-
PDF
- Split View
-
Views
-
Cite
Cite
Thomas Haselhorst, Helen Blanchard, Martin Frank, Mark J. Kraschnefski, Milton J. Kiefel, Alex J. Szyczew, Jeffery C. Dyason, Fiona Fleming, Gavan Holloway, Barbara S. Coulson, Mark von Itzstein, STD NMR spectroscopy and molecular modeling investigation of the binding of N-acetylneuraminic acid derivatives to rhesus rotavirus VP8* core, Glycobiology, Volume 17, Issue 1, January 2007, Pages 68–81, https://doi.org/10.1093/glycob/cwl051
Close - Share Icon Share
Abstract
The VP8* subunit of rotavirus spike protein VP4 contains a sialic acid (Sia)-binding domain important for host cell attachment and infection. In this study, the binding epitope of the N-acetylneuraminic acid (Neu5Ac) derivatives has been characterized by saturation transfer difference (STD) nuclear magnetic resonance (NMR) spectroscopy. From this STD NMR data, it is proposed that the VP8* core recognizes an identical binding epitope in both methyl α-D-N-acetylneuraminide (Neu5Acα2Me) and the disaccharide methyl S-(α-D-N-acetylneuraminosyl)-(2→6)-6-thio-β-D-galactopyranoside (Neu5Ac-α(2,6)-S-Galβ1Me). In the VP8*–disaccharide complex, the Neu5Ac moiety contributes to the majority of interaction with the protein, whereas the galactose moiety is solvent-exposed. Molecular dynamics calculations of the VP8*–disaccharide complex indicated that the galactose moiety is unable to adopt a conformation that is in close proximity to the protein surface. STD NMR experiments with methyl 9-O-acetyl-α-D-N-acetylneuraminide (Neu5,9Ac2α2Me) in complex with rhesus rotavirus (RRV) VP8* revealed that both the N-acetamide and 9-O-acetate moieties are in close proximity to the Sia-binding domain, with the N-acetamide's methyl group being saturated to a larger extent, indicating a closer association with the protein. RRV VP8* does not appear to significantly recognize the unsaturated Neu5Ac derivative [2-deoxy-2,3-didehydro-D-N-acetylneuraminic acid (Neu5Ac2en)]. Molecular modeling of the protein–Neu5Ac2en complex indicates that key interactions between the protein and the unsaturated Neu5Ac derivative when compared with Neu5Acα2Me would not be sustained. Neu5Acα2Me, Neu5Ac-α(2,6)-S-Galβ1Me, Neu5,9Ac2α2Me, and Neu5Ac2en inhibited rotavirus infection of MA104 cells by 61%, 35%, 30%, and 0%, respectively, at 10 mM concentration. NMR spectroscopic, molecular modeling, and infectivity inhibition results are in excellent agreement and provide valuable information for the design of inhibitors of rotavirus infection.
Introduction
Rotaviruses are members of the Reoviridae family and are recognized as the single most important cause of severe gastroenteritis in the infants of a wide range of mammals including humans (Estes 2001). Rotavirus infection afflicts millions of young children worldwide and is estimated to be responsible for 33% of all hospitalizations due to diarrhea in infants. Globally, over 440 000 children aged less than 5 years die each year from diarrhoea due to rotavirus, mainly in developing countries (Parashar et al. 2003). The high morbidity and mortality associated with rotavirus, as well as the economic burden (including the loss of productivity in the livestock industry) demonstrate an urgent need for the development of new methods for the treatment of this disease (Parashar et al. 1998). Although there are several rotavirus vaccines in clinical trials (Fischer et al. 2004; Svennerholm and Steele 2004), the safety and efficacy of rotavirus vaccines in general has been questioned (Franco and Greenberg 2001).
The outermost layer of the virion is composed of two proteins, VP4 and VP7, which are involved in the initial interaction between the virus and the host cell (Ludert et al. 1996; Coulson et al. 1997; Zarate et al. 2000; Estes 2001; Arias et al. 2002). Cellular integrins are also involved in this recognition process (Coulson et al. 1997; Hewish et al. 2000; Ciarlet et al. 2002). This outer layer is lost during cell entry, leading to the transcriptionally active double-layered particle. VP4 is an 88-kDa protein that forms spikes extending approximately 100 Å from the virion surface. The base of VP4 interacts with VP7 (Shaw et al. 1993; Yeager et al. 1994) and contains a large globular domain below the VP7 layer that interacts with VP6 (Shaw et al. 1993). Although VP4 has been identified as the protein responsible for receptor binding and cell penetration (Ludert et al. 1996; Zarate et al. 2000; Arias et al. 2002), the precise role of VP7 has not been completely defined, although it has been proposed that it modulates the functions of VP4 (Beisner et al. 1998) and interacts with cell surface molecules after the initial attachment of the virus through VP4 (Greenberg et al. 1983; Coulson et al. 1997; Mendez et al. 1999). Both VP4 and VP7 independently elicit neutralizing antibodies and are determinants of virulence and induce protective immunity (Greenberg et al. 1983).
VP4 is cleaved by trypsin into the N-terminal VP8* and C-terminal VP5* fragments, both of which remain associated with the virion (Clark et al. 1981; Dormitzer et al. 2001). Proteolysis of VP4 results in an enhancement of rotavirus infectivity in cell culture. Expressed VP8* from some virus strains has been shown to be responsible for hemagglutination, whereas VP5* contains an internal hydrophobic region and a putative cell fusion region similar to that observed in enveloped viruses. Tihova et al. (2001) have shown by electron cryo microscopy and difference map analysis that neutralizing monoclonal antibodies are directed against the sialic acid (Sia)-binding domain located in VP8* and that the membrane permeabilization domain is within VP5*. As a consequence, a model for cell entry by rotavirus, wherein regions within both VP8* and VP5* bind to receptors on the host-cell surface, was proposed (Tihova et al. 2001). The X-ray crystallographic structures of rhesus rotavirus (RRV) VP8* (Dormitzer, Sun, Wagner, et al. 2002) and of CRW-8 VP8* (Scott et al. 2005) in complex with methyl α-D-N-acetylneuraminide (Neu5Acα2Me), a Sia glycoside, support the notion of the host cell Sia recognition role of VP8* (Dormitzer, Sun, Wagner, et al. 2002).
The binding of rotavirus to host cells has generally been considered as being either dependent on, or independent of, the presence of Sia associated with the host cell surface glycoconjugates. Although several reports are somewhat contradictory in this regard (Fukudome et al. 1989; Ciarlet et al. 2001), the most recent evidence tends to support the conclusion that Sia are highly likely to be involved in the recognition process by most, if not all, strains of rotavirus. Generally it is considered that strains of rotavirus whose binding is sensitive to bacterial or viral sialidase treatment of cells bind to terminal Sia residues in glycoconjugates, whereas rotavirus strains insensitive to sialidase treatment recognize internal Sia residues such as that found in GM1, which are resistant to the action of these exo-sialidases (Guo et al. 1999; Delorme et al. 2001).
In order to provide further insight into the nature of the interaction between RRV VP8* and cell surface glycoconjugates and possible inhibitors, in the present study, we have investigated the binding of a number of N-acetylneuraminic acid-based ligands (Figure 1) including Neu5Acα2Me, methyl S-(α-D-N-acetylneuraminosyl)-(2→6)-6-thio-β-D-galactopyranoside (Neu5Ac-α(2,6)-S-Galβ1Me), methyl 9-O-acetyl-α-D-N-acetylneuraminide (Neu5,9Ac2α2Me), and the well-known sialidase inhibitor 2-deoxy-2,3-didehydro-D-N-acetylneuraminic acid (Neu5Ac2en) in complex with RRV VP8* core by nuclear magnetic resonance (NMR) spectroscopy and computational chemistry. Specifically, we have used one-dimensional 1H and two-dimensional saturation transfer difference NMR (STD NMR) spectroscopy (Mayer and Meyer 1999) and molecular dynamics (MD) simulations to study the interaction(s) of the selected N-acetylneuraminic acid derivatives with RRV VP8*. A dynamic analysis by NMR spectroscopy and MD simulations of RRV VP8*-complexes are in particular interesting because it was previously noted that the protein may have some flexibility within the Sia-binding domain (Dormitzer, Sun, Wagner, et al. 2002). The effect of treatment with the selected N-acetylneuraminic acid derivatives on RRV rotavirus infectivity of MA104 cells was assessed in relation to the VP8*-binding requirements of these derivatives.
