-
PDF
- Split View
-
Views
-
Cite
Cite
Madeleine J. H. van Oppen, Ciro Rico, George F. Turner, Godfrey M. Hewitt, Extensive Homoplasy, Nonstepwise Mutations, and Shared Ancestral Polymorphism at a Complex Microsatellite Locus in Lake Malawi Cichlids, Molecular Biology and Evolution, Volume 17, Issue 4, April 2000, Pages 489–498, https://doi.org/10.1093/oxfordjournals.molbev.a026329
Close - Share Icon Share
Abstract
Recent studies have suggested that size homoplasy is a prevalent feature of microsatellites and is expected to increase with time of divergence among populations and taxa. In this study, we performed sequence analysis of alleles from a complex microsatellite locus (Pzeb4, initially isolated from Pseudotropheus (Maylandia) zebra) from 1 midwater-feeding and 10 rock-dwelling cichlid fish species from Lake Malawi, East Africa, to investigate how widespread size homoplasy is among closely related taxa at this locus. All cichlid fishes endemic to this lake are believed to have originated within the last 700,000 years, and some species may be less than 200 years old. The number of eletromorphs found per species varied from 3 to 13. Sequence analysis of 95 cloned Pzeb4 PCR products (representing 18 electromorphs) revealed 13 new alleles. Ten of the 13 electromorphs (77%) were found to show size homoplasy due to either single nucleotide substitutions/indels or large indels. To investigate how well this locus fits the single-step mutation model (SMM), the minimum number of mutations required to explain the length differences between pairs of alleles was plotted against their size differences. Of the 300 comparisons, 166 (55.3%) corresponded to SMM expectations and 86 (28.7%) required a smaller number of mutations, and for 48 (16.0%) pairwise comparisons, a larger number of mutations were required to explain the length differences as compared with SMM expectations. Finally, a large deletion in the microsatellite sequence observed in the three rock-dwelling species Pseudotropheus lucerna, Pseudotropheus (Tropheops) ‘band,’ and Pseudotropheus (Tropheops) ‘rust’ and the midwater-feeding species Copadichromis sp. is believed to represent a shared ancestral polymorphism.
Introduction
Microsatellites are tandem repeats of simple short sequences (1–6 bp in length) that are scattered throughout the nuclear genomes of most eukaryotes (reviewed in Jarne and Lagoda 1996 ). They have been found in plant chloroplast (Powell et al. 1995 ; Provan et al. 1996 ; Whitton, Rieseberg, and Ungerer 1997 ; Procaccini and Mazzella 1998 ) and mitochondrial genomes as well (Hoelzel et al. 1994 ; Soranzo, Provan, and Powell 1999 ). In general, microsatellites are highly polymorphic neutral markers exhibiting many alleles per locus and high heterozygosities. For this reason, they are often the preferred markers for population genetic, forensic, gene mapping, and parentage studies.
Estimates of population parameters such as structure, number of migrants, and effective population size are highly dependent on the mutational model assumed for the molecular markers of choice. For microsatellites in particular, this dependence is expected to be very strong due to the high mutation rate at these loci (Estoup and Angers 1998 ). Conventionally, two models of mutation have been considered for microsatellites. Classical divergence estimates for neutral loci, which follow the infinite-allele model (IAM; Kimura and Crow 1964 ) assume that each mutation creates a novel allele. However, Slatkin (1995) and Goldstein et al. (1995a, 1995b ) have suggested that the IAM is not appropriate for microsatellites because of high mutation rates and mutation processes that retain memory of the ancestral allelic states. Instead, these authors suggest that a stepwise mutation model (SMM; Kimura and Ohta 1978 ) would describe the evolution of these loci much more accurately. More recently, two additional models have been considered. Di Rienzo et al. (1994) introduced a two-phase model (TPM) which allows loss and gain of several repeats following a geometric distribution. Another classical model, the K-allele model (KAM) (Crow and Kimura 1970 ), assumes a finite number of K allelic possibilities with a constant probability (μ/K − 1) of mutating toward any other K − 1 allelic state (Estoup and Cornuet 1999 ).
Characterization of a single microsatellite locus in different organisms has led to inconclusive results. For example, all 10 de novo mutations at a (GA)n locus found in the ant Camponotus consobrinus represented increases or decreases of a single step (Crozier et al. 1999 ). However, both single-step mutations (four out of a total of nine mutations) and larger mutations of at least three repeat units (five of the nine mutations) were observed in one dinucleotide locus in Drosophila melanogaster (Schlötterer et al. 1998 ), and 7 out of 44 mutations involved 2–5 repeat units in one tetranucleotide locus in the barn swallow, while the majority (37 out of 44) were single-step mutations (Primmer et al. 1998 ). More recently, Jones et al. (1999) observed that in the pipefish Syngnathus typhle, 23 out of 26 mutations in one tetranucleotide locus differed from their progenitors by a single repeat unit, 2 by two units, and 1 by three units, supporting the general view that some mutations do not conform to SMM.
