Abstract
The paper reports on the production of a panel of seven new monoclonal antibodies (MAbs) capable of specifically binding to the NS11–124 fragment of the influenza virus A/Puerto Rico/8/1934 (H1N1) NS1 protein in indirect enzyme-linked immunosorbent assay (ELISA), as well as to the full-size native NS1 protein in cells infected with different subtypes of the influenza A virus (A(H1N1), A(H1N1)pdm09, A(H2N2), A(H3N2), and A(H9N2)). A sensitive variant of the microcultural ELISA based on the use of the generated MAbs is proposed; it may be utilized to detect NS1 protein in infected cells and to study its antigenic variability. The results of the performed microcultural ELISA demonstrate type- and subtype-specific antigenic variability of the influenza virus NS1 protein.
Similar content being viewed by others
INTRODUCTION
The coevolution of viruses and their hosts has resulted, on one hand, in a full range of mechanisms used by the immune system of the infected organs (e.g., interferon production) to protect it from infection. On the other hand, it also resulted in the emergence of viruses that are able to counteract the host immune system response directed against them. The NS1 protein plays the key role in one of the adaptation mechanisms used by influenza viruses.
NS1 is a nonstructural protein of the influenza A virus. It consists of 217–237 amino acid residues (aa) constituting two functional domains: the N-terminal RNA-binding domain (aa 1–73), which interacts with the double-stranded RNAs that are produced in the course of virus replication and trigger the antivirus response, and the C-terminal effector domain (starting at the aa 74), which is able to form complexes with a number of cellular proteins.
NS1 protein performs a number of regulatory functions and thus plays an important role in the pathogenesis of influenza viral infection. It is involved in the control of virus replication, regulating the activity of viral RNA-polymerase and stimulating the translation of specific viral mRNAs, and is a potent antagonist of the host interferon system. NS1 can block interferon (Inf) production by the infected cells in at least two ways: inhibition of the activation of the interferon regulatory factor 3 (IFR3) and interaction with the cellular Cleavage and Polyadenylation Specificity Factor (CPSF30), which is involved in mRNA processing. This interaction results in the formation of the CPSF30/NS1 complex, which promotes the accumulation of immature pre-mRNAs in the nucleus and inhibits the production of mature mRNAs in the cytoplasm, including the interferon mRNAs [1, 2].
The influenza virus NS1 protein controls, directly or indirectly, the expression of a number of cytokines. For example, cell infection with the influenza virus with the deletion of the NS gene suppressed the synthesis of the IL-4 mRNA, while it activated the synthesis of the IL-10 mRNA [3].
Influenza viruses lacking the NS1 gene are characterized by reduced replication and pathogenic properties [4].
At the same time, the function of anti-NS1 antibodies in the infected organism remains fully unexplored. NS1 is a nonstructural intracellular protein and is thus less accessible to antibodies than the protein components of the virus particle. Nevertheless, high titers of antibodies against NS1 were detected in the blood of the patients infected by the virus A(H5N1) [5]. Additionally, the genome sites encoding the NS1 antigenic determinants are subject to positive selection, which points to a certain immune pressure [6].
It was demonstrated in model experiments that the interaction between antibodies and certain amino acid residues in the effector domain of the NS1 protein hinders NS1 interaction with double-stranded RNA [7] or cellular components, such as CPSF30 and eIF4G1 (Eukaryotic Translation Initiation Factor 4 Gamma I), which play an important role in mRNA processing [8], thereby significantly inhibiting the replication of the influenza A viruses.
The facts described above clearly show that it is necessary to investigate further the roles of the NS1 protein and the antibodies interacting with it in influenza pathogenesis. Monoclonal antibodies (MAbs) may serve as an efficient tool in such studies. MAbs are successfully used to map NS1 epitopes and to characterize the polymorphism of its structure, as well as to study the functions of this protein [7, 9–11].
The purpose of the current work was to generate and describe the panel of monoclonal antibodies against the influenza A virus NS1 protein.
MATERIALS AND METHODS
Viruses, Cells, and Monoclonal Antibodies
The following wild-type strains of the influenza virus containing the full-size unmodified NS1 gene were used: A/Puerto Rico/8/1934/Mount Sinai (A/PR/8/1934) (H1N1), A/Beijing/262/1995 (H1N1), A/New Caledonia/20/1999 (H1N1), A/California/07/2009 (H1N1)pdm09, A/Singapore/1/1957 (H2N2), A/Panama/2007/1999 (H3N2), A/Wisconsin/67/2005 (H3N2), A/Brisbane/10/2007 (H3N2), A/Texas/50/2012 (H3N2), A/Hong Kong/1/1968 (H3N2), A/duck/Potsdam/1402-6/1986 (H5N2), A/mallard/Netherlands/12/2000 (H7N3), A/Hong Kong/1073/99 (H9N2), B/Brisbane/60/2008 (Victoria evolutionary lineage), and B/Massachusetts/02/2012 (Yamagata evolutionary lineage). The indicated strains were provided by the Fransis Crick Institute (London). The A/Saint Petersburg/5/2009 (H1N1)pdm and A/Kaliningrad/53/2015 (H1N1)pdm strains were obtained from the collection of the Museum of Viruses of the Research Institute of Influenza.
