129, 359–370 (2007).
Cayuso, J. et al. EphrinB1/EphB3b Coordinate Bidirectional Epithelial-Mesenchymal Interactions Controlling Liver Morphogenesis and Laterality. Dev Cell. 39, 316–328 (2016).
Sheleg, M. et al. Decreased maternal behavior and anxiety in ephrin-A5-/- mice. Genes Brain Behav. 10.1111/gbb.12319 (2016).
Martin-Almedina, S. et al. EPHB4 kinase-inactivating mutations cause autosomal dominant lymphatic-related hydrops fetalis. J Clin Invest. 126, 3080–3088 (2016).
Kwak, H. et al. Sinusoidal ephrin receptor EPHB4 controls hematopoietic progenitor cell mobilization from bone marrow. J Clin Invest. 126, 4554–4568 (2016).
Luo, H. et al. Receptor tyrosine kinase Ephb6 regulates vascular smooth muscle contractility and modulates blood pressure in concert with sex hormones. J Biol Chem. 287, 6819–6829 (2012).
Wang, Y. et al. EPHB4 Protein Expression in Vascular Smooth Muscle Cells Regulates Their Contractility, and EPHB4 Deletion Leads to Hypotension in Mice. J Biol Chem. 290, 14235–14244 (2015).
Wu, Z. et al. Possible role of Efnb1 protein, a ligand of Eph receptor tyrosine kinases, in modulating blood pressure. J Biol Chem. 287, 15557–15569 (2012).
Wang, Y. et al. Estrogen and testosterone in concert with EFNB3 regulate vascular smooth muscle cell contractility and blood pressure. Am J Physiol Heart Circ Physiol. 310, H861–72 (2016).
Wang, Y. et al. Reduced blood pressure after smooth muscle EFNB2 deletion and the potential association of EFNB2 mutation with human hypertension risk. Eur J Hum Genet. 24, 1817–1825 (2016).
Wang, Y. et al. The role GRIP1 and EFNB3 signaling in blood pressure control and vascular smooth muscle cell contractility. Sci Rep. 6, 38976 (2016).
Advance-trial. Available at: www.advance-trial.com/patients
Colagiuri, S. et al. Prevention of vascular outcomes in type 2 diabetes: benefits of intensive blood pressure and blood glucose control with Preterax and Diamicron MR, Issue III. (Wolters Kluwer Health France, 2008).
Sverre, K. et al. Updated National and International Hypertension Guidelines: A Review of Current Recommendations. Drugs. 74, 2033–2051 (2014)
Marchini, J. & Howie, B. Genotype imputation for genome-wide association studies. Nat Rev Genet. 11, 499–511 (2010).
Snptest: https://mathgen.stats.ox.ac.uk/genetics_software/snptest/old/snptest.html (2014).
Price, A. L. et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 38, 904–909 (2006).
LDSelect-Multipopulation: Download and Documentation: http://droog.gs.washington.edu/multiPopTagSelect.html (2014).
Ammann, P., Pfisterer, M., Fehr, T. & Rickli, H. Raised cardiac troponins. BMJ. 328, 1028–1029 (2004).
Gonick, H. C. & Buckalew, V. M. Editorial: Natriuretic Hormones. Front Endocrinol (Lausanne). 6, 108 (2015).
Pickering, T. G. et al. How common is white coat hypertension? JAMA. 259, 225–228 (1988).
O’Brien E. et al. European Society of Hypertension position paper on ambulatory blood pressure monitoring. J Hypertens. 31, 1731–1768 (2013).
International Consortium for Blood Pressure Genome-Wide Association, S. et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 478, 103–109 (2011).
Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 447, 661–678 (2007).
Org, E. et al. Genome-wide scan identifies CDH13 as a novel susceptibility locus contributing to blood pressure determination in two European populations. Hum Mol Genet. 18, 2288–2296 (2009).
Levy, D. et al. Genome-wide association study of blood pressure and hypertension. Nat Genet. 41, 677–687 (2009).