Multilocus analyses have yielded similar results. Shriver et al. (1993) studied the evolution of human microsatellites and found that none of the tri- to pentanucleotide repeats examined were different from the SMM simulations for three summary statistics (allelic size range, number of modes [alleles that are more frequent than both the allele one step larger and the allele one step smaller], and number of alleles), while 35% of the dinucleotide repeats were different for at least one of the three summary statistics. Valdes, Slatkin, and Freimer (1993) , however, found that allele frequencies at 108 human dinucleotide loci are consistent with the SMM in a population of constant size, and direct studies of mutations in human families have demonstrated that almost all mutants differ from their ancestors by one or two repeat units (e.g., Mahtani and Willard 1993 ; Weber and Wong 1993 ; Bowcock et al. 1994 ). Additionally, Di Rienzo et al. (1994) found that allele frequency distributions at 10 microsatellite loci in a Sardinian human population had a better fit with the TPM, and Colson and Goldstein (1999) found in an extensive survey of microsatellite evolution in Drosophila that in only 7 out of 19 loci size variation among species was consistent with the occurrence of strictly stepwise mutations. To date, the adequacy of any mutation model for microsatellite allelic distributions in natural populations has not been unequivocally validated. This presumably reflects a far more complex mutation process than is assumed by the available models and that these processes vary greatly among loci (see Estoup and Cornuet [1999] for a detailed review on microsatellite evolution).
An additional concern when using microsatellite frequencies to estimate population parameters is size homoplasy. Microsatellite alleles of the same size (electromorphs) can arise from mutational events outside of the repeat or by interrupting a perfect repeat producing alleles that are not identical by descent. For example, Estoup et al. (1995b ) performed sequence analysis of electromorphs from two interrupted microsatellite loci in two bee species and reported extensive homoplasy due to mutations, which did not involve gain or loss of repeats. Similarly, Angers and Bernatchez (1997) performed sequence analysis of 78 alleles of a microsatellite locus at various taxonomic levels among salmonid fishes and showed that in addition to changes in the repeat number, large indels as well as single base substitutions within the repeat region have occurred frequently among very diverged species, as well as within species. It has also been found that the flanking regions tend to be more variable in close proximity to the repeat region (Blanquer-Maumont and Crouau-Roy 1995 ; Garza, Slatkin, and Freimer 1995 ; Zardoya et al. 1996 ; Grimaldi and Crouau-Roy 1997) . Therefore, the occurrence of size homoplasy should be taken into account when estimating population parameters. For example, Viard et al. (1998) demonstrated the effect of size homoplasy on the resolution of population structure in three invertebrate species. When sequencing information was considered in the definition of alleles, several single-locus pairwise tests of population differentiation became significant, and nonstepwise estimators of genetic structure were larger.
Since size homoplasy is expected to increase with time of divergence among populations and taxa (Estoup and Cornuet 1999 ), the above findings are not completely unexpected because most of the species studied have long divergence times. However, studies documenting size homoplasy among recently evolved taxa are less common (e.g., Metzgar et al. 1998 ; Taylor, Sanny, and Breden 1999 ). In this study, we performed sequence analysis of alleles from a complex microsatellite locus (Pzeb4; van Oppen et al. 1997a ) among 11 closely related species, including 1 midwater-feeding and 10 rock-dwelling cichlid fish species from Lake Malawi, East Africa. All cichlid fishes endemic to Lake Malawi have evolved over the last 700,000 years (Meyer 1993 ). Primarily, we wanted to investigate how widespread size homoplasy is at this locus in order to get a better understanding of its mode of evolution. Second, we aimed to test the effect of this locus on the population structure estimates based on the IAM and the SMM. A final interest was to find out whether any potentially phylogenetically informative mutations are present within this locus, as has been previously reported for microsatellite flanking regions (Zardoya et al. 1996 ).
Materials And Methods
Sampling
Ten rock-dwelling cichlid species (mbuna) were included in the present study. All of these species occur sympatrically at Nkhata Bay, situated on the western shore of the lake. In a previous study of nine of these species, population genetic analysis of six microsatellite loci in combination with behavioral observations showed that most of these species are true biological species. Significant genetic differences exist among all pairs of species, but the differences are small (van Oppen et al. 1998 ). Nine of the rock-dwelling cichlid species (mbuna) used in this study belong to two closely related subgenera, Pseudotropheus (Maylandia) and P. (Tropheops). A tenth species that was studied, Pseudotropheus lucerna, is thought to be more distantly related to both P. (Maylandia) and P. (Tropheops) than these subgenera are related to each other (Ribbink et al. 1983 ). Finally, a monospecific shoal of a midwater zooplankton feeder of the cichlid genus Copadichromis—called “utaka” by the local people (species identity unknown)—was included to make a comparison with a slightly more distantly related taxon. Copadichromis is believed to be a sister group to the mbuna (Albertson et al. 1999 ).