The study also involved the following recombinant viruses obtained with reverse genetics techniques: A/Puerto Rica/8/1934 delNS1 (A/PR-/8/34 delNS1) (with the deletion of the entire NS1 protein-coding sequence), A/Puerto Rico/8/1934 (which encodes a truncated NS1 protein containing only first 124 aa residues and heterologous Micobacterium tuberculosis ESAT-6 protein) (A/PR/8(1–124)-ESAT), as well as 6 : 2 virus A/Hong Kong/1/1968 reassortant (which contains the internal genes from the virus A/Puerto Rico/8/1934, truncated NS1 protein, and inserted coding sequence for the heat shock protein (A/H-K‑PR/8(1–124)-HspX). The three indicated strains are a proprietary design and were kindly provided by the Laboratory of Molecular Virology and Genetics Engineering of the Research Institute of Influenza.
The MDCK (London Line) cell culture was obtained from the IRR collection (United States, no. FR-58), and the WHO Vero cell culture was obtained from the European Cell Culture Collection (Great Britain, ECACC, no. 88020401).
Commercial MAbs 1AF10 and 1A4A (Bialexa, Moscow) and ab181436 (Abcam, Great Britain) against the NS1 protein of the influenza A virus were used to study the properties of the new MAbs. Type-specific MAbs against the NP protein of influenza A (MAbs 6D11) and influenza B (MAbs 2/3) viruses obtained from the Research Institute of Influenza were used to analyze viral reproduction in the cell culture.
Production of the Recombinant rNS1124Protein of the Influenza Virus A/Puerto Rico/8/1934 (H1N1)
The synthesis, optimization for subsequent E. coli expression, and ligation with the pET22b vector of the nucleotide sequence encoding the recombinant rNS1124 protein (first 124 aa residues of the influenza virus A/PR/8/1934 NS1 protein) or the Ebola virus GP protein fragment was performed by Evrogen (Moscow). To obtain the producer strain, E. coli BL21(DE) Codon plus cells (Evrogen) were transformed with the pET22b/rNS1124 plasmid (Evrogen). The transformed bacterial cells were grown in LB broth (Becton Dickinson, France) containing ampicillin (Sintez, Russia) to OD600 = 0.6–0.7, and recombinant protein production was then induced by the addition of 1mM IPTG (Thermo Scientific, Lithuania). After 16 h of growth, the cells were pelleted and treated with lysis buffer, pH 8.5, containing 50 mM Na2HPO4 (LenReactiv, Saint Petersburg), 500 mM NaCl (LenReactiv), 8 M urea (Sigma, United States), 5 mM imidazole (Sigma), and 1% E. coli protease inhibitors (Protease Inhibitor Cocktail for His-tagged proteins) (Sigma). After 20 min of incubation at room temperature, Triton X-114 (Sigma) and lysozyme (Serva, Germany) were added to concentrations of 0.01% and 1 mg/mL, respectively.
After incubation for 1 h, the mixture was centrifuged for 1 h at 13 000 g. Recombinant proteins were purified from the cell lysate by metal-affinity chromatography on a 5-mL HisTrap FF Crude column containing Ni sorbent (GE Health Care, Sweden). The starting buffer was the lysis buffer of the composition indicated above. A starting buffer containing 0.1% Triton X-110 (Sigma) was used to wash from endotoxins. At the second washing step (to wash from Triton X-110), starting buffer with 0.1% Tween-20 (Sigma) was used. The third solution used to wash the column was starting buffer containing 10 mM imidazole. Starting buffer containing 500 mM imidazole was used to elute proteins. The flow rate at each stage of chromatographic protein purification was 5 mL/min. All purification procedures were performed with a BioLogic DuoFlow device (BioRad Laboratories, Inc, United States).
Generation of MAbs Specific to NS1 Protein
BALB/c mice (obtained from the Stolbovaya Branch of the Scientific Center for Biomedical Technologies of the Federal Medical and Biological Agency, Moscow Region) were immunized twice with a 3-week interval with the recombinant rNS1124 protein of the influenza virus A/Puerto Rico/8/1934. Protein solution in 0.1 M PBS, pH 7.2 (BioloT, Saint Petersburg), was administered intramuscularly to animals at a dose of 40 μg protein/mouse, together with 500 μg/mouse of the adjuvant, aluminium hydroxide (Brenntag, Denmark). A boost immunization with rNS124 protein at a dose of 30 μ/mouse was performed three weeks after the second immunization, and, three days later, the splenocytes of the immunized mice were hybridized with cells of the murine myeloma line X63Ag8.653 in the presence of 50% PEG 1000 (Sigma).