Adeyemo, A. et al. A genome-wide association study of hypertension and blood pressure in African Americans. PLoS Genet. 5, e1000564 (2009).
Hiura, Y. et al. A genome-wide association study of hypertension-related phenotypes in a Japanese population. Circ J. 74, 2353–2359 (2010).
Padmanabhan, S. et al. Genome-wide association study of blood pressure extremes identifies variant near UMOD associated with hypertension. PLoS Genet. 6, e1001177 (2010).
Slavin, T. P., Feng, T., Schnell, A., Zhu, X. & Elston, R. C. Two-marker association tests yield new disease associations for coronary artery disease and hypertension. Hum Genet. 130, 725–733 (2011).
Guo, Y. et al. A genome-wide linkage and association scan reveals novel loci for hypertension and blood pressure traits. PloS one. 7, e31489 (2012).
Frances, T. F. & Marshall, B. D. A Manual of Laboratory and Diagnostic Tests (ed. Marshall, B. D. ) 407 (Lippincott Williams and Wilkins, 2009).
Traish, A. M. et al. Testosterone deficiency. Am J Med. 124, 578–587 (2011)
Zitzmann, M. Testosterone deficiency, insulin resistance and the metabolic syndrome. Nat Rev Endocrinol. 5, 673–681 (2009).
Liu, P. Y., Death, A. K. & Handelsman, D. J. Androgens and cardiovascular disease. Endocr Rev. 24, 313–340 (2003).
Svartberg, J. et al. Association of endogenous testosterone with blood pressure and left ventricular mass in men. The Tromsø Study. Eur J Endocrinol. 150, 65–71 (2004).
Garcia-Cruz E. et al. Hypertension, dyslipidemia and overweight are related to lower testosterone levels in a cohort of men undergoing prostate biopsy. Int J Impot Res. 24, 110–113 (2012).
Reckelhoff, J. F. & Granger, J. P. Role of androgens in mediating hypertension and renal injury. Clin Exp Pharmacol Physiol. 26, 127–131 (1999).
Torkler, S. et al. Inverse association between total testosterone concentrations, incident hypertension and blood pressure. Aging Male. 14, 176–182 (2011).
Colby, S. et al. Exome sequencing reveals germline gain-of-function EGFR mutation in an adult with Lhermitte-Duclos disease. Cold Spring Harb Mol Case Stud. 2, a001230 (2016).
Acknowledgements
The authors thank Jerry Pelletier (Biochemistry, McGill) for help in analyzing RNA motifs in the 3′ UTR of EFNB3. This work was supported by grants from the Canadian Institutes of Health Research to J.W. (MOP57697, MOP69089 and MOP 123389), H.L. (MOP97829), JT and P.H (ISO106797). It was also financed by grants from the Natural Sciences and Engineering Research Council of Canada (203906-2012), Juvenile Diabetes Research Foundation (17-2013-440), Fonds de recherche du Quebec-Santé (Ag-06) and the J.-Louis Levesque Foundation to J.W. It is financed also by grants from OPTI-THERA to J.T. and P.H.
Author information
Authors and Affiliations
Contributions
J.T., J.C., M.W., S.H. and P.H. were principal investigators of the ADVANCE genetic study from which the data used in this project are derived. Y.W., Z.W. H.L., and J.W. generated the concept and initiated this project. J.R., F.M. and E.B. conducted the statistical analysis. J.T., J.R., P.H. and J.W. drafted the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Tremblay, J., Wang, Y., Raelson, J. et al. Evidence from single nucleotide polymorphism analyses of ADVANCE study demonstrates EFNB3 as a hypertension risk gene. Sci Rep 7, 44114 (2017). https://doi.org/10.1038/srep44114
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep44114
This article is cited by
-
Analysis of the association of EPHB6, EFNB1 and EFNB3 variants with hypertension risks in males with hypogonadism
Scientific Reports (2018)
-
EPHA6 rs4857055 C > T polymorphism associates with hypertension through triglyceride and LDL particle size in the Korean population
Lipids in Health and Disease (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.