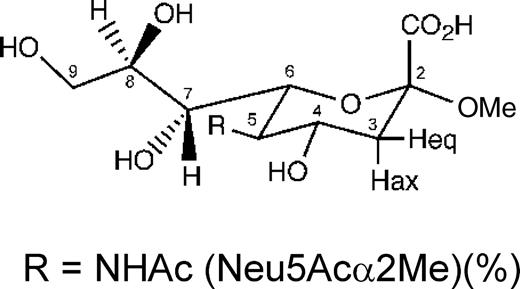
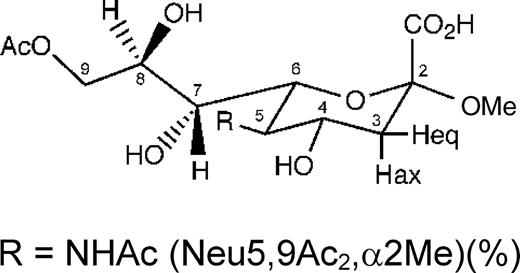
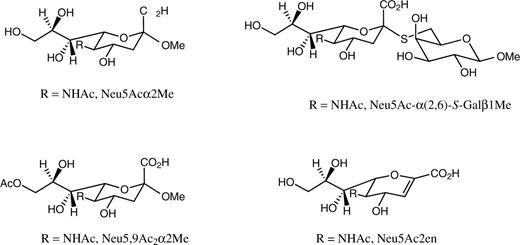
Structures of the N-acetylneuraminic acid derivatives Neu5Acα2Me, Neu5Ac-α(2,6)-S-Galβ1Me, Neu5,9Ac2α2Me, and Neu5Ac2en, which were used in the analysis.
Results
STD NMR experiments of Neu5Acα2Me in complex with RRV VP8*
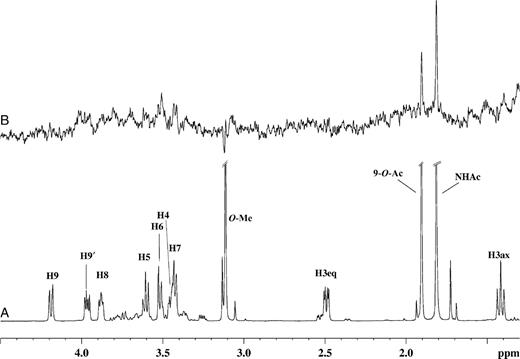
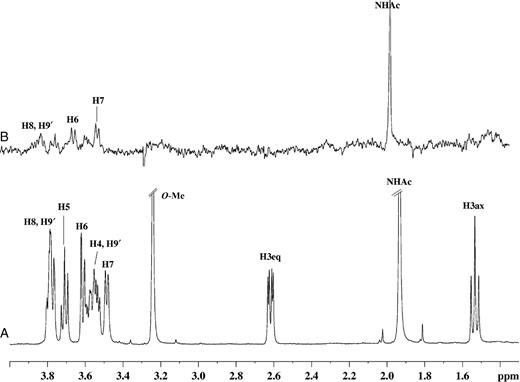
The 1H NMR and one-dimensional STD NMR spectra of RRV VP8* in the presence of Neu5Acα2Me are shown in Figure 2A and B, respectively. Comparison of these spectra clearly shows that Neu5Acα2Me binds to the protein, as indicated by strong STD NMR signals. The largest STD effect was observed for the methyl group of the N-acetamide moiety at approximately 1.95 ppm and consequently this signal was used as a reference and set to 100%. The relative %STD effects for individual protons of Neu5Acα2Me bound to RRV VP8* were calculated accordingly and are shown in Table I. Interestingly, the signal for the methyl aglycon at approximately 3.24 ppm is not detectable in the STD NMR spectrum (Figure 2B), indicating that the aglycon unit is not in close proximity to the protein surface and thus apparently not significantly involved in binding interactions of Neu5Acα2Me. In contrast, significant STD effects of 62%, 88%, 65%, and 77% for the glycerol side chain protons H9′, H9, H8, and H7, respectively, were observed (Table I). An apparent STD NMR effect for the H4 proton of Neu5Acα2Me was determined to be 62%, but owing to severe overlapping, this effect may also be attributed to some contribution from the H9′ proton and warranted further investigation. To further resolve these signals and to investigate which proton was in closer contact with the protein surface, a two-dimensional STD total correlation spectroscopy (TOCSY) spectrum of Neu5Acα2Me in complex with RRV VP8* was acquired. Figure 3 shows a standard TOCSY experiment (Figure 3A and C) of Neu5Acα2Me and the corresponding STD TOCSY experiment (Figure 3B and D). Figure 3C shows the expansion of the TOCSY spectrum of the glycerol side chain protons of Neu5Acα2Me. The corresponding STD TOCSY NMR spectrum of the same region of Neu5Acα2Me in complex with RRV VP8* is shown in Figure 3D. As can be seen, the cross signals for H8, H9/H7 and H8, H8/H9′ show strong intensities in the STD TOCSY spectrum. The H5/H4 and H5/H6 proton cross signals also receive saturation from the protein but are relatively weak STD signal intensities compared with the STD TOCSY signals of the H9 and H8 protons. This spectrum provides strong evidence that the STD NMR effect determined for the H9′/H4 signals is mainly contributed by the STD effect of the H9′ proton and only to a lesser extent the H4 proton. Furthermore, the STD TOCSY spectrum reveals an STD TOCSY cross signal for the proton pair H4/H6.
(A) 1H NMR spectra of 46 µM of RRV VP8* complex with Neu5Acα2Me at a mole ratio of 1:100. (B) STD 1H NMR spectrum with saturation frequency of the protein on-resonance at 7.13 ppm and off-resonance at 33 ppm using a Gaussian pulse cascade (40 Gaussian pulses of 50 ms duration, each with a delay of 100 µs in between each pulse) with a total saturation time of 2 s. All spectra were acquired in 20 mM phosphate buffer, pH 7.1, containing 10 mM NaCl, at 288 K and 600 MHz.
Two-dimensional TOCSY [(A) and (C)] and STD TOCSY [(B) and (D)] NMR spectra of 0.2 mM of RRV VP8* complexed with Neu5Acα2Me at a mole ratio of 1:100. Saturation of the protein was achieved with a Gaussian pulse cascade (40 Gaussian pulses of 50 ms duration, each with a delay of 100 µs in between each pulse) with a total saturation time of 2 s. The on-resonance saturation frequency was set to 7.13 ppm and off-resonance to 33 ppm; 300 t1 increments were collected in total with 160 scans per increment and a Malcolm Levitt's composite-pulse decoupling sequence (MLEV) mixing time of 70 ms. All spectra were acquired in 20 mM phosphate buffer, pH 7.1, containing 10 mM NaCl, at 298 K and 600 MHz.
Relative STD NMR effectsa for Neu5Acα2Me and Neu5,9Acα2Me in complex with RRV VP8*
| . |  .
. |  .
. |
|---|---|---|
| H3ax (N)b | 0 | 0 |
| H3eq (N) | NQ | NQ |
| H4 (N) | 62c | <30 |
| H5 (N) | 31 | <30 |
| H6 (N) | 70 | <30 |
| H7 (N) | 77 | <30 |
| H8 (N) | 65 | <30 |
| H9 (N) | 88 | <30 |
| H9′ (N) | 62c | <30 |
| NHAc (N) | 100 | 100 |
| O-Me | 0 | 0 |
| 9-O-Ac | — | 56 |
| . |  .
. |  .
. |
|---|---|---|
| H3ax (N)b | 0 | 0 |
| H3eq (N) | NQ | NQ |
| H4 (N) | 62c | <30 |
| H5 (N) | 31 | <30 |
| H6 (N) | 70 | <30 |
| H7 (N) | 77 | <30 |
| H8 (N) | 65 | <30 |
| H9 (N) | 88 | <30 |
| H9′ (N) | 62c | <30 |
| NHAc (N) | 100 | 100 |
| O-Me | 0 | 0 |
| 9-O-Ac | — | 56 |
aSTD effects were calculated according to the formula ASTD=(I0−Isat)/I0−ISTD/I0. All STD effects are given relative to the strongest STD NMR effect of the methyl group of the N-acetamide.
bN, N-acetylneuraminic acid.
cOverlapping signals: H9′ and H4 of Neu5Acα2Me.
NQ, not quantified.
Relative STD NMR effectsa for Neu5Acα2Me and Neu5,9Acα2Me in complex with RRV VP8*
| . |  .