The obtained hybrid cells were grown in selective HAT medium (Sigma). Conventional indirect ELISA was used for primary testing of theh obtained clones. The analyzed culture liquid was placed into plate wells sensitized with purified rNS124 protein or E. coli cell lysate as a negative control. Hybrid clones within the desired range were subcloned two to four times. Stable MAb-producing clones were used to obtain ascytes. To this end, hybrid cells at a dose of 5–10 million cells/mouse were administered intraperitoneally to BALB/c mice preliminarily primed with pristane (0.5 mL/mouse, Sigma). Ascytic liquid was collected 2–3 weeks later. The supernatant obtained from centrifugation of the ascytic liquid served as a Mab source, while the hybridoma cells that underwent passaging in mice were stored under low temperatures.
The study was performed in full accordance with the Recommendations for Animal Experimentation (Ministry of Health Care of the Russian Federation order no. 266, issued June 19, 2003).
MAb Properties Assessment by Nondirect ELISA
Plates for ELISA (Medpolimer, Saint Petersburg) were sensitized with antigens (rNS124, or recombinant Ebola virus GP protein as negative control) diluted with 0.05 M carbonate–bicarbonate buffer, pH 9.5 (LenReactiv), to a concentration of 1 μg/mL for 18 h at 4°C. After the unbound antigenic material was washed away with 0.01 M PBS, pH 7.2 (BioloT) containing 0.05% Tween-20 (Serva) (PBS-T), different dilutions of MAbs in PBS-T were applied to the plate wells and incubated for 1 h at 37°C. Bound MAbs were detected by addition of peroxidase-conjugated, antimouse IgG antibodies (Sigma) diluted 1 : 5000 with PBS-T and incubation for 1 h at 37°C. The peroxidase reaction was performed via the addition of a substrate mixture containing 0.1 mg/mL of TMB (Sigma) and 0.02% H2O2 solution in 0.1 M acetate–citrate buffer, pH 5.0 (ZAO LenReactiv). The reaction was stopped by the addition of 2 N H2SO4 (ZAO LenReactiv), and the OD was measured with a Multiskan MS photometer (Labsystems, Finland) at a wavelength of about 450 nm.
Mab-Isotype Identification
MAb isotypes were determined with the use of a set of goat anti-isotype, antimouse Ig sera and peroxidase-conjugated rabbit antibodies against goat IgG (Sigma-Aldrich, United States). rNS124 at a concentration of 1 μg/mL was used as an antigen to sensitize the plates.
Western Blotting Analysis of the Obtained MAbs
To assess the antigenic specificity of the obtained MAbs, cell lysates of MDCK cells infected with the influenza A virus at a dose of 0.1–1 TCID50/cell were used as an antigen in Western blotting analysis. Cells were lysed in the buffer containing 5% β-ME (Amresco, United States) 24 h after infection, and the lysate was boiled for 2 min. The negative control was the cell lysate of intact MDCK cells treated in the same way. SDS-PAGE of the proteins in the lysates was performed under reducing conditions according to Laemmli [12] in a 8–20% gradient of polyacrylamide gel (Serva). Upon electrophoresis, proteins were transferred from the gel onto the nitrocellulose membrane (NM) (BioRad, Germany). Nonoccupied sites on the membrane were blocked with 1.5% BSA (Sigma) in PBS-T (blocking solution PBS-BSA) for 18 h at 4°C. Viral antigens on NM were incubated with MAbs diluted with PBS-BSA to a concentration of 5–10 μg/mL for two hours at 37°C. NM was further incubated with peroxidase-conjugated, antimouse IgG antibodies (Life Technologies, United States) diluted 1 : 1000 with blocking buffer for 2 h at 37°C. After the membrane was washed, the peroxidase reaction was performed via the addition of TMB-containing Immnublot solution (Life Technologies). The MW of the proteins with which MAbs interacted on the membrane was determined with a calibration curve constructed with the use of the Kaleidoscope protein molecular weight standard (BioRad).
Microcultural ELISA (mcELISA) for Assessment of the Interaction between MAbs and Viral Antigens in Infected Cells
MDCK or Vero cells in a monolayer grown in 96‑well cultural plates (Thermo Scientific–Nunc, Danmark) were infected with influenza viruses at MOI 0.01. After 24 h of incubation in a CO2 incubator (at 37°C for influenza A viruses and at 34°C for influenza B viruses), cells were fixed in wells with 80% cooled acetone (Vekton, Saint Petersburg) for 20 min. MAbs diluted with 5% dry skimmed milk solution (SI Project, St. Petersburg) in PBS (PBS-M) were added to wells containing fixed and washed cells, and the mixture was incubated for 2 h at 37°C. All MAbs used, both against the influenza A NS1 protein and NP protein, were obtained at the Research Institute of Influenza. After the wells were washed, bound MAbs were detected by the addition of peroxidase-conjugated, goat antimouse IgG antibodies (Sigma) diluted 1 : 5000 with PBS-M. The plates were incubated for 1 h at 37°C, and the peroxidase reaction was performed as indicated above for indirect ELISA.
