Abstract
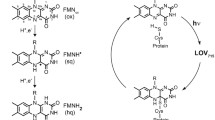
Light-Oxygen-Voltage (LOV) domains are conserved parts of photoreceptors in plants, bacteria and fungi that bind flavins as chromophores and detect blue light. In the past, LOV domain variants have been developed as fluorescent reporter proteins (called flavin-based fluorescent proteins; FbFPs), which due to their ability to fluoresce under anaerobic conditions, fast folding kinetics and a small size of ~12–16 kDa are a promising reporter system for quantitative real-time analysis of biological processes. Here, we present a small thermostable flavin-based fluorescent protein CagFbFP derived from a soluble LOV domain-containing histidine kinase from the thermophilic bacterium Chloroflexus aggregans. CagFbFP is composed of 107 amino acids with a molecular weight of 11.6 kDa and consists only of the conserved LOV core domain. The protein is thermostable with a melting point of about 68 °C. It crystallizes easily and its crystals diffract to 1.07 Å. Both the crystal structure and small angle scattering data show that the protein is a dimer. Unexpectedly, gluta-mine 148, which in LOV photoreceptor proteins is the key residue responsible for signal transduction, occupies two conformations. Molecular dynamics simulations show that the two conformations interconvert rapidly. The crystal structure of the wild-type Chloroflexus aggregans LOV domain determined at 1.22 Å resolution confirmed the presence of two alternative conformations of the glutamine 148 side chain. Overall, this protein, due to its stability and ease of crystallization, appears to be a promising model for ultra-high resolution structural studies of LOV domains and for application as a fluorescent reporter.
Similar content being viewed by others
References
J. Herrou and S. Crosson, Function, structure and mechanism of bacterial photosensory LOV proteins, Nat. Rev. Microbiol., 2011, 9, 713–723.
B. D. Zoltowski and K. H. Gardner, Tripping the light fantastic: blue-light photoreceptors as examples of environmentally modulated protein-protein interactions, Biochemistry, 2011, 50, 4–16.
K. S. Conrad, C. C. Manahan and B. R. Crane, Photochemistry of flavoprotein light sensors, Nat. Chem. Biol., 2014, 10, 801–809.
A. Losi, C. Mandalari and W. Gärtner, The Evolution and Functional Role of Flavin-based Prokaryotic Photoreceptors, Photochem. Photobiol., 2015, 91, 1021–1031.
J. T. Henry and S. Crosson, Ligand-Binding PAS Domains in a Genomic, Cellular, and Structural Context, Annu. Rev. Microbiol., 2011, 65, 261–286.
S. T. Glantz, E. J. Carpenter, M. Melkonian, K. H. Gardner, E. S. Boyden, G. K.-S. Wong and B. Y. Chow, Functional and topological diversity of LOV domain photoreceptors, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, E1442–E1451.
A. Losi and W. Gärtner, Solving Blue Light Riddles: New Lessons from Flavin-binding LOV Photoreceptors, Photochem. Photobiol., 2017, 93, 141–158.
T. Fettweiss, K. Röllen, J. Granzin, O. Reiners, S. Endres, T. Drepper, D. Willbold, K.-E. Jaeger, R. Batra-Safferling and U. Krauss, Mechanistic basis of the fast dark recovery of the short LOV protein DsLOV from Dinoroseobacter shibae, Biochemistry, 2018, 57, 4833–4847.
M. Salomon, J. M. Christie, E. Knieb, U. Lempert and W. R. Briggs, Photochemical and mutational analysis of the FMN-binding domains of the plant blue light receptor, phototropin, Biochemistry, 2000, 39, 9401–9410.
A. M. Buckley, J. Petersen, A. J. Roe, G. R. Douce and J. M. Christie, LOV-based reporters for fluorescence imaging, Curr. Opin. Chem. Biol., 2015, 27, 39–45.
A. Mukherjee and C. M. Schroeder, Flavin-based fluorescent proteins: emerging paradigms in biological imaging, Curr. Opin. Biotechnol., 2015, 31, 16–23.
T. Drepper, T. Eggert, F. Circolone, A. Heck, U. Krauss, J.-K. Guterl, M. Wendorff, A. Losi, W. Gärtner and K.-E. Jaeger, Reporter proteins for, in vivo fluorescence without oxygen, Nat. Biotechnol., 2007, 25, 443–445.