. |  .
. |
|---|---|---|
| H3ax (N)b | 0 | 0 |
| H3eq (N) | NQ | NQ |
| H4 (N) | 62c | <30 |
| H5 (N) | 31 | <30 |
| H6 (N) | 70 | <30 |
| H7 (N) | 77 | <30 |
| H8 (N) | 65 | <30 |
| H9 (N) | 88 | <30 |
| H9′ (N) | 62c | <30 |
| NHAc (N) | 100 | 100 |
| O-Me | 0 | 0 |
| 9-O-Ac | — | 56 |
| . |  .
. |  .
. |
|---|---|---|
| H3ax (N)b | 0 | 0 |
| H3eq (N) | NQ | NQ |
| H4 (N) | 62c | <30 |
| H5 (N) | 31 | <30 |
| H6 (N) | 70 | <30 |
| H7 (N) | 77 | <30 |
| H8 (N) | 65 | <30 |
| H9 (N) | 88 | <30 |
| H9′ (N) | 62c | <30 |
| NHAc (N) | 100 | 100 |
| O-Me | 0 | 0 |
| 9-O-Ac | — | 56 |
aSTD effects were calculated according to the formula ASTD=(I0−Isat)/I0−ISTD/I0. All STD effects are given relative to the strongest STD NMR effect of the methyl group of the N-acetamide.
bN, N-acetylneuraminic acid.
cOverlapping signals: H9′ and H4 of Neu5Acα2Me.
NQ, not quantified.
These findings support the relative STD effects determined from the 1H STD NMR spectrum (Figure 2B), particularly in that the H7, H8, H9, and H9′ protons of the Neu5Acα2Me glycerol side chain are in close proximity to the surface of RRV VP8* and appear to play a key role in the binding of Neu5Acα2Me. These NMR data and conclusions are in good agreement with previously reported X-ray data (Dormitzer, Sun, Wagner, et al. 2002). A small (weak) STD NMR effect (Figure 2B) was observed for the H3equatorial (eq) (δ approximately 2.61 ppm) of Neu5Acα2Me, though it could not be quantified as a result of the low signal-to-noise ratio. No STD NMR effect was observed for the H3axial (ax) (δ approximately 1.55 ppm) proton of Neu5Acα2Me.
STD NMR experiments of Neu5Ac-α(2,6)-S-Galβ1Me in complex with RRV VP8*
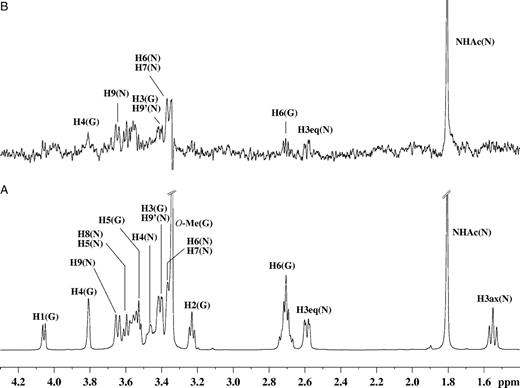
The interaction(s) of a disaccharide containing an N-acetylneuraminic acid moiety with RRV VP8* were also investigated. It has been proposed in a recent study that the aglycon unit, in general, does not play a significant role in the binding of sialosylglycosides to RRV VP8* (Dormitzer, Sun, Blixt, et al. 2002). We have performed an NMR spectroscopic-based investigation of the interactions of the synthetic disaccharide Neu5Ac-α(2,6)-S-Galβ1Me, which contains a galactose aglycon, with RRV VP8*. The 1H NMR and STD NMR spectra of Neu5Ac-α(2,6)-S-Galβ1Me in complex with RRV VP8* are shown in Figure 4A and B, respectively. From the STD NMR spectrum (Figure 4B), it can be clearly seen that ligand Neu5Ac-α(2,6)-S-Galβ1Me binds to VP8*, with an intense signal for the methyl group of the N-acetamide moiety at δ approximately equal to 1.82 ppm, which has the strongest STD NMR effect. This spectrum further revealed some protein-derived background signals. A control STD NMR spectrum on the RRV VP8* protein without ligand, using the identical experimental conditions (data not shown), was acquired and this spectrum revealed some protein background signals. In an attempt to reduce protein background signals, the strength of the applied spin lock filter was increased; however, only increased subtraction artifacts were observed. Although these background signals prevented quantification of the STD NMR effects of the VP8*–Neu5Ac-α(2,6)-S-Galβ1Me complex, some qualitative trends were apparent. The analysis of the STD NMR spectrum (Figure 4B) further reveals that the galactose methyl aglycon (δ approximately 3.40 ppm) shows no apparent STD NMR signal and is comparable with the spectrum obtained for Neu5Acα2Me in complex with VP8* (Figure 2B). Very weak STD NMR effects were detected for the galactose unit, even though some signal overlap with resonances of the N-acetylneuraminic acid moiety did occur. To further explore the interaction of Neu5Ac-α(2,6)-S-Galβ1Me with VP8*, a two-dimensional STD TOCSY experiment, in a similar manner to that described for ligand Neu5Acα2Me, was performed. Figure 5 shows the reference TOCSY spectrum of Neu5Ac-α(2,6)-S-Galβ1Me in complex with RRV VP8* (Figure 5A and C) and the corresponding STD TOCSY spectrum (Figure 5B and D). Signals corresponding to Neu5Ac-α(2,6)-S-Galβ1Me protons in close proximity to the Sia-binding domain surface are clearly observed in the STD TOCSY experiment. From these spectra, it is immediately evident that the proton cross-peaks of the N-acetylneuraminic acid unit are the predominant STD TOCSY signals. As anticipated from the one-dimensional STD NMR experiments, the protons of the glycerol side chain of the N-acetylneuraminic acid moiety in Neu5Ac-α(2,6)-S-Galβ1Me show strong STD TOCSY NMR signals (Figure 5B and D), suggesting that these protons are in close proximity to the Sia-binding domain surface of RRV VP8*. The STD TOCSY NMR spectrum (Figure 5B and D) also allows us to discriminate between the H3 of the galactose moiety (H3 G) and H9′ of the N-acetylneuraminic acid (H9′ N) protons. This spectrum clearly reveals that the H3 proton of the galactose unit does not show any STD effect and provides further supporting evidence to suggest that the apparent H3 (G) signal in the 1H STD NMR spectrum at δ approximately equal to 3.55 ppm is the result of the overlapped signals of H3 (G) and H9′ (N). Thus, the observed STD effect can be attributed exclusively to the H9′ (N) proton.
(A) 1H NMR spectra of 46 µM of VP8* complex with Neu5Ac-α(2,6)-S-Galβ1Me at a mole ratio of 1:100. (B) STD 1H NMR spectrum with saturation frequency of the protein on-resonance at 7.13 ppm and off-resonance at 33 ppm using a Gaussian pulse cascade (40 Gaussian pulses of 50 ms duration, each with a delay of 100 µs in between each pulse) with a total saturation time of 2 s. All spectra were acquired in 20 mM phosphate buffer, pH 7.1, containing 10 mM NaCl, at 288 K and 600 MHz.
Two-dimensional TOCSY [(A) and (C)] and STD TOCSY [(B) and (D)] NMR spectra of 0.2 mM of VP8* complexed with Neu5Ac-α(2,6)-S-Galβ1Me at a mole ratio of 1:100. Saturation of the protein was achieved with a Gaussian pulse cascade (40 Gaussian pulses of 50 ms duration, each with a delay of 100 µs in between each pulse) with a total saturation time of 2 s. The on-resonance saturation frequency was set to 7.13 ppm and the off-resonance to 33 ppm; 300 t1 increments were collected in total with 160 scans per increment and an MLEV mixing time of 70 ms. All spectra were acquired in 20 mM phosphate buffer, pH 7.1, containing 10 mM NaCl, at 298 K and 600 MHz.