Analysis of the Amino Acid Sequences of NS1 Proteins from Different Influenza Viruses
NS1 protein sequences of the influenza virus were taken from the GenBank database (AAM75163.1, ACF41872.1, AFO65032.1, YP_009118625.1, ACF54482.1, DQ487336.1, ABW81757.1, AHX37622.1, AIE52530.1, GU052544.1, AGY42287.1, NP_859034.1, AFH57915.1, and KC891763.1). The alignment was performed with the AlignX software implemented in the Vector NTI Advance 11 software package (Invitrogen Corporation, United States).
RESULTS AND DISCUSSION
We generated a panel of seven MAbs (2C5, 1A11, 5G8, 2G11, 5G11, 5D2, and 1H7) that were shown to specifically bind the recombinant rNS1124 protein in ELISA. We did not observe cross-reactivity with the heterologous protein (Ebola virus GP protein fragment) obtained with the same expression system for any of the obtained antibodies. All MAbs belong to the same immunoglobulin isotype, IgG1.
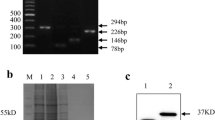
The antigenic specificity of the new MAbs was tested by Western blotting with the lysates of MDCK cells infected with different influenza A virus strains as the source of the antigen. After the lysate proteins were separated by electrophoresis under the conditions indicated above, immunoblotting was performed. It showed that all of the obtained MAbs reacted specifically with the NS1 immobilized in the membrane. The interaction was observed not just in those cases in which the lysates of cells infected with recombinant influenza A viruses encoding truncated NS1 were used as an antigen source; all of the obtained MAbs also interacted with the full-size NS1 protein from cells infected with wild-type viruses A(H1N1) and A(H3N2). The results obtained with the MAbs 1H7 are shown as an example (Fig. 2a). The same results were obtained for other MAbs.
Antigenic specificity comparison for MAbs 1H7 generated at the Research Institute of Influenza (a), and commercially available MAbs 1AF10 (Bialexa) (b) and ab181436 (Abcam) (c). The interaction between the indicated antibodies and the NS1 protein in the infected cells was assessed (by the results of Western blotting): 1—virus A/HK-PR/8(1–124)-HspX (expected MW of the NS1 protein containing the heterologous insert 30.5 KDa) antigens: 2—virus A/PR/8(1–124)/-ESAT (NS1 MW with, or without the heterologous insert 25.9 and 15.9 KDa, respectively) antigens; 3—virus A/Puerto Rico/8/1934 (wild type, NS1MW 25.9 KDa) antigens; 4—virus A/Hong Kong/1/1968 (wild type, NS1 MW 26.9 KDa) antigens; 5—antigens in uninfected cell lysate.
For the purpose of comparison, we tested commercially-obtained MAbs against NS1 by immunoblotting, using the same antigens as above. Unlike the MAbs generated in our work, MAbs 1AF10 (Bialexa) bound with the full-size NS1 protein (MW 26.9 KDa) from cells infected with the wild-type influenza A virus but not with its truncated form, which contains aa residues 1–124 (MW 15.9 KDa) (Fig. 2b, compare with the lanes 2 and 4 in Fig. 2a). Apparently, MAbs 1AF10 are specific for the C-terminal end of the protein. It should be noted that, when we used MAbs ab181436 (Abcam), we observed, in addition to the specific band corresponding to the protein with MW of 26 KDa (Fig. 2c), multiple bands, including the 15.9 KDa band in the wild-type virus sample, where such a NS1 fragment is absent (Fig. 2c, lane 4). This result speaks for the low antigenic specificity of MAbs ab 181436.
The MAbs generated in this work showed a sufficiently high sensitivity to be used in an analysis of the dynamics of the virus NS1 protein synthesis in infected cells. To this end, we used the mcELISA technique and infected Vero cells (African green monkey kidney epithelial cell culture). Unlike other cell lines originating from normal mammalian cells, including canine kidney epithelial MDCK cells, Vero cells do not produce Inf α/β [13]. Only Inf-deficient cells allow normal replication of influenza A viruses with deleted NS1 protein, which is an antagonist of the Inf type I system.
Figure 3 presents the results of NS1 detection at different stages of the viral replicative cycle in Vero cells infected with the wild-type virus A/Puerto Rico/8/1934 and the recombinant A/Puerto Rico/8/1934 delNS strain by mcELISA. NS1 was detected with the use of three specific MAbs (2C5, 1A11, and 1H7) that showed the highest efficiency when used in this assay (see Fig. 3a). Both viruses reproduced successfully, as evidenced by the active production of the NP protein as early as 6 h after infection, which was detected with MAbs 6D11 against the influenza A virus NP protein. The indicator of the specificity of the analyzed MAbs was the absence of interaction with viral antigens in cells infected with the A/Puerto Rico/8/1934 delNS strain (see Fig. 3b).
Dynamics of interaction between the tested MAbs and the wild-type influenza virus A/Puerto Rico/8/1934 (a) or recombinant A/Puerto Rico/8/1934 delNS strain (b) antigens in Vero cells infected at MOI = 0.01 (microcultural ELISA results). K—average OD450 in the case of MAb interaction with uninfected Vero cells (negative control).