Samples were collected from four rocky patches within Nkhata Bay (in 1995 and 1997) and from three more northerly sites, Cape Manulo, Ruarwe, and Mara Rocks (in 1997), in Lake Malawi as described in van Oppen et al. (1997b ). Preliminary analysis indicated the presence of a deletion of a low-variability region of the microsatellite and that this deletion was shared by Pseudotropheus (Tropheops) ‘band’ and P. lucerna (see Results). This was unexpected, as previous authorities had not considered these species particularly closely related. Thus, we collected additional samples of these two species from Nkhata Bay, screening a further 49 P. ‘band’ and 54 P. lucerna for short alleles. Of these, sequences were obtained for 10 ‘band’ and 7 P. lucerna originally identified as carriers of the small alleles from both homozygous and heterozygous individuals, with the exception of 6 individuals (three P. ‘band’ and three P. lucerna) for which no good sequences could be produced. In addition, we examined short alleles discovered in mbuna samples collected at more northerly sites. From the Nkhata Bay samples, a range of alleles were cloned and sequenced, whereas only alleles from individuals that carried short alleles (<119 bp) were sequenced from the three northerly sites. Longer alleles from the northerly sites come from heterozygous individuals. DNA was extracted and PCR-amplifications were performed as described in van Oppen et al. (1997a) . For cloning purposes, PCR was carried out in 50-μl reaction volumes and using high-fidelity Bio-X-act (Bioline, London, England) instead of BioTaq (Bioline), to minimize PCR errors.
T-Vector Cloning of PCR Products
PCR products were run on a 1.2% TBE-agarose gel stained with ethidium bromide (EtBr) and were excised from the gel on the UV-box using a razor blade to remove nucleotides, primers, and primer dimers. The gel slices were placed in 0.5-ml punctured eppendorf tubes filled with nonabsorbent cotton wool. The 0.5-ml eppendorf tubes were placed in 1.5-ml eppendorf tubes and were spun in an eppendorf centrifuge for ca. 5 min at 12,000 rpm. The DNA in TBE was recovered at the bottoms of the 1.5-ml eppendorf tubes, while the agarose remained in the cotton wool. The DNA was precipitated and resuspended in 10 μl H2O. If some agarose had coprecipitated, the samples were heated to 45°C for 3–5 min. One microliter was run on a gel to check the product size and to obtain a rough estimate of its concentration.
Approximately 10 μg of M13mp18 vector (Pharmacia, St. Albans, Herts, England) was digested overnight at 37°C with 2 U of SmaI (Gibco BRL, Life Technologies, Paisley, Scotland). The vector was then precipitated with 10 μl 2 M NaCl and 250 μl 100% ethanol at −20°C for 30 min, followed by a centrifugation step of 20 min at 12,000 rpm in an eppendorf centrifuge. The pellet was washed once with ice-cold 70% EtOH, air-dried, and resuspended in 86.8 μl H2O. To add the T-overhang, 2 μl 100 mM dTTP (Boehringer Mannheim Ltd., Lewes, England), 10 μl 10 × KCL-buffer (Bioline), and 1.2 μl BioTaq (5 U/μl; Bioline) was added to the vector. Seventy-five microliters of mineral oil was added as an overlay, and the mixture was incubated at 70°C for 2 h in a heating block. The oil was then removed by running the sample off a strip of parafilm, and the vector was purified using the Wizard DNA clean-up system (Promega Biotec, Madison, Wis.) according to the manufacturer's protocol. DNA was eluted in sterile H2O and run on a 1% EtBr-stained TBE-agarose gel against 100 ng of undigested M13mp18 to check whether the digestion had been complete and to estimate the DNA concentration.
The PCR product was ligated into M13mp18 by mixing 2 μl of PCR product (0.1–0.2 pmol), 1 μl of T-vector (∼100 ng), 2 μl of T4 DNA ligase (1 U/μl, Gibco BRL, Life Technologies) and 2 μl 5 × ligation buffer (Gibco BRL, Life Technologies). Three microliters of sterile H2O were added to bring the total reaction volume up to 10 μl, and the mixture was incubated overnight at 15°C. Transformation and plating were performed using standard procedures (Sambrook, Fritsch, and Maniatis 1989 ). From each plate, two to four transparent plaques were picked and placed in separate 1.5-ml eppendorf tubes containing 1 ml of LB medium using sterile Pasteur pipettes. The tubes were vortexed vigorously for 1 min. One microliter of this solution was used for PCR amplification to check for insert size. PCR was carried out in 11-μl volumes consisting of 1 μl of the M13 clone, 0.2 μl M13 universal primer (1.5 μM; Pharmacia), 0.2 μl M13 reverse primer (2.1 μM; Pharmacia), 1.1 μl 10 × KCl buffer (Bioline), 1.1 μl dNTPs (2 mM; Boehringer Mannheim Ltd.), 0.05 μl BioTaq (5 U/μl; Bioline), and 7.35 H2O. The mixture was overlaid with 10 μl of oil. The PCR profile used on an Omnigene Thermal Cycler (Hybaid) was as follows: 3 min at 94°C, followed by 30 cycles of 30 s at 94°C, 30 s at 55°C, and 30 s at 72°C. PCR products were then run against the 123 size marker (Gibco BRL, Life Technologies) on 1.2% TBE-agarose gels stained with EtBr to check for insert size.