STD NMR experiments of Neu5,9Ac2α2Me in complex with RRV VP8*
In a number of reports (Willoughby et al. 1990; Kiefel et al. 1996), acetylated Sia, including 9-O-acetylated Neu5Ac derivatives (Kiefel et al. 1996), have been found to inhibit infection by some rotavirus strains in a cell-based assay. Therefore, to examine the potential influence of the 9-O-acetate (9-O-Ac) in the N-acetylneuraminic acid derivative Neu5,9Ac2α2Me on binding with RRV VP8*, an STD NMR spectroscopic-based investigation was undertaken. Figure 6A and B shows a 1H NMR spectrum of Neu5,9Ac2α2Me in complex with RRV VP8* and the corresponding STD NMR at 288 K, respectively. A strong STD NMR signal (δ approximately 1.85 ppm) for the methyl group of the N-acetamide moiety is observed, confirming that Neu5,9Ac2α2Me binds to RRV VP8*. Interestingly, a smaller STD NMR effect (56%, relative to the STD NMR effect of the methyl group of the N-acetamide moiety) for the methyl group of the 9-O-Ac (δ approximately 1.90 ppm) was observed. The STD NMR spectrum at 298 K of the VP8*–Neu5,9Ac2α2Me complex (data not shown) was comparable with the 288-K spectrum, except that no STD effect for the methyl protons of the 9-O-Ac was observed. This observed temperature effect is indicative of a higher flexibility; consequently, apparent weaker association of this group may have led to the observed variations of STD effects acquired at different temperatures.
(A) 1H NMR spectra of 46 µM of VP8* complex with Neu5,9Ac2α2Me at a mole ratio of 1:100. (B) STD 1H NMR spectrum with saturation frequency of the protein on-resonance at 7.13 ppm and off-resonance at 33 ppm using a Gaussian pulse cascade (40 Gaussian pulses of 50 ms duration, each with a delay of 100 µs in between each pulse) with a total saturation time of 2 s. All spectra were acquired in 20 mM phosphate buffer, pH 7.1, containing 10 mM NaCl, at 288 K and 600 MHz.
As expected, the methyl aglycon moiety (δ approximately 3.15 ppm) did not show any STD NMR effects, consistent with the notion that the aglycon moiety is solvent-exposed and is not in close contact with the protein surface. The carbohydrate ring and glycerol side chain protons received saturation via the protein resulting in an approximate total STD effect of <30% and indicates that these protons do not make significant contact with the protein surface.
STD NMR experiments of Neu5Ac2en in the presence of RRV VP8*
Our interest in the Sia family, various Sia derivatives, and sialylmimetics has led us to also investigate the interactions and affinity of the unsaturated N-acetylneuraminic acid Neu5Ac2en with VP8*. To compare potential binding modes of the Neu5Ac2en with the other derivatives Neu5Acα2Me, Neu5Ac-α(2,6)-S-Galβ1Me, and Neu5,9Ac2α2Me, an STD NMR investigation of Neu5Ac2en in the presence of VP8* was performed. The STD NMR analysis (data not shown) clearly revealed no STD NMR signal intensities for Neu5Ac2en, demonstrating that RRV VP8* has no binding affinity for Neu5Ac2en.
Molecular modeling studies of Neu5Ac-α(2,6)-S-Galβ1Me, Neu5,9Ac2α2Me, and Neu5Ac2en in complex with RRV VP8*
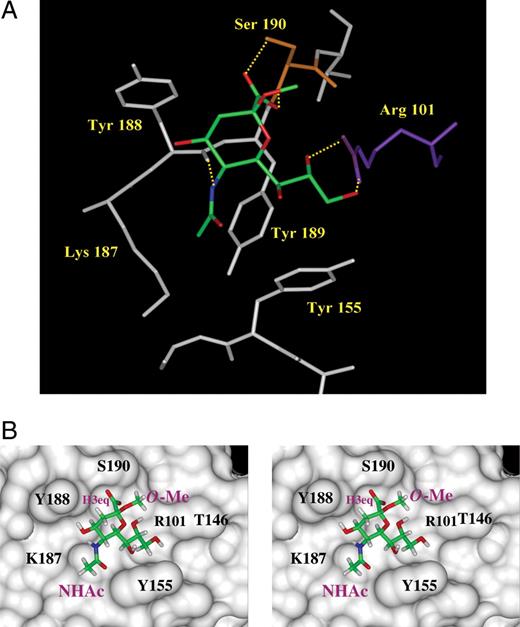
To further investigate the structural basis for the observed experimental STD NMR effects, we performed molecular modeling investigations for ligands Neu5Ac-α(2,6)-S-Galβ1Me, Neu5,9Ac2α2Me, and Neu5Ac2en in complex with VP8*. It has been previously noted that Neu5Acα2Me (Dormitzer, Sun, Wagner, et al. 2002) makes a series of interactions with the Sia-binding domain (Figure 7A and B).
The Sia-binding domain of RRV VP8* with bound Neu5Acα2Me (PDB access code=1KQR, Dormitzer, Sun, Wagner, et al. 2002). The ligand Neu5Acα2Me is displayed in stick representation with standard atom colors. Note that waters in this region of the structure are omitted from the figure. (A) The molecular details of the Sia-binding site. Direct hydrogen bond interactions between the ligand and protein are depicted (in yellow). (B) Stereo diagram showing the solvent accessible surface of the Sia-binding domain of RRV-VP8* with bound ligand. (B) is in the same orientation as (A), and the functional groups N-acetamide (NHAc) and methoxyl (O-Me) are indicated as is the H3eq, which is directed toward Tyr188.
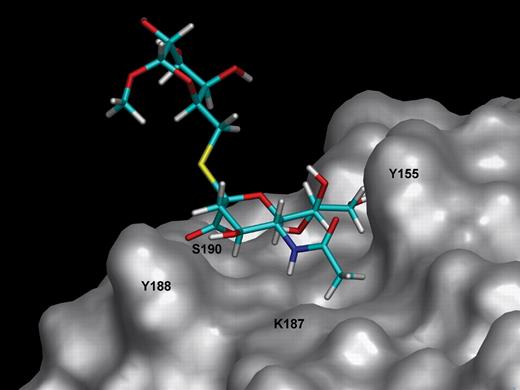
To investigate VP8*–Neu5Ac-α(2,6)-S-Galβ1Me interaction(s), an MD approach was employed. In the first instance, the potential bound conformations of the disaccharide Neu5Ac-α(2,6)-S-Galβ1Me, in complex with RRV VP8*, were sampled by a high-temperature (1000 K), in vacuo MD simulation. In the MD approach, the conformation of the Neu5Acα2Me from the X-ray crystal structure (Dormitzer, Sun, Wagner, et al. 2002) was used as a template to build the thiosialoside ligand Neu5Ac-α(2,6)-S-Galβ1Me. During the high-temperature MD calculation, the protein and the N-acetylneuraminic acid moiety were kept fixed in order to maintain the predominant binding mode of the sugar residue within the Sia-binding domain of VP8*. The thio-α(2,6)-linkage was allowed to be flexible during the MD simulation. This approach is not unreasonable because the presented STD NMR experiments show comparable STD effects for the N-acetylneuraminic acid moiety of Neu5Acα2Me and Neu5Ac-α(2,6)-S-Galβ1Me and led us to conclude that an identical binding epitope for these two ligands is recognized by RRV VP8*. Figure 8 illustrates a selected conformation of Neu5Ac-α(2,6)-S-Galβ1Me in complex with RRV VP8*. It is immediately obvious that in this selected conformation, the galactose unit appears to have little, if any, interaction with the protein surface. In fact, our MD calculations revealed that the H4 (G) and H5 (G) protons may interact with the protein surface, but such interactions are extremely rare due to the high energy of the corresponding conformations.
Molecular model of Neu5Ac-α(2,6)-S-Galβ1Me bound to the Sia-binding domain of RRV VP8*. The protein and the N-acetamide moiety were kept fixed during the simulation. The galactose moiety has no direct contacts with the protein surface. Molecular dynamic simulations were carried out using DISCOVER/CVFF at 1000 K and 1000 ps.