S. Chapman, C. Faulkner, E. Kaiserli, C. Garcia-Mata, E. I. Savenkov, A. G. Roberts, K. J. Oparka and J. M. Christie, The photoreversible fluorescent protein iLOV outperforms GFP as a reporter of plant virus infection, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 20038–20043.
J. M. Christie, K. Hitomi, A. S. Arvai, K. A. Hartfield, M. Mettlen, A. J. Pratt, J. A. Tainer and E. D. Getzoff, Structural tuning of the fluorescent protein iLOV for improved photostability, J. Biol. Chem., 2012, 287, 22295–22304.
A. Mukherjee, J. Walker, K. B. Weyant and C. M. Schroeder, Characterization of flavin-based fluorescent proteins: an emerging class of fluorescent reporters, PLoS One, 2013, 8, e64753.
X. Song, Y. Wang, Z. Shu, J. Hong, T. Li and L. Yao, Engineering a more thermostable blue light photo receptor Bacillus subtilis YtvA LOV domain by a computer aided rational design method, PLoS Comput. Biol., 2013, 9, e1003129.
A. Mukherjee, K. B. Weyant, U. Agrawal, J. Walker, I. K. O. Cann and C. M. Schroeder, Engineering and characterization of new LOV-based fluorescent proteins from Chlamydomonas reinhardtii and Vaucheria frigida, ACS Synth. Biol., 2015, 4, 371–377.
M. Wingen, K.-E. Jaeger, T. Gensch and T. Drepper, Novel Thermostable Flavin-binding Fluorescent Proteins from Thermophilic Organisms, Photochem. Photobiol., 2017, 93, 849–856.
S. Hanada, A. Hiraishi, K. Shimada and K. Matsuura, Chloroflexus aggregans sp. nov., a filamentous phototrophic bacterium which forms dense cell aggregates by active gliding movement, Int. J. Syst. Bacteriol., 1995, 45, 676–681.
R. D. Finn, T. K. Attwood, P. C. Babbitt, A. Bateman, P. Bork, A. J. Bridge, H.-Y. Chang, Z. Dosztányi, S. El-Gebali, M. Fraser, J. Gough, D. Haft, G. L. Holliday, H. Huang, X. Huang, I. Letunic, R. Lopez, S. Lu, A. Marchler-Bauer, H. Mi, J. Mistry, D. A. Natale, M. Necci, G. Nuka, C. A. Orengo, Y. Park, S. Pesseat, D. Piovesan, S. C. Potter, N. D. Rawlings, N. Redaschi, L. Richardson, C. Rivoire, A. Sangrador-Vegas, C. Sigrist, I. Sillitoe, B. Smithers, S. Squizzato, G. Sutton, N. Thanki, P. D. Thomas, S. C. E. Tosatto, C. H. Wu, I. Xenarios, L.-S. Yeh, S.-Y. Young and A. L. Mitchell, InterPro in 2017—beyond protein family and domain annotations, Nucleic Acids Res., 2017, 45, D190–D199.
C. P. Zschiedrich, V. Keidel and H. Szurmant, Molecular Mechanisms of Two-Component Signal Transduction, J. Mol. Biol., 2016, 428, 3752–3775.
M. P. Bhate, K. S. Molnar, M. Goulian and W. F. DeGrado, Signal Transduction in Histidine Kinases: Insights from New Structures, Structure, 2015, 23, 981–994.
I. Gushchin and V. Gordeliy, Transmembrane Signal Transduction in Two-Component Systems: Piston, Scissoring, or Helical Rotation?, BioEssays, 2018, 40, 1700197.
K. Röllen, J. Granzin, R. Batra-Safferling and A. M. Stadler, Small-angle X-ray scattering study of the kinetics of lightdark transition in a LOV protein, PLoS One, 2018, 13(7), e0200746.
J. S. Lamb, B. D. Zoltowski, S. A. Pabit, B. R. Crane and L. Pollack, Time-resolved dimerization of a PAS-LOV protein measured with photocoupled small angle X-ray scattering, J. Am. Chem. Soc., 2008, 130, 12226–12227.
U. Heintz and I. Schlichting, Blue light-induced LOV domain dimerization enhances the affinity of Aureochrome 1a for its target DNA sequence, eLife, 2016, 5, e11860.