Under the indicated experimental conditions (MOI = 0.01), NS1 protein synthesis in cells infected with the wild-type virus could not be observed earlier than 17 h after infection (see Fig. 3a). At a higher virus infection dose (MOI = 3), NS1 protein could be clearly detected as early as 6 h after infection (data presented below). At MOI = 3, the OD450 values at the indicated time point were 0.715, 0.214, and 0.281 for the MAbs 1A11, 2C5, and 1H7, respectively, while the control value (K) was lower than 0.1.
Hence, the proposed mcELISA variant may be utilized to detect NS1 protein in cells infected with the influenza A virus, with its sensitivity being as high as that of the detection methods proposed by other authors. For example, a research group from Canada, using such techniques as Western blot and immunofluorescence with the original MAbs, demonstrated that, at MOI = 5, NS1 protein first appears in cells infected with the virus A/Puerto Rico/8/1934 no earlier than 5–8 h after infection [10].
The NS1 protein structure varies to a certain extent. The homology between the amino acid sequences of this protein in influenza A viruses belonging to the same or different subtypes may vary from 62 to 100%. The N-terminal RNA-binding region is more conservative than the effector C-terminal region [9, 10, 14, 15]. It was demonstrated that the structural characteristics of the NS1 protein affect the efficiency of its interaction with cellular regulatory factors (NF-κB, PI3K, IRF3, CPSF30, and others) [16–19]. NS1 polymorphism provides the basis for the strain and subtype-specific differences in the virus replicative cycle, virus pathogenicity rate, and the degree of its impact on the antivirus immune response (Inf system blockage and proinflammatory cytokine synthesis). This concept was experimentally proved in the case of the avian influenza H5 and H7 viruses [17, 20], as well as virus A(H1N1)pdm09 [16, 21, 22].
Monoclonal antibodies are a practical tool for the study of the antigenic variability of proteins. In this view, one of the objectives of our work was to assess how broad the interaction spectrum is for the obtained MAbs. To this end, we infected MDCK cells in a monolayer (MOI = 0.01) with different wild-type influenza viruses with the unmodified NS gene. The mcELISA results point to the type- and subtype-specific antigenic variability of the influenza virus NS1 protein (Fig. 4).
It should be noted that the OD450 values measured for the interaction between the anti-NS1 MAbs with the virus may seem to be low in those cases in which viral proteins are produced at a low level, i.e., when the virus reproduction level is low, which is observed at low virus infectious doses. However, in our case, viral reproduction was quantitatively assessed by interaction with the type-specific MAbs 6D11 or MAbs 2/3, which recognize the NP protein of all viruses belonging to type A or type B, respectively. The OD450 values in the case of the indicated antibodies were almost the same for all analyzed strains, which implies that the levels (amounts) of other virus proteins produced in the cell are comparable in all of the analyzed strains. Therefore, the low OD450 values for NS1-specific MAbs do not necessarily represent the low NS1 protein content, but they may be a consequence of its structural characteristics, resulting in a reduced affinity for interaction between the protein and the specific Ab.
As expected, all MAbs most efficiently recognized the NS1 protein of the virus A/ Puerto Rico/8/1934. However, the antibodies interacted with the native intracellular NS1 protein in infected cells (mcELISA) and the recombinant rNS124 protein used for the immunization of mice in order to obtain MAbs (indirect ELISA) in somewhat different ways. For example, MAb 5G8 showed a significantly weaker reaction with the native protein as compared to the recombinant protein (compare Figs. 1 and 4a), which may result from the conformational differences between the analyzed protein forms.
MAbs 2G11, 5D2, and 1H7 most efficiently interacted with the NS1 protein of the viruses A/Beijing/262/1995 and A/New Caledonia/20/1999 (see Figs. 4b and 4c) belonging to the subtype A(H1N1) as well, which circulated broadly in the human population until the emergence of the pandemic virus A(H1N1)pdm in 2009. The lower activity of MAbs 2G11 and 5D2 provide evidence that the structure of the NS1 protein domain encompassing aa residues 1–124 is altered in pandemic influenza A(H1N1)pdm viruses as compared to the preceding seasonal influenza A(H1N1) viruses, which circulated in the 1990s (see Figs. 4b–4f). A certain variation in the interaction with MAbs 1A11, 5G11, and 1H7 points to the presence of the interstrain differences in the structure of the NS1 protein antigenic determinants among the pandemic influenza viruses (see Figs. 4d–4f).
While we observed a general decrease in the efficiency of interaction between the MAbs and the subtype A(H3N2) influenza viruses as compared to the virus A/Puerti Rico/8/1934, some strain-specific variation of the MAb activity was also detected (see Figs. 4g–4j). Most antibodies showed no activity toward the influenza virus A(H2N2), with weak interaction detected only in the case of the MAbs 1A11 and 5G11 (Fig. 4k). Interestingly, several MAbs (1A11, 5G11, and 1H7) interacted with the virus A(H9N2) as efficiently as with the virus A/Puerto Rico/8/1934 (Fig. 4n).