Sequencing
Clones containing the correct insert were grown in LB medium (Sambrook, Fritsch, and Maniatis 1989 ) and purified using the QIAprep spin M13 single-stranded DNA purification kit (Qiagen Ltd., Dorking, England). One microgram of M13 ss-DNA was used for sequencing using the Thermo Sequenase cycle sequencing kit with 7-deaza-dGTP for fluorescent labeled primers (Amersham International plc, Little Chalfont, England) following the manufacturer's instructions. In addition to the fluorescein-labeled M13 universal primer (Pharmacia), a newly designed fluorescein-labeled M13 forward primer (5′-ACTGTTGGGAAGGGCGATCGGTGCGG-3′) was used for sequencing, because the 5′ end of the insert was sometimes missed when using the M13 universal primer. The cycle sequencing profile used on a Perkin Elmer/Cetus DNA thermal cycler 480 was as follows: 2 min at 95°C, followed by 20 cycles of 45 s at 50°C for universal primer (60°C for newly designed primer), 1 min at 72°C, and 45 s at 95°C. Sequence products were run on the A.L.F. sequencer (Pharmacia) using Sequagel Extended (National Diagnostics Ltd., Hessle, England).
Data Analysis
Sequences were aligned by eye using a text editor and divided into five parts for ease of analysis (see Results). The smallest number of mutations between pairs of alleles from the Nkhata Bay samples (25 alleles) was calculated under the assumption that: (1) Length differences originate from single- step additions or deletions (i.e., a single mutation can cause an increase or decrease in length of one repeat unit only). (2) The deletion of part 3 (see fig. 3 ) is caused by one (or zero and four for comparisons with alleles 127b and 141b, respectively) slippage mutations plus one deletion of the remaining CGTGTC(TG)5. Based on our data set, it is impossible to know the precise mechanism by which part 3 became deleted. However, the fact that copy number variability exists in the (TG)n region—i.e., (TG)5, (TG)6, and (TG)9—but alleles with less than five TG repeats are absent makes it most likely that slippage is responsible for length variation between CGTGTC(TG)5 and CGTGTC(TG)9, while the deletion of CGTGTC(TG)5 is a single event. Under the alternative assumption that the deletion of part 3 is caused by five (or four and eight for comparisons with alleles 127b and 141b, respectively) slippage mutations plus one deletion of the remaining CGTGTC(TG)1, the results of the analysis to examine the fit conformance of locus Pzeb4 to the SMM expectations did hardly change (55.0% of the pairwise comparisons corresponded to SMM expectations, while for 25.7% and 19.3% of the comparisons, a smaller and a larger number of mutations, respectively, was required). (3) The deletion of four bases in part 3 is caused by a single event; the fact that no alleles with one, two, or three nucleotides deleted were observed suggests that a single deletion of four nucleotides is the most plausible scenario for this part of the repeat. (4) The deletion of part 4 is caused by a single deletion; the presence of the duplication of CG(TG)2 in allele 127c suggests that this part is behaving as a whole unit. Point mutations were not taken into account. In this manner, 300 comparisons were made and plotted against the difference in size between pairs of alleles.
The effects of Pzeb4 on the population structure estimates based on the IAM and the SMM were evaluated by calculating fixation indices with and without the data from this locus for all pairwise comparisons of nine P. (Maylandia) and P. (Tropheops) taxa. The data set used for this analysis is the same as the one reported in van Oppen et al. (1998) , which includes four additional uninterrupted loci and one compound locus. Due to the effort that would be required for sequencing all alleles for the 650 individuals genotyped at this locus in the above study, it was beyond the scope of this analysis to estimate population structure considering sequencing information in the definition of alleles (e.g., Viard et al. 1998 ). The extent of population subdivision between samples was investigated by calculating fixation indices using nonparametric permutational procedures to test for significance. The computer program Arlequin 1.1 was used to calculate Weir and Cockerham's (1984) unbiased estimates of Wright's (1951)F-statistics (𝛉) and ϕst (as defined by Michalakis and Excoffier 1996 ), an analog of Slatkin's (1995)Rst. The significance of genetic subdivision under the assumptions of both the IAM and the SMM was assessed using 1,000 permutations and bootstraps for all loci and sample pairwise comparisons. To correct for multiple simultaneous comparisons, sequential Bonferroni corrections were applied using a global significance level of 0.05 (Rice 1989 ).
Results
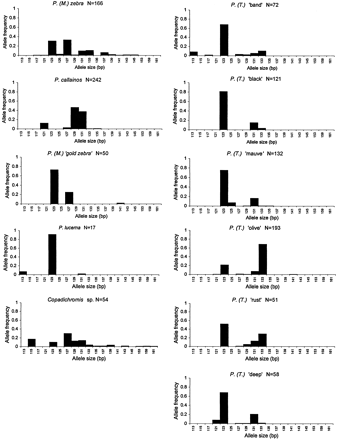
Figure 1 shows the allele frequency distribution of locus Pzeb4 for the 11 investigated taxa using the samples collected at Nkhata Bay in 1995. For P. lucerna, the 1995 sample consisted of only 17 individuals and thus may not reflect the real allele frequency distribution for this species. For the 1997 samples, we concentrated only on individuals possessing short alleles; hence, we could not increase the sample sizes for allele frequency distributions of P. lucerna and P. (Tropheops) ‘band’ using these samples. The number of alleles found for each of the 11 taxa varied from 3 to 13. The 105-bp allele was not present among these samples but occurred in a single individual collected in 1997. This is probably a very rare allele in the Nkhata Bay populations.