A. Banerjee, E. Herman, T. Kottke and L.-O. Essen, Structure of a Native-like Aureochrome 1a LOV Domain Dimer from Phaeodactylum tricornutum, Structure, 2016, 24, 171–178.
K. S. Conrad, A. M. Bilwes and B. R. Crane, Light-induced subunit dissociation by a light-oxygen-voltage domain photoreceptor from Rhodobacter sphaeroides, Biochemistry, 2013, 52, 378–391.
M. Wingen, J. Potzkei, S. Endres, G. Casini, C. Rupprecht, C. Fahlke, U. Krauss, K.-E. Jaeger, T. Drepper and T. Gensch, The photophysics of LOV-based fluorescent proteins–new tools for cell biology, Photochem. Photobiol. Sci., 2014, 13, 875–883.
M. D. Davari, B. Kopka, M. Wingen, M. Bocola, T. Drepper, K.-E. Jaeger, U. Schwaneberg and U. Krauss, Photophysics of the LOV-Based Fluorescent Protein Variant iLOV-Q489 K Determined by Simulation and Experiment, J. Phys. Chem. B, 2016, 120, 3344–3352.
J. Jancarik and S.-H. Kim, Sparse matrix sampling: a screening method for crystallization of proteins, J. Appl. Crystallogr., 1991, 24, 409–411.
J. Torra, C. Lafaye, L. Signor, S. Aumonier, C. Flors, X. Shu, S. Nonell, G. Gotthard and A. Royant, Tailing miniSOG: structural bases of the complex photophysics of a flavinbinding singlet oxygen photosensitizing protein, Sci. Rep., 2019, 9, 2428.
A. Pudasaini, J. S. Shim, Y. H. Song, H. Shi, T. Kiba, D. E. Somers, T. Imaizumi and B. D. Zoltowski, Kinetics of the LOV domain of ZEITLUPE determine its circadian function in Arabidopsis, eLife, 2017, 6, e21646.
V. Arinkin, J. Granzin, K. Röllen, U. Krauss, K.-E. Jaeger, D. Willbold and R. Batra-Safferling, Structure of a LOV protein in apo-state and implications for construction of LOV-based optical tools, Sci. Rep., 2017, 7, 42971.
B. D. Zoltowski, C. Schwerdtfeger, J. Widom, J. J. Loros, A. M. Bilwes, J. C. Dunlap and B. R. Crane, Conformational switching in the fungal light sensor Vivid, Science, 2007, 316, 1054–1057.
M. G. Khrenova, A. V. Nemukhin and T. Domratcheva, Theoretical Characterization of the Flavin-Based Fluorescent Protein iLOV and its Q489 K Mutant, J. Phys. Chem. B, 2015, 119, 5176–5183.
B. D. Zoltowski, B. Vaccaro and B. R. Crane, Mechanismbased tuning of a LOV domain photoreceptor, Nat. Chem. Biol., 2009, 5, 827–834.
J. Lokhandwala, R. I. Silverman, Y. de la Vega, H. C. Hopkins, C. W. Britton, A. Rodriguez-Iglesias, R. Bogomolni, M. Schmoll and B. D. Zoltowski, A Native Threonine Coordinates Ordered Water to Tune Light-Oxygen-Voltage (LOV) Domain Photocycle Kinetics and Osmotic Stress Signaling in Trichoderma reesei ENVOY, J. Biol. Chem., 2016, 291, 14839–14850.
F. Kawano, Y. Aono, H. Suzuki and M. Sato, Fluorescence imaging-based high-throughput screening of fast- and slow-cycling LOV proteins, PLoS One, 2013, 8, e82693.
J. M. Christie, S. B. Corchnoy, T. E. Swartz, M. Hokenson, I.-S. Han, W. R. Briggs and R. A. Bogomolni, Steric interactions stabilize the signaling state of the LOV2 domain of phototropin 1, Biochemistry, 2007, 46, 9310–9319.
G. Rivera-Cancel, W. Ko, D. R. Tomchick, F. Correa and K. H. Gardner, Full-length structure of a monomeric histidine kinase reveals basis for sensory regulation, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 17839–17844.
J. Lokhandwala, H. C. Hopkins, A. Rodriguez-Iglesias, C. Dattenböck, M. Schmoll and B. D. Zoltowski, Structural biochemistry of a fungal LOV domain photoreceptor reveals an evolutionarily conserved pathway integrating light and oxidative stress, Structure, 2015, 23, 116–125.