None of the generated MAbs interacted with antigens of the influenza viruses A(H5N2) and A(H7N3) (see Figs. 4l and 4m). No interaction was also observed in the case of the influenza B viruses belonging to either the Yamagata or Victoria lineages (see Figs. 4o and 4p). The observed interaction patterns remained unchanged, even when the MOI was increased by 100 times as compared to the experimental conditions represented in Fig. 4.
The observed differences in the efficiency of the interaction between the studied MAbs and the viruses provide evidence for significant structural heterogeneity of the NS protein fragment (NS11–124) observed at the type, subtype, and species levels and suggest the possibility of using the obtained MAbs to study the antigenic variability of the influenza A viruses.
The current literature provides a substantial body of data on certain aa residues in the influenza A virus NS1 protein, which are involved in the interaction with the cell components and are responsible for the protein’s effect on the Inf system of the host organism [2, 18, 23]. At the same time, there is only very small body of experimentally-proven data on the composition of B-cell and T-cell epitopes in the influenza virus NS1 protein. There is almost no research on the relationship between the antigenic properties of this protein and its nucleotide sequence. In the current work we attempted to compare the observed antigenic variability of the NS11–124 protein fragment used to generate the MAb panel and its genetic structure.
Figure 5 shows the aa sequences for the NS11–124 protein fragment of the influenza viruses obtained from the GenBank database that were used in our work. The obtained alignment revealed considerable genetic variability of the primary NS1 protein structure. In most cases, we detected a correlation between the substitutions in the NS1 protein sequences in the analyzed strains and changes in the antigenic characteristics as compared to the virus A/Puerto Rico/8/1934, which were measured by the decreased efficiency of MAb binding to the NS1 protein from different viruses as compared to the NS1 from the indicated virus as detected by mcELISA. For example, the highest structural homology with the NS11–124 protein fragment of the virus A/Puerto Rico/8/1934 (88–99%) was demonstrated for the NS1 proteins of the viruses A(H1N1), which circulated before the 2009 pandemics, and of the virus A(H9N2) (Table 1). The activity of 3–4 MAbs was comparable with their activity towards the A/Puerto Rico/8/1934 strain only towards these strains. The interaction between these viruses and the other antibodies was much weaker (see Fig. 4). It should be noted that the aa sequences of the NS11–124 protein from the virus A/Puerto Rico/8/1934 and virus A/New Caledonia/20/1999 differ by just a single position. The virus A/New Caledonia/20/1999 has a substitution of the negatively charged glutamic acid for the positively charged lysine (E55K) in its NS1 protein. It may be speculated that the aa residue at position 55 of NS1 is the part of a B-cell epitope and that the presence of the negative charge at this position is important for the binding of certain Abs to the antigen. The E55K substitution was also observed in all of the analyzed virus A(H3N2) strains. In virus A(H9N2), the glutamic acid at position 55 is substituted by the positively charged arginine (E55R).
Amino acid sequences of the NS11–124 protein fragment of different viruses. Figures in parenthesis indicate the number of the first aa residue in the line, since, when aligned, the NS1 aa sequences from the influenza A and influenza B viruses become shifted relative to each other. The gray background highlights the amino acid residues that differ from those in the amino acid sequence of the NS1 protein of the influenza virus A/Puerto Rico/8/1934.
Among the analyzed influenza A viruses, the lowest structural homology with the NS11–124 of the virus A/ Puerto Rico/8/1934 (65%) was demonstrated by the viruses A(H5N2) and A(H7N3), which corresponds well with the absence of interaction between these viruses and all of the tested MAbs (see Fig. 4). These viruses also showed the presence of the E55R substitution. In addition, the strains A/Duck/Potsdam/1402-6/1986 and A/mallard/Netherlands/12/2000 contain an important E71S substitution. It is well known that E71 is a key aa residue that stabilizes NS1 protein conformation [15]. Hence, changes in the protein conformation resulting from a substitution at position 71 may be the cause of weak antigen–antibody interaction in this case.
Virus A/California/07/2009 (H1N1)pdm and virus A/Puerto Rico/8/1934 show 81% homology between their NS11–124 proteins and significant antigenic differences, although the indicated pandemic influenza virus contain no substitutions at positions 55 and 71.
Immediately after the emergence of the new pandemic influenza virus in 2009, significant differences were found in the antigenic properties of the B-cell epitopes of its hemagglutinin (HA) molecule and the hemagglutinine molecules of the seasonal influenza viruses A(H1N1), which circulated previously. The amino acid sequences of HA from the virus A/California/07/2009 (H1N1)pdm and virus A/New Caledonia/ 20/1999 (H1N1) showed a 79% similarity [24]. The level of homology between the amino acid sequences of the NS11–124 protein in these viruses (81%) is comparable with that for the HA sequences, which is considered the most variable influenza virus protein. The increase in the number of differences in the genetic structure of the pandemic influenza virus A/California/07/2009 NS1 protein as compared to that of the seasonal virus A/New Caledonia/20/1999 correlated with a decrease in the activity of the MAbs 5G8, 2G11, 5D2, and 1H7 towards this protein (see Fig. 4 and Table 1).