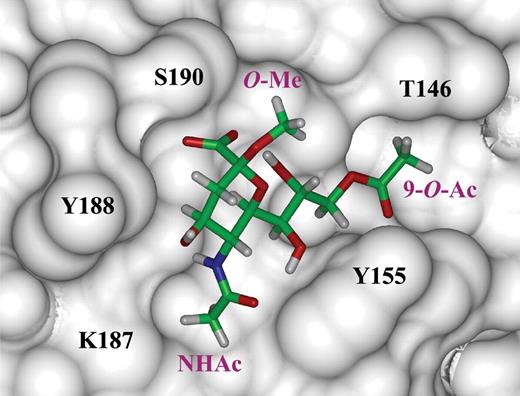
Solvent-based MD simulations were carried out for the ligand Neu5,9Ac2α2Me in complex with rotavirus VP8* to establish potential binding modes of Neu5,9Ac2α2Me and to enable the determination of key interactions of the ligand with the VP8* Sia-binding domain. A direct comparison of these data is also possible with the observed STD NMR experimental data. On the basis of this MD simulation, two potential binding modes for Neu5,9Ac2α2Me were determined. The binding conformation shown in Figure 9 indicates that Neu5,9Ac2α2Me can maintain key hydrophobic interactions with the VP8* Sia-binding domain and hydrogen bond interactions of the Neu5Ac carboxylate. The methyl protons of the 9-O-acetyl group are in close contact with the Sia-binding domain and are specifically directed toward Thr-146. Furthermore, the 9-O-Ac's carbonyl group is in hydrogen-bonding distance to Tyr-155. The MD simulation further revealed an alternative conformation of Neu5,9Ac2α2Me bound to VP8*, indicating an overall similar orientation except that the 9-O-Ac carbonyl has rotated to form a hydrogen bond with Arg-101 (data not shown).
One potential conformation of Neu5,9Ac2α2Me bound to the Sia-binding domain of RRV VP8*. This particular model shows that Neu5,9Acα2Me can adopt a similar conformation to Neu5Acα2Me and maintain key protein contacts, but the hydrogen bonds from the C8 and C9 hydroxyl groups to Arg-101 are disrupted. The methyl protons of the 9-O-acetate (9-O-Ac) moiety participate in hydrophobic interactions with Thr-146. The 9-O-Ac carbonyl participates in a hydrogen bond with the Tyr-155 hydroxyl group. Molecular dynamic simulations of the Neu5,9Ac2α2Me–VP8* complex were carried out using AMBER/GAFF at 300 K and 300 ps. The ligand Neu5,9Ac2α2Me and protein were kept flexible during the simulation.
To investigate the possibility of Neu5Acα2Me-like binding modes of Neu5Ac2en, we positioned the ligand manually into the RRV VP8* Sia-binding domain by superimposing Neu5Ac2en on Neu5Acα2Me, as positioned in the crystal structure (Dormitzer, Sun, Wagner, et al. 2002). One possible binding orientation of Neu5Ac2en was derived by the superimposition of the Neu5Ac2en N-acetamide moiety on the Neu5Acα2Me N-acetamide moiety. In this orientation, although the hydrophobic interactions of the methyl group of the N-acetamide moiety in Neu5Ac2en with VP8* are maintained, other key interactions between the ligand's carboxylic acid and Ser-190 are lost. A second orientation was derived by the superimposition of the Neu5Ac2en carboxylate moiety onto the Neu5Acα2Me carboxylate moiety. This potential bound conformation revealed that although the hydrogen bonds between the ligand's carboxylate moiety and Ser-190 are maintained, the hydrophobic interactions made by the methyl group of the N-acetamide moiety are not possible. More importantly, the hydrogen bond formed by the N–H of the N-acetamide moiety in Neu5Ac2en with the backbone carbonyl of Tyr-188 would be disrupted. The unsaturated N-acetylneuraminic acid derivative Neu5Ac2en has a distinct half-boat conformation owing to the presence of the geometrically defining double bond. This ligand is conformationally distinct from Neu5Acα2Me in that the latter adopts a chair conformation (Figure 7), and as a consequence, the unsaturated ligand cannot interact to the same extent as Neu5Acα2Me. The loss of key interactions between Neu5Ac2en and VP8*, and therefore affinity, is further supported by the lack of any apparent STD NMR effects.
Effects of Neu5Acα2Me, Neu5Ac-α(2,6)-S-Galβ1Me, Neu5,9Ac2α2Me, and Neu5Ac2en on rotavirus infection
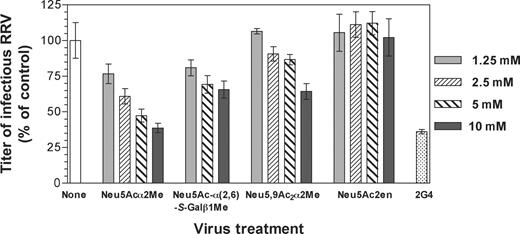
The ability of these N-acetylneuraminic acid derivatives to inhibit RRV rotavirus infection of permissive cells was assessed. As shown in Figure 10, Neu5Acα2Me, Neu5Ac-α(2,6)-S-Galβ1Me, and Neu5,9Ac2α2Me inhibited RRV infection of MA104 cells in dose-dependent fashion to a maximum at 10 mM of 61%, 35% and 36%, respectively. In contrast, Neu5Ac2en did not affect RRV infectivity at any concentration up to 10 mM
Effect of selected N-acetylneuraminic acid derivatives on RRV infection of MA104 cells. RRV was reacted with Neu5Acα2Me, Neu5Ac-α(2,6)-S-Galβ1Me, Neu5,9Acα2Me, or Neu5Ac2en prior to infection of cells at concentrations ranging from 1.25 to 10 mM, as described in Materials and methods section. As a positive control, RRV was reacted with neutralizing monoclonal antibody 2G4, directed to VP4, at a 1:5000 dilution of ascites fluid (2G4). This antibody does not completely neutralize RRV infection at this dilution. The infectivity titer of virus treated with ligand or antibody is expressed as a percentage of the titer obtained in the absence of ligand or antibody (none). The RRV titer obtained in the absence of ligand or antibody was 5.1×104 fluorescent cell-forming units per milliliter.
Discussion
Solution STD NMR spectroscopy provides an exciting and complementary technique to X-ray crystallography, which allows a direct investigation of the dynamic interactions of ligands with proteins of interest. Such studies can provide valuable information about what key ligand functionalities are essential for the ligand to successfully bind to the protein. In the present study, we have clearly demonstrated that epitope mapping by STD NMR spectroscopy provides information about the solution-binding mode of a number of N-acetylneuraminic acid derivatives to VP8* and that the NMR data for Neu5Acα2Me-VP8* complex are in very good agreement with the X-ray crystallographic structure (Dormitzer, Sun, Wagner, et al. 2002). Dormitzer, Sun, Wagner, et al. (2002) have identified the key amino acids that appear to be essential for the sialoside Neu5Acα2Me binding to the Sia-binding domain of VP8*. A number of these amino acid residues form either direct or indirect (water-mediated) hydrogen bonds or make van der Waals contacts with Neu5Acα2Me, which determines its overall binding conformation (Figure 7A). A solvent-accessible surface constructed over the binding site of RRV VP8* with bound Neu5Acα2Me (Figure 7B) clearly demonstrates that the Neu5Acα2Me sialoside is oriented to take advantage of a number of amino acids that create the Sia-binding domain. The methyl aglycon group of the sialoside Neu5Acα2Me, which substitutes for the penultimate residue of a glycoprotein's oligosaccharide side chain, was found in the X-ray structure of Neu5Acα2Me in complex with VP8* (Dormitzer, Sun, Wagner, et al. 2002) to face the open channel of the Sia-binding domain, which points away from the protein into the bulk solvent. The outcomes of the solution NMR spectroscopic investigation lead us to conclude that the methyl aglycon in both Neu5Acα2Me and Neu5,9Ac2α2Me does not receive saturation from the protein and confirms that the aglycon moiety of these sialosides is not in close contact with the protein surface (Figure 7B). Furthermore, the NMR studies also revealed that the glycerol side chain is important in binding, as evidenced through strong STD NMR effects for the protons H6, H7, H8, H9, and H9′ (Table I). In general, a good agreement between the presented STD NMR experimental data (one-dimensional and two-dimensional TOCSY STD NMR) and the X-ray crystal structure (Dormitzer, Sun, Wagner, et al. 2002) was found. For example, hydrogen bonds between the Neu5Acα2Me C8 and C9 hydroxyl groups with Arg-101 are reported (Dormitzer, Sun, Wagner, et al. 2002) and positions the associated glycerol side chain C8 and C9 C–H protons in close contact with the protein surface, made up in part by Tyr-155 (Figure 7A). The presented NMR data clearly show strong STD NMR effects for the Neu5Acα2Me C8 and C9 C–H protons and confirm that these protons are in close contact with the protein surface in the solution Neu5Acα2Me–VP8* complex. The adjacent Tyr-155 also contributes to hydrophobic interactions with the backbone C–H protons of the Neu5Acα2Me glycerol side chain C7 C–H proton and with the methyl group of the N-acetamide moiety that displays the most significant STD NMR effect. The strong STD effect observed for the N-acetamide's methyl group is readily explained by the observation that this group is in close contact with the hydrophobic aromatic rings Tyr-155 and Tyr-189 and the hydrophobic alkyl chain of Lys-187 (Figure 7B). A weak STD NMR effect was observed for the H3eq proton in contrast to no effect found for the H3ax proton of Neu5Acα2Me. An analysis of the X-ray structure (Dormitzer, Sun, Blixt, et al. 2002) reveals that the H3eq proton faces toward the protein surface, specifically residue Tyr 188, whereas the H3ax proton is directed into the solvent with no direct protein contact, supporting the presented NMR data and interpretation.