J. Rinaldi, M. Gallo, S. Klinke, G. Paris, H. R. Bonomi, R. A. Bogomolni, D. O. Cicero and F. A. Goldbaum, The β-scaffold of the LOV domain of the Brucella light-activated histidine kinase is a key element for signal transduction, J. Mol. Biol., 2012, 420, 112–127.
A. Möglich and K. Moffat, Structural basis for light-dependent signaling in the dimeric LOV domain of the photosensor YtvA, J. Mol. Biol., 2007, 373, 112–126.
A. S. Halavaty and K. Moffat, N- and C-terminal flanking regions modulate light-induced signal transduction in the LOV2 domain of the blue light sensor phototropin 1 from Avena sativa, Biochemistry, 2007, 46, 14001–14009.
F. W. Studier, Protein production by auto-induction in high-density shaking cultures, Protein Expression Purif., 2005, 41, 207–234.
L. G. Whitby, A new method for preparing flavin-adenine dinucleotide, Biochem. J., 1953, 54, 437–442.
M. Kataoka, S. Shimizu and H. Yamada, Purification and characterization of a novel FMN-dependent enzyme. Membrane-bound L-(+)-pantoyl lactone dehydrogenase from Nocardia asteroides, Eur. J. Biochem., 1992, 204, 799–806.
M. R. Wilkins, E. Gasteiger, A. Bairoch, J. C. Sanchez, K. L. Williams, R. D. Appel and D. F. Hochstrasser, Protein identification and analysis tools in the ExPASy server, Methods Mol. Biol., 1999, 112, 531–552.
M. Kaschner, A. Loeschcke, J. Krause, B. Q. Minh, A. Heck, S. Endres, V. Svensson, A. Wirtz, A. von Haeseler, K.-E. Jaeger, T. Drepper and U. Krauss, Discovery of the first light-dependent protochlorophyllide oxidoreductase in anoxygenic phototrophic bacteria, Mol. Microbiol., 2014, 93, 1066–1078.
C. E. Blanchet, A. Spilotros, F. Schwemmer, M. A. Graewert, A. Kikhney, C. M. Jeffries, D. Franke, D. Mark, R. Zengerle, F. Cipriani, S. Fiedler, M. Roessle and D. I. Svergun, Versatile sample environments and automation for biological solution X-ray scattering experiments at the P12 beamline (PETRA III, DESY), J. Appl. Crystallogr., 2015, 48, 431–443.
D. Franke, M. V. Petoukhov, P. V. Konarev, A. Panjkovich, A. Tuukkanen, H. D. T. Mertens, A. G. Kikhney, N. R. Hajizadeh, J. M. Franklin, C. M. Jeffries and D. I. Svergun, ATSAS 2.8: a comprehensive data analysis suite for small-angle scattering from macromolecular solutions, J. Appl. Crystallogr., 2017, 50, 1212–1225.
H. Fischer, O. Neto, M. De, H. B. Napolitano, I. Polikarpov and A. F. Craievich, Determination of the molecular weight of proteins in solution from a single small-angle X-ray scat-tering measurement on a relative scale, J. Appl. Crystallogr., 2010, 43, 101–109.
A. Hoffmann and S. Grudinin, NOLB: Nonlinear Rigid Block Normal-Mode Analysis Method, J. Chem. Theory Comput., 2017, 13, 2123–2134.
S. Grudinin, M. Garkavenko and A. Kazennov, Pepsi-SAXS: an adaptive method for rapid and accurate computation of small-angle X-ray scattering profiles, Acta Crystallogr., Sect. D: Struct. Biol., 2017, 73, 449–464.
W. Kabsch, XDS, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2010, 66, 125–132.
P. Evans, Scaling and assessment of data quality, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2005, 62, 72–82.
A. Vagin and A. Teplyakov, Molecular replacement with MOLREP, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2009, 66, 22–25.
M. Källberg, H. Wang, S. Wang, J. Peng, Z. Wang, H. Lu and J. Xu, Template-based protein structure modeling using the RaptorX web server, Nat. Protoc., 2012, 7, 1511–1522.
P. Emsley, B. Lohkamp, W. G. Scott and K. Cowtan, Features and development of Coot, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2010, 66, 486–501.