The NS1 protein of the influenza B viruses showed extremely low homology with the NS11–124 protein fragment of the virus A/Puerto Rico/8/1934 (15%). This has a natural impact on the structure of the antigenic determinants, which results in the absence of interaction with the MAbs specific for the influenza A virus.
CONCLUSIONS
The obtained data indicate that the nonstructural protein NS1 of the influenza virus is characterized by significant genetic and antigenic variability.
The generated panel of MAbs makes it possible to detect not only type- and subtype-specific but also strain-specific differences among a wide number of influenza viruses circulating in nature. With the use of fluorescence-based and mcELISA techniques in cell cultures infected with virus-containing material, high-affinity, NS1-specific MAbs may be used to detect influenza A viruses in clinic material, such as epithelial cells in nasal brushings, at early disease stages.
REFERENCES
Shchelkanov, M.Yu., Popov, A.F., Simakova, A.I., et al., Pathogenesis of influenza: mechanisms of modulation by the pathogen proteins, Zh. Infektol., 2015, vol. 7, no. 2, pp. 31–46. doi 10.22625/2072-6732-2015-7-2-31-46
Krug, R.M., Functions of the influenza A virus NS1 protein in antiviral defense, Curr. Opin. Virol., 2015, vol. 12, pp. 1–6. doi 10.1016/j.coviro.2015.01.007
Plotnikova, M.A., Vasin, A.V., Klotchenko, S.A., et al., A comparison of the cytokine mRNA expression pattern in epithelial A-549 cells infected with influenza viruses A/ H1N1pdm09, A/H3N2, and A/H5N1, Tsitokiny Vosp., 2013, vol. 1, pp. 57—65. http://www.cytokines.ru/ 2013/1/Art9.php.
Romanova, J., Krenn, B.M., Wolschek, M., et al., Preclinical evaluation of a replication-deficient intranasal DeltaNS1 H5N1 influenza vaccine, PLoS One, 2009, vol. 4, no. 6. e1000049. doi 10.1371/journal.pmed.1000049
Khurana, S., Suguitan, A.L., Rivera, Y., et al., Antigenic fingerprinting of H5N1 avian influenza using convalescent sera and monoclonal antibodies reveals potential vaccine and diagnostic targets, PLoS Med, 2009, vol. 6, no. 4. e1000049. doi 10.1371/journal.pmed.1000049
Arunachalam, R., Detection of site-specific positive Darwinian selection on pandemic influenza A/H1N1 virus genome: integrative approaches, Genetics, 2013, vol. 141, nos. 4–6, pp. 143–155. doi 10.1007/s10709-013-9713-x
Wu, J., Mok, C.K., Chow, V.T., et al., Biochemical and structural characterization of the interface mediating interaction between the influenza A virus non-structural protein-1 and a monoclonal antibody, Sci. Rep, 2016, vol. 6, p. 33382. doi 10.1038/srep33382
Yodsheewan, R., Maneewatch, S., Srimanote, P., et al., Human monoclonal ScFv specific to NS1 protein inhibits replication of influenza viruses across types and subtypes, Antiviral Res., 2013, vol. 100, no. 1, pp. 226–237. doi 10.1016/j.antiviral.2013.07.019
Tan, Z., Akerstrom, S., Wee, B.Y., et al., A new panel of NS1 antibodies for easy detection and titration of influenza A virus, J. Med. Virol., 2010, vol. 82, no. 3, pp. 467–475. doi 10.1002/jmv.21709
Rahim, M.N., Selman, M., Sauder, P.J., et al., Generation and characterization of a new panel of broadly reactive anti-NS1 mAbs for detection of influenza A virus, J. Gen. Virol., 2013, vol. 94, no. 3, pp. 593–605. doi 10.1099/vir.0.046649-0
Sun, J., Wang, X., Wen, X., et al., Identification of a highly conserved epitope on avian influenza virus non-structural protein 1 using a peptide microarray, PLoS One, 2016, vol. 11, no. 3. e0149868. doi 10.1371/journal.pone.0149868
Laemmi, U.K., Cleveage of structural proteins during the assembly of the head of bacteriophage T 4, Nature, 1970, vol. 227, no. 5259, pp. 680–685. PMID: 5432063
Desmyter, J., Melnick, J.L., and Rawls, W.E., Defectiveness of interferon production and of rubella virus interference in a line of African green monkey kidney cells (Vero), J. Virol., 1968, vol. 2, no. 10, pp. 955–961. PMID: 4302013
Ludwig, S., Schultz, U., Mandler, J., et al., Phylogenetic relationship of the nonstructural (NS) genes of influenza A viruses, Virology, 1991, vol. 183, pp. 566–577. PMID: 1830182