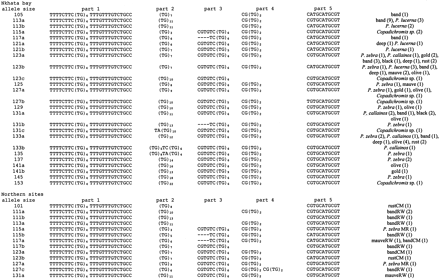
The sequences were divided into five different parts in order to facilitate the comparison between alleles (fig. 2 ). Parts 1 and 5 represent the flanking regions. Part 1 was monomorphic in all species, while part 5 presented two different transitions in the first two nucleotides, which resulted in three different sequences. Part 3 of the microsatellite sequence was absent from alleles 101, 105, 111a, 111b, 113a, and 113b. These alleles were present only in P. (Tropheops) ‘band,’ P. (Tropheops) ‘rust,’ and P. lucerna. Alleles 101 (P. (Tropheops) ‘rust’) and 111b (P. (Tropheops) ‘band’) are further characterized by an additional deletion of part 4 of the sequence. The deletion of part 4 (but not part 3) is also found in alleles 123c (Copadichromis sp.) and 117b (P. (Tropheops) ‘band’). We were unable to obtain the DNA sequence of allele 117 from Pseudotropheus (Maylandia) zebra, because repeated cloning of this fragment was unsuccessful.
Sequence analysis of all 95 clones containing alleles of Pzeb4 (representing 18 electromorphs) revealed 13 new alleles. We obtained two or more sequences for 13 different electromorphs. Of these, 10 (77%) were found to show size homoplasy (111, 113, 115, 117, 121, 123, 127, 131, 133, and 141). Some cases of size homoplasy were due to one or more point mutations between alleles, but moderately large indels were present as well. For example, alleles 101, 111b, 117b, and 123c have a 6-nt-long deletion (i.e., these lack part 4), and the 127 alleles have different repeat lengths in parts 2, 3, and 4, which have a 6-bp insertion in 127c. The same is true for the 141 alleles.
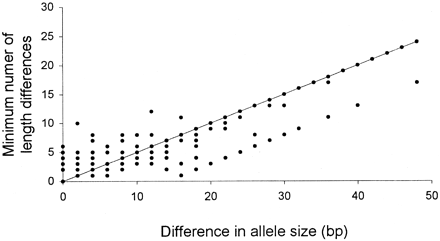
To investigate how well this locus fits the SMM, the minimum number of mutations required to explain the length differences between pairs of alleles was plotted against their size difference (fig. 3 ). Some of the points in figure 3 represent several pairwise comparisons. The solid line in the graph shows the expected number of mutations between alleles under a perfect fit of the SMM. Of the 300 comparisons, 166 (55.3%) correspond to SMM expectations and 86 (28.7%) require a smaller number of mutations, and for 48 (16.0%) pairwise comparisons, a larger number of mutations is required to explain the length differences as compared with SMM expectations.
Population structure estimates based on the IAM and the SMM were evaluated by calculating fixation indices for all pairwise population comparisons with and without the allele data from locus Pzeb4. As expected, the effects of this locus on the significance of 𝛉 values were important. Contrary to the analysis of fixation indices based on the IAM (table 1 ), when the allele data from this locus were excluded from the data set, the significance of six pairwise comparisons changed. Five previously nonsignificant fixation indices based on SMM became significant, and only one significant comparison became nonsignificant (table 1 ). On the other hand, the effects of this locus on the significance of 𝛉 values were less important, as only the significance of two pairwise distances between P. (Tropheops) ‘band’ with P. (Tropheops) ‘black’ and P. (Tropheops) ‘rust’ changed (became nonsignificant) when the allele data from Pzeb4 were excluded from the data set. Furthermore, ϕst values were generally lower than 𝛉 values for all significant comparisons.
Discussion
Our analysis of the locus Pzeb4 further substantiates the notion that size homoplasy should be considered when estimating population parameters based on microsatellite allele frequencies. In addition, it suggests that this concern is likely to be equally important for recently evolved taxa as for more distantly related species. This conclusion is based on the observation that several large indels and point mutations were present among these closely related taxa in the same way that Angers and Bernatchez (1997) detected complex mutations among several salmonid species that are known to have evolved several million years ago. Our results further indicate that it is essential to demonstrate conformance to SMM for microsatellite loci used in studies on both distantly related and closely related taxa when using SMM-based distances (e.g., Goldstein et al. 1995a ). Finally, this study shows that DNA sequence analysis of flanking microsatellite sequences for the purpose of phylogenetic analyses among recently diverged species may prove disappointing because insufficient time has probably passed for alleles to reach reciprocal monophyly.