To explore the binding epitope of a Neu5Ac-containing disaccharide and particularly the role of the penultimate carbohydrate moiety in binding to RRV VP8*, we turned to a sialosylglycoside that contains a galactose residue as this moiety. The STD NMR data reveal that VP8* binds the sialoside Neu5Ac-α(2,6)-S-Galβ1Me via the N-acetylneuraminic acid component of the disaccharide. Interestingly, the observed STD effects for the N-acetylneuraminic acid moiety of Neu5Acα2Me and Neu5Ac-α(2,6)-S-Galβ1Me are comparable and lead us to the conclusion that RRV VP8* engages the ligands predominantly via the N-acetylneuraminic acid moiety. The chemical shift of one of the H9 protons of the N-acetylneuraminic acid moiety [H9′ (N)] in the ligand Neu5Ac-α(2,6)-S-Galβ1Me shows overlap with the chemical shift of the H3 (G) proton of the galactose unit. Although this overlap confounded interpretation of these data, a further two-dimensional STD TOCSY NMR experiment unequivocally demonstrated that the STD effect in this particular chemical shift regime originates almost purely from the H9′ (N) proton of the N-acetylneuraminic acid moiety and not from the H3 proton of the galactose unit. Interestingly, the galactose H6 (G) protons, which are adjacent to the sulfur linkage of the thiosialoside, receive saturation via the protein only to a minor extent, showing a residual weak STD NMR effect. We investigated this further by an MD simulation that examined the overall flexibility and the dynamics of the α(2,6) sulfur linkage in order to ascertain whether a conformation of ligand Neu5Ac-α(2,6)-S-Galβ1Me can be adopted with a close contact between the galactose moiety and the protein. The position of the N-acetylneuraminic acid moiety of Neu5Ac-α(2,6)-S-Galβ1Me was superimposed with bound ligand Neu5Acα2Me, as positioned in the published (Dormitzer, Sun, Wagner, et al. 2002) crystal structure. We believe that this approach is appropriate because our STD TOCSY NMR experiments revealed that Neu5Ac moiety of Neu5Ac-α(2,6)-S-Galβ1Me binds in a similar manner to RRV VP8* when compared with ligand Neu5Acα2Me. Figure 8 illustrates the results of the MD calculations of Neu5Ac-α(2,6)-S-Galβ1Me bound to VP8* in one potential conformation. None of the calculated conformations of Neu5Ac-α(2,6)-S-Galβ1Me (data not shown) allow contact of the protons of the galactose unit with the protein surface. This is in excellent agreement with our experimental STD NMR analysis revealing only very weak STD effects for H4 (G), H5 (G), and H6 (G) in Neu5Ac-α(2,6)-S-Galβ1Me in complex with RRV VP8*. In summary, this analysis clearly shows that the galactose-associated protons in Neu5Ac-α(2,6)-S-Galβ1Me are not in close proximity to the protein surface and are not involved in binding of this ligand to the protein. We have previously reported that synthetic thiosialosides with either an α(2,6) or an α(2,4) linkage between the N-acetylneuraminic acid and galactose units are equipotent inhibitors of in vitro rotavirus infection (Kiefel et al. 1996). Rolsma et al. (1994) have reported that α(2,3)- and α(2,6) sialyllactose are also equipotent inhibitors of in vitro rotavirus infection. Dormitzer, Sun, Wagner, et al. (2002) determined by NMR spectroscopy a KD value of 1.2 mM for 3′-sialyllactose and found that 6′-sialyllactose bound with comparable affinity. These data when taken together with the present study strongly support the notion that the RRV VP8* core recognizes a range of α-glycosides of N-acetylneuraminic acid and is independent of linkage. Furthermore, it would appear that the aglycon unit (penultimate carbohydrate unit and other associated carbohydrate, in the case of sialyllactose, galactose, and glucose, respectively) does not play a significant role in the binding of the saccharide chain. In the present investigation of ligand–RRV VP8* interactions by STD NMR spectroscopy and MD simulations, we have revealed, for the first time, direct evidence that the penultimate carbohydrate galactose moiety of Neu5Ac-α(2,6)-S-Galβ1Me does not interact strongly with the protein. Finally, in the present study, the majority of sampled conformations found for Neu5Ac-α(2,6)-S-Galβ1Me from the molecular modeling experiments confirm the solvent-exposed nature of the galactose sugar residue.
In our investigation of the ligand Neu5,9Ac2α2Me in complex with RRV VP8*, we observed the strongest STD NMR signal (100%) for the methyl group of the N-acetamide moiety compared with a STD NMR signal (56%) for the methyl group of the 9-O-Ac. As expected, on the basis of our previous observations, the methyl aglycon group did not receive any saturation, suggesting that this moiety is not in contact with the protein. To elucidate a potential binding mode of Neu5,9Ac2α2Me in more detail, we undertook an MD simulation of Neu5,9Ac2α2Me bound to VP8*. Two predominant bound conformations of Neu5,9Ac2α2Me were observed and they showed that a number of hydrophobic interactions occur between the methyl group of the N-acetamide moiety in Neu5,9Ac2α2Me, the aromatic rings of Tyr-155 and Tyr-189, and the alkyl chain of Lys-187 (Figure 9). In both of these observed conformations, the 9-O-Ac's carbonyl group is in hydrogen bonding distance to either Tyr-155 (Figure 9) or, alternatively, Arg-101. Furthermore, these bound structures also reveal a close proximity of the 9-O-Ac's methyl group with Thr-146 and is consistent with the STD NMR spectrum of Neu5,9Ac2α2Me complexed with VP8* at 298 K. This NMR spectrum revealed a relative STD NMR effect of 56% of the 9-O-Ac's methyl protons compared with the N-acetamide's methyl group in Neu5,9Ac2α2Me. The observation that there is a factor of approximately 2 difference in the STD NMR effects for the 9-O-Ac and N-acetamide in Neu5,9Ac2α2Me is readily explained by the fact the N-acetamide methyl group of Neu5,9Ac2α2Me in complex with RRV VP8* is located in a pocket within the binding cavity and is surrounded by, and in contact with, several hydrophobic motif-containing amino acids. In complete contrast, the 9-O-acetyl methyl group is predicted to interact only with Thr-146 and in addition appears to have a higher flexibility as shown by the MD simulation. A comparison of the bound structure of Neu5Acα2Me and Neu5,9Ac2α2Me within the Sia-binding domain of RRV VP8* reveals that the presence of a 9-O-acetyl group disrupts the hydrogen bond between the C9 hydroxyl group of Neu5Acα2Me and Arg-101 in the VP8*–Neu5,9Ac2α2Me complex. This partial disruption of interaction appears to, even though some additional hydrophobic interaction is acquired, contribute to a weaker affinity and overall less-intense STD NMR effects for Neu5,9Ac2α2Me. The inhibition of RRV infection by Neu5,9Ac2α2Me supports this hypothesis because this ligand showed only 36% inhibition of RRV infection, whereas Neu5Acα2Me showed 61% inhibition. This 1.7-fold decrease in inhibition of infection may be directly correlated with the lower affinity of Neu5,9Ac2α2Me for VP8*. To further investigate the flexibility of the 9-O-Ac moiety in Neu5,9Ac2α2Me bound to VP8*, an STD NMR study at a higher temperature was undertaken. The influence of probe temperature on STD NMR effects has been reported elsewhere by us (Haselhorst et al. 2004) and others (Wen et al. 2005). In an STD NMR experiment, performed at 298 K, STD effects for the carbohydrate ring protons and the N-acetamide methyl group were observed. However, there was complete loss of the STD effect for the 9-O-Ac's methyl group. This observation suggests that the 9-O-Ac moiety is indeed flexible. It is therefore not surprising that the flexibility of the additional 9-O-Ac influences the overall affinity of Neu5,9Ac2α2Me to RRV VP8*, resulting in a decreased interaction of this group with the Sia-binding domain and consequently a decrease in inhibition of RRV infection of MA104 cells. Further evidence that Neu5,9Ac2α2Me may bind weaker than the parent ligand Neu5Acα2Me to RRV VP8* is provided by the fact that the STD NMR signal intensities originating from the glycerol side chain of Neu5,9Ac2α2Me (Figure 6B) are not observed. For the related de-O-acetylated parent ligand Neu5Acα2Me, strong STD NMR effects for H7, H8, and H9 protons could be detected, which is in accord with the X-ray structure (Dormitzer, Sun, Wagner, et al. 2002).