Carrillo, B., Choi, J.M., Bornholdt, Z.A., et al., The influenza A virus protein NS1 displays structural polymorphism, J. Virol., 2014, vol. 88, no. 8, pp. 4113–4122. doi 10.1128/JVI.03692-13
Kuo, R.L., Zhao, C., Malur, M., and Krug, R.M., Influenza A virus strains that circulate in humans differ in the ability of their NS1 proteins to block the activation of IRF3 and interferon-beta transcription, Virology, 2010, vol. 408, pp. 146–158. doi 10.1016/j.virol.2010.09.012
Spesock, A., Malur, M., Hossain, M.J., et al., The virulence of 1997 H5N1 influenza viruses in the mouse model is increased by correcting a defect in their NS1 proteins, J. Virol., 2011, vol. 85, pp. 7048–7058. doi 10.1128/JVI.00417-11
Ayllon, J., Hale, B.G., and Garcia-Sastre, A., Strain-specific contribution of NS1-activated phosphoinositide 3-kinase signaling to influenza A virus replication and virulence, J. Virol., 2012, vol. 86, pp. 5366–5370. doi 10.1128/JVI.06722-11
Hartmann, B.M., Thakar, J., Albrecht, R.A., et al., Dendritic cell response signatures distinguish 1918, pandemic, and seasonal H1N1 influenza viruses, J. Virol., 2015, vol. 89, no. 20, pp. 10190–10205. doi 10.1128/JVI.01523-15
Ma, W., Brenner, D., Wang, Z., et al., The NS segment of an H5N1 highly pathogenic avian influenza virus (HPAIV) is sufficient to alter replication efficiency, cell tropism, and host range of an H7N1 HPAIV, J. Virol., 2010, vol. 84, pp. 2122–2133. doi 10.1128/JVI.01668-09
Hale, B.G., Steel, J., Medina, R.A., et al., Inefficient control of host gene expression by the 2009 pandemic H1N1 influenza A virus NS1 protein, J. Virol., 2010, vol. 84, no. 14, pp. 6909–6922. doi 10.1128/JVI.00081-10
Menachery, V.D., Eisfeld, A.J., Schafer, A., et al., Pathogenic influenza viruses and coronaviruses utilize similar and contrasting approaches to control interferon-stimulated gene responses, MBio, 2014, vol. 5, no. 3. e01174-14. doi 10.1128/mBio.01174-14
Ozawa, M., Basnet, S., Burley, L.M., et al., Impact of amino acid mutations in PB2, PB1-F2, and NS1 on the replication and pathogenicity of pandemic (H1N1) 2009 influenza viruses, J. Virol., 2011, vol. 85, no. 9, pp. 4596–4601. doi 10.1128/JVI.00029-11
Rodriguez-Alvarez, M., Velasco-Velasco, A.M., Alvarez-Anell, N.J., et al., Identification of seasonal vaccine hemagglutinin subtype 1 (H1) epitopes in Mexican isolates of the new influenza A(H1N1) 2009 virus, Arch. Med. Res, 2009, vol. 40, no. 8, pp. 687–692. doi 10.1016/j.arcmed.2009.12.002
Author information
Authors and Affiliations
Corresponding author
Additional information
Translated by E. Martynova
Abbreviations: AA—amino acid; Ab—antibodies; BSA—bovine serum albumin; ELISA—enzyme-linked immunosorbent assay; mcELISA—microcultural ELISA; MW—molecular weight; MAbs—monoclonal antibodies; NM—nitrocellulose membrane; OD450—optical density at 450 nm; PEG—polyethylene glycol; PEG 100—PEG with an average molecular weight of 1000; LB broth—Luria–Bertani broth; TCID50—virus titer at which 50% of cells in the culture become infected; TMB—3,3’,5,5’-Teteramethylbenzidine; EF—electrophoresis; delNS—NS gene deletion; HAT—hypoxanthine–aminopterin–thymidine selection medium; Ig—immunoglobulin; IgG—class G immunoglobulin; IL-4—interleukin-4; Inf—interferon; MOI—multiplicity of infection; IPTG—isopropyl β-D-1-thiogalactopyranoside; NP—influenza virus nucleoprotein; NS1—one of the nonstructural proteins of the influenza virus; PBS—phosphate-buffered saline; PBS-BSA—PBS with added BSA; PBS-M—PBS with added skimmed milk; PBS-T—PBS with added Tween-20; rNS1124—recombinant protein containing the first 124 amino acid residues of the influenza virus A/Puerto Rico/8/1934 NS1 protein; SDS—sodium dodecyl sulfate; β‑ME—β-mercaptoethanol.
Rights and permissions
About this article
Cite this article
Krivitskaya, V.Z., Sorokin, E.V., Tsareva, T.R. et al. Generation and Characterization of the Monoclonal Antibody Panel Specific to the NS1 Protein of the Influenza A Virus. Appl Biochem Microbiol 54, 756–765 (2018). https://doi.org/10.1134/S0003683818070049
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0003683818070049