Evolution of Locus Pzeb4
The double interruption of the TG repeats in part 3 of the sequence seems to be a common feature of this locus. Of the 95 alleles that were sequenced, 70 showed two interruptions. Five other alleles (115b, 117a, and 131b) showed single interruptions. All interruptions were caused by a transition from T to C at the 5′ end of part 3 and by a transversion form G to C at the 3′ end. The remaining 20 alleles lacked part 3 entirely. The interruption of the (TG)n array in part 4 was present in all sequenced alleles. The (TG)4 repeat in part 1 was invariable for the 95 alleles sequenced, as was the (TG)2 in the noninterrupted section of part 4. Allele 127c contains two repeats of CG(TG)2 in part 4, whereas all other alleles have only a single copy of this sequence. The most parsimonious explanation is that this sequence was duplicated in a single event and may represent the beginning of a hexanucleotide microsatellite. Alternatively, slippage has reduced this hexanucleotide from two repeats to just one. The same was found at an avian microsatellite dinucleotide locus, for which it was hypothesized that a (GA)2 repeat mutated to a (GA)1 and a (GA)3 via slippage (Primmer and Ellegren 1998 ).
More mutations than expected under the SMM were required to explain the length difference between alleles in 16.0% of the comparisons, and fewer mutations were needed in 28.7% of pairwise comparisons, indicating that the SMM does not describe the evolution at locus Pzeb4 very well. Had point mutations between alleles been taken into account, an even larger deviation from the SMM would have been found. Other studies have also shown that interrupted loci generally do not fit the SMM very well (e.g., Estoup et al. 1995a ; Angers and Bernachez 1997 ). Of particular relevance is the study by Angers and Bernachez (1997) , who documented complex mutations at a microsatellite locus across genera, within genera, and within species among salmonid fishes. Most variation not involving loss or gain of repeats was found among genera or among Salvelinus species, while variation in the number of dinucleotide arrays was a more important source of allelic diversity at the intraspecific level. Nevertheless, up to 18% of all intraspecific allelic pairwise comparisons involved large deletions of repeated and nonrepeated sequences. The large deletions found within and between species complexes in P. (Maylandia) and P. (Tropheops) are similar to those found among salmonid species at higher taxonomic levels for species that diverged several million years ago. However, similar types of complex mutations were detected within recently diverged S. fontinalis populations. It is difficult to assert whether the temporal scale for the divergence of Salvelinus fontinalis populations is similar to that of P. (Maylandia) and P. (Tropheops) species, but these results confirm that complex, nonstepwise mutations may occur at a microsatellite locus among recently divergent populations and species. This calls for caution on the estimation of population parameters or inference of phylogenies using SMM-based distances using allele size information alone.
A further point we would like to stress is that large deletions, such as the deletion of part 4, can be responsible for the multimodal allele frequency distributions often observed at microsatellite loci. Bimodal allele frequency distributions caused by large indels followed by stepwise mutations, for instance, may be misinterpreted as evidence for introgression between two species.
Size Homoplasy and Population Genetics Parameters
The present study revealed that size homoplasy at microsatellite loci can occur between relatively closely related species (all species studied are believed to have diverged less than 700,000 years ago [Meyer 1993] ) and even within a single species. Because of this, the numbers of alleles, heterozygosities, and genetic diversities within and between species are underestimated if only allele sizes are considered (which is usually the case in microsatellite analyses).
Depending on the frequency of homoplastic alleles, genetic distances between populations and species may be inaccurately estimated from electromorph frequencies, as demonstrated by the effects of Pzeb4 on the genetic divergence among P. (Maylandia) and P. (Tropheops) taxa using five additional loci. Although we do not know to what extent the additional loci conform to a strict stepwise mutation model, it is clear that Pzeb4 has a more important effect on fixation indices based on SMM and little effect on those based on IAM. Even with the limited amount of results available from this data set, we can certainly suggest that it is important to demonstrate conformance to SMM for microsatellite loci to be used in the inference of population parameters using any SMM-based distance.
This conclusion has recently been substantiated through an extensive survey of microsatellites in Drosophila.Colson and Goldstein (1999) documented the frequent occurrence of polymorphism in 7 out of 17 microsatellite loci resulting from complex nonstepwise mutations in three closely related species of Drosophila. In addition, several alleles showed size homoplasy, confirming that great care should be taken when using microsatellite loci for population inference. These conclusions were reached through a moderate sequencing effort of alleles (≥4) for all loci, suggesting that a thorough analysis of allele sequences—such as the one performed here—is not necessary to allow selection of loci that show consistent mutation properties across species.
Caution has also to be taken when using this kind of data for phylogenetic reconstruction, especially when taxa are expected to be distantly related. The occurrence of homoplasy is expected to increase with time of divergence among populations and taxa, as well as with mutation rates (Estoup and Angers 1998 ). It is well recognized that size homoplasy is a major drawback of microsatellites as markers for the estimation of population parameters such as structure, number of migrants, and effective size (reviewed in Estoup and Cornuet 1999 ). Although, several recently developed population genetics statistics (e.g., Goldstein et al. 1995a, 1995b ; Slatkin 1995 ; Rousset 1996 ) take account of size homoplasy, this only applies to loci that follow a strict stepwise mutation model.
Hybridization or Ancestral Polymorphism?