G. N. Murshudov, P. Skubák, A. A. Lebedev, N. S. Pannu, R. A. Steiner, R. A. Nicholls, M. D. Winn, F. Long and A. A. Vagin, REFMAC5 for the refinement of macromolecular crystal structures, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2011, 67, 355–367.
M. H. M. Olsson, C. R. Søndergaard, M. Rostkowski and J. H. Jensen, PROPKA3: Consistent Treatment of Internal and Surface Residues in Empirical pKa Predictions, J. Chem. Theory Comput., 2011, 7, 525–537.
V. Hornak, R. Abel, A. Okur, B. Strockbine, A. Roitberg and C. Simmerling, Comparison of multiple Amber force fields and development of improved protein backbone parameters, Proteins: Struct., Funct., Bioinf., 2006, 65, 712–725.
W. D. Cornell, P. Cieplak, C. I. Bayly, I. R. Gould, K. M. Merz, D. M. Ferguson, D. C. Spellmeyer, T. Fox, J. W. Caldwell and P. A. Kollman, A Second Generation Force Field for the Simulation of Proteins, Nucleic Acids, and Organic Molecules, J. Am. Chem. Soc., 1995, 117, 5179–5197.
J. Wang, R. M. Wolf, J. W. Caldwell, P. A. Kollman and D. A. Case, Development and testing of a general amber force field, J. Comput. Chem., 2004, 25, 1157–1174.
D. A. Case, V. Babin, J. Berryman, R. M. Betz, Q. Cai, D. S. Cerutti, C. Iii, T. E. Cheatham, III, T. A. Darden, R. E. Duke, H. Gohlke, A. W. Goetz, S. Gusarov, N. Homeyer, P. Janowski, J. Kaus, I. Kolossváry, A. Kovalenko, T. S. Lee, S. LeGrand, T. Luchko, R. Luo, B. Madej, K. M. Merz, F. Paesani, D. R. Roe, A. Roitberg, C. Sagui, R. Salomon-Ferrer, G. Seabra, C. L. Simmerling, W. Smith, J. Swails, R. C. Walker, J. Wang, R. M. Wolf, X. Wu and P. A. Kollman, Amber 14, University of California, San Francisco, 2014.
W. L. Jorgensen, J. Chandrasekhar, J. D. Madura, R. W. Impey and M. L. Klein, Comparison of simple potential functions for simulating liquid water, J. Chem. Phys., 1983, 79, 926–935.
U. Essmann, L. Perera, M. L. Berkowitz, T. Darden, H. Lee and L. G. Pedersen, A smooth particle mesh Ewald method, J. Chem. Phys., 1995, 103, 8577–8593.
W. L. DeLano, The PyMOL molecular graphics system, 2002.
W. Humphrey, A. Dalke and K. Schulten, VMD Visual molecular dynamics, J. Mol. Graphics, 1996, 14, 33–38.
P. A. Karplus and K. Diederichs, Linking Crystallographic Model and Data Quality, Science, 2012, 336, 1030–1033.
M. A. Larkin, G. Blackshields, N. P. Brown, R. Chenna, P. A. McGettigan, H. McWilliam, F. Valentin, I. M. Wallace, A. Wilm, R. Lopez, J. D. Thompson, T. J. Gibson and D. G. Higgins, Clustal W and Clustal X version 2.0, Bioinformatics, 2007, 23, 2947–2948.
A. M. Waterhouse, J. B. Procter, D. M. A. Martin, M. Clamp and G. J. Barton, Jalview Version 2–a multiple sequence alignment editor and analysis workbench, Bioinformatics, 2009, 25, 1189–1191.
D. Liebschner, P. V. Afonine, N. W. Moriarty, B. K. Poon, O. V. Sobolev, T. C. Terwilliger and P. D. Adams, Polder maps: improving OMIT maps by excluding bulk solvent, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2017, 73, 148–157.
Author information
Authors and Affiliations
Corresponding author
Additional information
Electronic supplementary information (ESI) available: ESI Table S1 and Fig. S1–S5. See DOI: 10.1039/c9pp00067d
Rights and permissions
About this article
Cite this article
Nazarenko, V.V., Remeeva, A., Yudenko, A. et al. A thermostable flavin-based fluorescent protein from Chloroflexus aggregans: a framework for ultra-high resolution structural studies. Photochem Photobiol Sci 18, 1793–1805 (2019). https://doi.org/10.1039/c9pp00067d
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1039/c9pp00067d