In the case of the unsaturated N-acetylneuraminic acid derivative Neu5Ac2en, it was found that this ligand cannot maintain key interactions with the protein. As a result of the half-boat-like conformation, Neu5Ac2en cannot interact, through its carboxylate, with Ser-190 and simultaneously maintains the important hydrophobic interactions between the methyl group of the N-acetamide moiety and Tyr-189, Tyr-155, and Lys-187. The STD NMR data strongly support the notion that Neu5Ac2en should have little, if any, binding affinity for VP8*. Furthermore, the infection inhibition assay results with Neu5Ac2en (Figure 10) indicate that 10-mM Neu5Ac2en does not inhibit RRV infection of MA104 cells. The infection inhibition data are in excellent agreement with the presented STD NMR and modeling data and show that Neu5Ac2en, most likely as a consequence of an inability to maintain a number of key interactions that Neu5Acα2Me utilizes, does not affect RRV VP8* function.
In summary, NMR spectroscopy provides an excellent opportunity to undertake solution-based ligand–protein investigations that facilitate the rapid identification of ligand interactions with the protein of interest at the molecular level. Our STD NMR analysis and MD simulations of the interactions of N-acetylneuraminic acid derivatives in complex with RRV VP8* protein provide new information about the biologically relevant binding of N-acetylneuraminic acid derivatives to RRV VP8*. Here we present for the first time that N-acetylneuraminic acid glycosides bind to RRV VP8* in solution using an identical epitope. This information may contribute to the rational design of new N-acetylneuraminic acid-based compounds to inhibit rotavirus infection.
Materials and methods
Cloning, expression, and purification of RRV VP8*64–224
RRV VP8* was cloned and expressed using a modification of a previously described method (Dormitzer, Sun, Wagner, et al. 2002). RRV VP4 cDNA was used as a template for PCR, using the expand high-fidelity polymerase chain reaction (PCR) system (Roche Diagnostics, Castle Hill, Australia) to amplify the VP8* gene fragment for cloning into the bacterial expression vector pGEX-4T-1 (Amersham Biosciences, Castle Hill, Australia). A single round of PCR was performed using the forward primer 5′-CGCGGATCCGTACTTGATGGTCCTTATCAACC-3′ and the reverse primer 5′-GGAATTCTCATAACCCGTTATTAATGTACTCGG-3′ containing BamHI and EcoRI restriction sites (bold). Following digestion, PCR products were ligated into pGEX-4T-1, yielding pGEX-RRV-VP8*, which encoded amino acid residues 64–224 of RRV VP4, fused to the C-terminus of glutathione-S-transferase (GST). The integrity of the inserted VP8* gene fragment was assessed by DNA sequencing. The predicted amino acid sequence of pGEX-RRV-VP8* was identical with the published RRV VP4 sequence (Accession number AY033150, Entrez Database). An optimized protein expression protocol was developed from that previously reported (Dormitzer, Sun, Wagner, et al. 2002). Thus, Escherichia coli strain BL21 DE3, transformed with the pGEX-RRV-VP8* plasmid encoding the GST fusion protein, were grown at 37 °C in Luria-Bertani media supplemented with 150 µg/mL ampicillin. Once an OD600 of 0.6 had been reached, the cultures were incubated at 25 °C for 1 h and then induced with 1 mM isopropyl α-D-thiogalactopyranoside (BioVectra DCL, Templestowe, Australia). Cells were harvested 4 h after induction by centrifugation at 8000g for 10 min. Frozen cell pellets were thawed in phosphate buffered saline (pH 7.3) (comprising 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4) supplemented with 1 mM phenylmethylsulfonyl fluoride (PMSF, Roche Diagnostics) and lysed with lysozyme (1 mg/mL) supplemented with 1% wt/vol Triton X-100 and DnaseI (20 µg/mL). Cell debris was removed by centrifugation at 20 000g for 30 min and the supernatant was passed over a glutathione–Sepharose column (Amersham-Pharmacia Biotech). The column was washed with 20 mM Tris (pH 8.0), containing 100 mM NaCl and 1 mM CaCl2 (TNC), and bound GST-fusion protein was digested with 8 µg/mL TPCK-treated trypsin (Worthington Biochemical, Lakewood, NJ) for 2 h at room temperature. A benzamidine–Sepharose (Amersham-Pharmacia Biotech) column pre-equilibrated with TNC was connected in series with the glutathione–Sepharose column. The cleaved protein was eluted with 20 mM NaPO4 (pH 7.5), containing 1 M NaCl and 1 mM PMSF, and 2.5 mM benzamidine was added to the eluant. The protein was dialyzed against 20 mM NaPO4 (pH 7.0) and 10 mM NaCl and concentrated to approximately 10 mg/mL (DC Assay, Bio-Rad Laboratories, Regents Park, Australia) and analyzed for purity and homogeneity by SDS–PAGE and dynamic light scattering using a CoolBatch+90T instrument (Precision Detectors, Blackburn, Australia).
Sample preparation for NMR analysis
Neu5Acα2Me (Kononov et al. 1998) and Neu5Ac-α(2,6)-S-Galβ1Me were synthesized in-house as previously described (Kiefel et al. 1996). Neu5,9Ac2α2Me was synthesized from Neu5Acα2Meusing trimethylorthoacetate and p-toluenesulfonic acid in an analogous way to that described for other N-acetylneuraminides (Kiefel et al. 1996). Neu5Ac2en was prepared according to previously published procedures (Meindl and Tuppy 1969). Deuterium oxide (99.9%, deuterium) was purchased from (Novachem Pty Ltd., Collingwood, Australia). NMR samples were prepared by dissolving RRV VP8* protein (0.5 mg, 46 µM) and a quantity of ligand to give a protein:ligand mole ratio of 1:100 in 600 µL of 20 mM phosphate buffer (pH 7.1), containing 10 mM NaCl, for all ligands resulting in a final ligand concentration of 4.6 mM. NMR samples for STD TOCSY experiments were prepared in a similar fashion with a concentration of 0.2 mM RRV VP8* and 20 mM ligand Neu5Acα2Me and Neu5Ac-α(2,6)-S-Galβ1Me to give a protein:ligand mole ratio of 1:100 in 600 µL buffer.
Standard NMR experiments
All NMR experiments were performed on a (Bruker Avance, Alexandria, Australia) 600 MHz spectrometer, equipped with a 5-mm TXI probe with triple axis gradients. The measurements were performed at 298 and 288 K without sample spinning. 1H NMR spectra were acquired with 32 scans, a 2 s relaxation delay over a spectral width of 6000 Hz. Solvent suppression of the residual HDO peak was achieved by continuous low-power presaturation pulse during the relaxation delay. Data acquisition and processing were performed with XWINNMR software (version 3.1) running on a Silicon Graphics O2 workstation. Standard TOCSY NMR spectra were recorded with a 70-ms TOCSY mixing time, 16 scans per t1 increment. 512 t1 increments were collected, resulting in a total acquisition time of approximately 6 h.