Pseudotropheus (Tropheops) ‘band’ and P. (Tropheops) ‘rust’ sampled from a single rocky site at Nkhata Bay differ at six microsatellite loci by fixation indices of 0.017 (theta: IAM) and 0.067 (Rst: SMM) (van Oppen et al. 1998 ). These values are small but statistically significant, suggesting that some assortment occurs between the two taxa but that occasional hybridization cannot be excluded. In these cichlid fishes, males hold a mating territory where they court females. It is believed that females select males for mating based on male color and hue patterns alone (Hert 1991 ; Seehausen and van Alphen 1998 ). The females of these two taxa are essentially indistinguishable to us, and field observations suggest that ‘band’ and ‘rust’ males are not able to distinguish them reliably either (van Oppen et al. 1998 ). It is possible, however, that reproductive isolation is maintained by female recognition of males, as no females have been observed to respond to both types of males. Our sequencing results confirm this. The combined frequency of alleles missing part 3 (allele, 105, 113a, and 113b) in P. (Tropheops) ‘band’ at Nkhata Bay is ∼9% (8.7% for 113a and 113b combined, the frequency of 105 is unknown but is most probably low), whereas no such alleles were found in ‘rust’ from Nkhata Bay. Hence, it is extremely unlikely that hybridization between the two occurs at Nkhata Bay. At one of the northern sites (Cape Manulo), however, a single ‘rust’ individual was found which possesses a very similar allele lacking both part 3 and part 4 (allele 101). Allele 111b, present in ‘band’ from the northern site Ruarwe also lacks parts 3 and 4. Such alleles have never been observed in Nkhata Bay. This may indicate that occasional hybridization among ‘band’ and ‘rust’ takes place at the northern sites. An alternative explanation for the shared presence of alleles having such large deletions is that it represents a shared ancestral polymorphism. Given that the deletion of part 3 of the sequence is found in alleles from P. (Tropheops) ‘rust,’ P. (Tropheops) ‘band,’ and P. lucerna and that part 4 is deleted in alleles 117b and 123c from P. (Tropheops) ‘band’ (mbuna) and Copadichromis sp. (utaka), this is a more likely scenario, since deletions of the same blocks of sequence are unlikely to have occurred more than once or in different lineages. Moreover, shared ancestral polymorphism has previously been observed for mtDNA (Moran and Kornfield 1993, 1995 ; Parker and Kornfield 1997 ) and class II MHC loci (Klein et al. 1993 ).
In summary, this study of a complex microsatellite locus shows that approximately 45% of the length mutations are nonstepwise and that size homoplasy is common at locus Pzeb4. This may be true for other microsatellite loci as well and calls for caution in electromorph frequency analyses of microsatellite markers—even in closely related taxa. In fact, our results provide further evidence for the conclusions reached by Colson and Goldstein (1999) , which strongly suggest that it is essential to demonstrate conformance to SMM for microsatellite loci used for population and phylogenetic inference. DNA sequence analysis of complex microsatellite loci for the purpose of phylogenetic analyses among recently diverged species may prove disappointing, because insufficient time has probably passed for alleles to reach reciprocal monophyly.
Axel Meyer, Reviewing Editor
Present address: Department of Biochemistry and Molecular Biology, James Cook University, Townsville, Australia.
Keywords: microsatellite evolution stepwise mutation model infinite-allele model cichlids shared ancestral polymorphism
Address for correspondence and reprints: Ciro Rico, School of Biological Sciences, University of East Anglia, Norwich, Norfolk NR4 7TJ, United Kingdom. E-mail: c.rico@uea.ac.uk.
Table 1 Pairwise 𝛉 and ϕst Values Including Allele Data for Pzeb4 (lower half of diagonal) and Excluding Pzeb4 (upper half of diagonal) Between Pseudotropheus (Maylandia) and Pseudotropheus (Tropheops) Taxa at Nkukuti Point (Nkhata Bay)
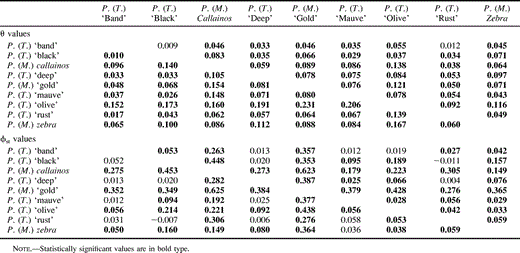
Table 1 Pairwise 𝛉 and ϕst Values Including Allele Data for Pzeb4 (lower half of diagonal) and Excluding Pzeb4 (upper half of diagonal) Between Pseudotropheus (Maylandia) and Pseudotropheus (Tropheops) Taxa at Nkukuti Point (Nkhata Bay)

Fig. 1.—Allele frequency distributions at locus Pzeb4 for the three Pseudotropheus (Maylandia) and six Pseudotropheus (Tropheops) color forms, the Pseudotropheus lucerna sample, and the Copadichromis shoal using the samples collected at Nkhata Bay in 1995. N = number of individuals sampled
Fig. 2.—Sequence alignment of the Pzeb4 alleles. Numbers in brackets indicate the number of times an allele was found in a particular species among the samples investigated. CM = Cape Manulo; MR = Mara Rocks; RW = Ruarwe
Fig. 3.—Minimum number of length mutations (based on the allele sequences) plotted against the size difference between alleles for 300 pairwise comparisons among all alleles sequenced from the Nkhata Bay samples
This project was funded by NERC and the Royal Society of London and the University of East Anglia. We thank the government of Malawi for permission to carry out research, the Department of Fisheries for providing facilities, and S. Chiotha, O. Msiska, M. Chiumia, H. Ngulube, R. L. Robinson, M. E. Knight, M. J. Genner, and P. Bouteillon for their assistance. We also thank L. Bernatchez, H. Lessios, and E. Bermingham for useful comments on the manuscript. Likewise, we are indebted to Axel Meyer and two anonymous referees for a number of valuable comments on this and a previous version of the manuscript.
literature cited
Albertson, R. C., J. A. Markert, P. D. Danley, and T. D. Kocher.
Angers, B., and L. Bernatchez.
Blanquer, A., and B. Crouau-Roy.
Bowcock, A. M., A. Ruiz-Linares, J. Tomfohrde, E. Minch, J. R. Kidd, and L. L. Cavalli-Sforza.
Colson, I., and D. B. Goldstein.
Crow, J. F., and M. Kimura.
Crozier, R. H., B. Kaufmann, M. E. Carew, and Y. C. Crozier.
Di Rienzo, A., A. C. Peterson, J. C. Garza, A. M. Valdes, M. Slatkin, and N. B. Freimer.
Estoup, A., and B. Angers.
Estoup, A., and J.-M. Cornuet.
Estoup, A., L. Garnery, M. Solignac, and J.-M. Cornuet. 1995a. Microsatellite variation in honey bee (Apis mellifera L.) populations: hierarchical genetic structure and test of the infinite allele and stepwise mutation models. Genetics 140:679–695.
Estoup, A., C. Tailliez, J. M. Cornuet, and M. Solignac. 1995b. Size homoplasy and mutational processes of interrupted microsatellites in two bee species, Apis mellifera and Bombus terrestris (Apidae). Mol. Biol. Evol. 12:1074–1084.
Garza, J. C., M. Slatkin, and N. B. Freimer.
Goldstein, D. B., A. R. Linares, M. W. Feldman, and L. L. Cavalli-Sforza. 1995a. An evaluation of genetic distances for use with microsatellite loci. Genetics 139:463–471.
———. 1995b. Genetic absolute dating based on microsatellites and the origin of modern humans. Proc. Natl. Acad. Sci. USA 92:6723–6727.
Grimaldi, M. C., and B. Crouau-Roy.
Hert, E.
Hoelzel, A. R., J. V. Lopez, G. A. Dover, and S. J. O'Brien.
Jarne, P., and P. J. L. Lagoda.
Jones, A. G., G. Rosenquist, A. Berglund, and J. C. Avise.
Kimura, M., and J. F. Crow.
Kimura, M., and T. Ohta.
Klein, D., H. Ono, C. O'huigin, V. Vincek, T. Goldschmidt, and J. Klein.
Mahtani, M. M., and H. F. Willard.
Metzgar, D., D. Field, R. Haubrich, and C. Wills.
Meyer, A.
Michalakis, Y., and L. Excoffier.
Moran, P., and I. Kornfield.
———.
Parker, A., and I. Kornfield.
Powell, W., M. Morgante, C. Andre, J. W. McNicol, G. C. Machray, J. J. Doyle, S. V. Tingey, and J. A. Rafalski.
Primmer, C. R., and H. Ellegren.
Primmer, C. R., N. Saino, A. P. Moller, and H. Ellegren.
Procaccini, G., and L. Mazzella.
Provan, J., G. Corbett, R. Waugh, J. W. McNicol, M. Morgante, and W. Powell.
Ribbink, A. J., B. A. Marsh, A. C. Marsh, A. C. Ribbink, and B. J. Sharp.
Rousset, F.
Sambrook, J., E. F. Fritch, and T. Maniatis.
Schlötterer, C., R. Ritter, B. Harr, and G. Brem.
Seehausen, O., and J. J. van Alphen.
Shriver, M. D., L. Jin, R. Chakraborty, and E. Boerwinkle.
Slatkin, M.
Soranzo, N., J. Provan, and W. Powell.
Taylor, J. S., P. Sanny, and F. Breden.
Valdes, A. M., M. Slatkin, and N. B. Freimer.
van Oppen, M. J. H., C. Rico, J. Deutsch, G. F. Turner, and G. M. Hewitt. 1997a. Isolation and characterisation of microsatellite loci in the cichlid fish Pseudotropheus zebra. Mol. Ecol. 6:387–388.
van Oppen, M. J. H., G. F. Turner, C. Rico, J. C. Deutsch, K. M. Ibrahim, R. L. Robinson, and G. M. Hewitt. 1997b. Unusually fine-scale genetic structuring found in rapidly speciating Malawi cichlid fishes. Proc. R. Soc. Lond. B Biol. Sci. 264:1803–1812.
van Oppen, M. J. H., G. F. Turner, C. Rico, R. L. Robinson, J. C. Deutsch, M. J. Genner, and G. M. Hewitt.
Viard, F., P. Franck, M.-P. Dubois, A. Estoup, and P. Jarne.
Weber, J. L., and C. Wong.
Weir, B. S., and C. C. Cockerham.
Whitton, J., L. H. Rieseberg, and M. C. Ungerer.