One-dimensional STD
In the STD NMR experiments of Neu5Acα2Me, Neu5Ac-α(2,6)-S-Galβ1Me, and Neu5,9Ac2α2Me in complex with VP8*, the protein was saturated at 7.13 ppm in the aromatic/amide region of the spectrum and off-resonance at 33 ppm with a cascade of 40 selective Gaussian-shaped pulses of 50 ms duration (50 dB), which correlates to a strength of 190 Hz. A 100 µs delay between each soft pulse was applied, resulting in a total saturation time of 2 s. In the STD NMR experiment of a VP8*–Neu5Ac2en complex, the protein was saturated at 7.32 ppm as a consequence of the H3 proton resonance of Neu5Ac2en being too close to the original saturation frequency and to ensure that no ligand signals were effected by the saturation pulse cascade. Data were obtained with an interspersed acquisition of pseudo-two-dimensional on-resonance and off-resonance spectra in order to minimize the effects of temperature and magnet instability. On- and off-resonance spectra were processed separately, and the final STD NMR spectrum was obtained by subtracting the individual on- and off-resonance spectra, resulting in less subtraction artifacts. The WATERGATE pulse train was used to suppress HDO signals. Relative STD effects were calculated according to the equation ASTD=(I0−Isat)/I0= ISTD/I0 by comparing the intensity of the signals in the STD NMR spectrum (ISTD) with signal intensities of a reference spectrum (I0). The STD signal with the highest intensity was set to 100% and other STD signals were calculated accordingly. A spin lock field of 10 ms was applied to remove unwanted background protein signals. Increased spin lock fields resulted in artifacts and reduced ligand signal intensities. Control STD NMR experiments were performed using an identical experimental setup and the same ligand concentration but in the absence of the protein
Saturation transfer difference-total correlation spectroscopy
STD TOCSY spectra (Haselhorst et al. 2001; Mayer and Meyer 2001) were recorded with a mixing time of 70 ms, 160 scans per t1 increment; 300 t1 increments were collected resulting in a total acquisition time of approximately 30 h. The spectra were multiplied with a square cosine bell function in both dimensions and twice zero-filled. The protein was saturated in the aromatic and amide proton regions of the spectrum at 7.13 ppm and off-resonance at 33 ppm, with a cascade of 40 selective Gaussian-shaped pulses of 50 ms duration, with a 100 µs delay between each pulse, resulting in a total saturation time of 2 s. STD NMR spectra were recorded at 298 K. The WATERGATE pulse train was used to suppress HDO signals.
Molecular modeling studies of ligands Neu5Ac-α(2,6)-S-Galβ1Me and Neu5,9Ac2α2Me in complex with RRV VP8*
The initial structures of the ligands Neu5Ac-α(2,6)-S-Galβ1Me and Neu5,9Ac2α2Me were built and bond order and atom types assigned using Insight II (Accelrys, San Diego, CA) and were oriented into the VP8*-binding site by superimposition of the corresponding heavy atoms of the N-acetylneuraminic acid residue as revealed in the previously reported X-ray crystallographic structure (Dormitzer, Sun, Wagner, et al. 2002). MD simulations of Neu5Ac-α(2,6)-S-Galβ1Me were performed at 1000 K using the consistent valence force field (Dauber-Osguthorpe et al. 1988) as implemented in the DISCOVER 2.98 program (Accelrys, San Diego, CA). MD simulations of Neu5,9Ac2α2Me were performed at 300 K and 300 ps in explicit solvent using the AMBER 1999 force field (Wang et al. 2000) for the protein and the GAFF parameters (Wang et al. 2004) for the ligand as implemented in the AMBER 8 distribution (Pearlman et al. 1995; Case et al. 2004).
Assay for inhibition of rotavirus infection
The propagation of rotavirus strain RRV and performance of this assay were as previously described (Kiefel et al. 1996). In summary, serial dilutions of the ligands were incubated with RRV at a multiplicity of infection of 0.02 for 1 h at 37 °C. Virus–ligand mixtures were inoculated onto confluent MA104 cell monolayers and incubated for 1 h at 37 °C. After removal of inoculum and incubation for a further 15 h, virus infectivity was determined using indirect immunofluorescence staining of infected cells. Virus titers are expressed as the number of fluorescent cell-forming units per milliliter.
Acknowledgments
MJK, HB, BSC, and M von I gratefully acknowledge the financial support of the Australian Research Council. T.H. thanks Peter Barron, Bruker Australia for technical support. Erich Mackow, Department of Medicine, Stony Brook University, Stony Brook, NY, is thanked for kindly providing the RRV VP4 plasmid. B.S.C. is a senior research fellow of the National Health and Medical Research Council of Australia. M. von I. also thanks the Australian Research Council for the award of an Australian federation fellowship and the National Health and Medical Research Council for financial support.
Conflict of interest statement
None declared.
Abbreviations
- ax
axial
- eq
equatorial
- GST
glutathione-S-transferase
- MD
molecular dynamics
- MLEV
Malcolm Levitt's composite-pulse decoupling sequence
- Neu5Ac2en
2-deoxy-2,3-didehydro-D-N-acetylneuraminic acid
- Neu5Acα2Me
methyl α-D-N-acetylneuraminide
- Neu5,9Ac2α2Me
methyl 9-O-acetyl-α-D-N-acetylneuraminide
- Neu5Ac-α(2,6)-S-Galβ1Me
methyl S-(α-D-N-acetylneuraminosyl)-(2→6)-6-thio-β-D-galactopyranoside
- NMR
nuclear magnetic resonance
- PCR
polymerase chain reaction
- PMSF
phenylmethylsulfonyl fluoride
- Sia
sialic acid
- STD
saturation transfer difference
- TOCSY
total correlation spectroscopy
- 9-O-Ac
9-O-acetate.
References
Author notes
These authors contributed equally to this work.



![Two-dimensional TOCSY [(A) and (C)] and STD TOCSY [(B) and (D)] NMR spectra of 0.2 mM of RRV VP8* complexed with Neu5Acα2Me at a mole ratio of 1:100. Saturation of the protein was achieved with a Gaussian pulse cascade (40 Gaussian pulses of 50 ms duration, each with a delay of 100 µs in between each pulse) with a total saturation time of 2 s. The on-resonance saturation frequency was set to 7.13 ppm and off-resonance to 33 ppm; 300 t1 increments were collected in total with 160 scans per increment and a Malcolm Levitt's composite-pulse decoupling sequence (MLEV) mixing time of 70 ms. All spectra were acquired in 20 mM phosphate buffer, pH 7.1, containing 10 mM NaCl, at 298 K and 600 MHz.](https://oup.silverchair-cdn.com/oup/backfile/Content_public/Journal/glycob/17/1/10.1093/glycob/cwl051/2/m_cwl05103.jpeg?Expires=1716436491&Signature=pFWQG6xydjXbAOyFWw37~fmyI6omCnlkIl-8IQPDPM0N0cefj~mRSJt1vd7pvvY-9z~eokJEYB6bbgQl3I3LBmr-c04YW6adnwVniMhMcxitPlrH~Qx99eIp7bv40Ss9iy2BZg9oBhNJC~5ezo4c90znLchs8KMHWzf2ZbpOFg493BIf2xpVsxBvMKTga76TP7Mq9b4mZqVmGsIZVJ~qNniXmsYLWTHnl4QilAfU7UPfr6MH2fZnyy8EsPAy4fwpPbjSnetDKAwCFEiNwlryfD41WcjtjdsNnrfiMGMhswXoC3uP5p2cFe77GEPY1l6sviAHh06LvvbJIit~iZaTyw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Two-dimensional TOCSY [(A) and (C)] and STD TOCSY [(B) and (D)] NMR spectra of 0.2 mM of VP8* complexed with Neu5Ac-α(2,6)-S-Galβ1Me at a mole ratio of 1:100. Saturation of the protein was achieved with a Gaussian pulse cascade (40 Gaussian pulses of 50 ms duration, each with a delay of 100 µs in between each pulse) with a total saturation time of 2 s. The on-resonance saturation frequency was set to 7.13 ppm and the off-resonance to 33 ppm; 300 t1 increments were collected in total with 160 scans per increment and an MLEV mixing time of 70 ms. All spectra were acquired in 20 mM phosphate buffer, pH 7.1, containing 10 mM NaCl, at 298 K and 600 MHz.](https://oup.silverchair-cdn.com/oup/backfile/Content_public/Journal/glycob/17/1/10.1093/glycob/cwl051/2/m_cwl05105.jpeg?Expires=1716436491&Signature=do0nmTBXqi0UjpTv-9GhDfuO9HzGNf7c9Eb~OTfg1-NOuVzAnkhwIL6Knkae0I16n7biq5xHrSOtaqoCwbkX9z9t2l7rRq5qcx7tntHlqBq-7oiv~F6ZumRc8mbwdjOFj~Wa6TTFHscUm8g2rjXZYdWH3adrhwT6MAwT4HGObo09KGqL5Lrl5QjGlLDTfWbFCUhVpr1sx1wYr7E6Z~Usdvu66Zx2NpTtXp4eHF1dU~TdQs5E-C4RG9zEtw7l2e7k~eR3hUazAGHvyYx~rn0QPTLACA5~lY4A87to3mzhgOfjPJ-W1edWsGJjG33bku1~mLfPd0gSXhrEu8pa4bbf4w__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)