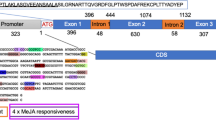
Abstract
Previous studies have shown that the late embryogenesis abundant (LEA) group 3 proteins significantly respond to changes in environmental conditions. However, reports that demonstrate their biological role, especially in Arabidopsis, are notably limited. This study examines the functional roles of the Arabidopsis LEA group 3 proteins AtLEA3-3 and AtLEA3-4 in abiotic stress and ABA treatments. Expression of AtLEA3-3 and AtLEA3-4 is upregulated by ABA, high salinity, and osmotic stress. Results on the ectopic expression of AtLEA3-3 and AtLEA3-4 in E. coli suggest that both proteins play important roles in resistance to cold stress. Overexpression of AtLEA3-3 in Arabidopsis (AtLEA3-3-OE) confers salt and osmotic stress tolerance that is characterized during germination and early seedling establishment. However, AtLEA3-3-OE lines show sensitivity to ABA treatment during early seedling development. These results suggest that accumulation of AtLEA3-3 mRNA and/or proteins may help heterologous ABA reinitiate second dormancy during seedling establishment. Analysis of yellow fluorescent fusion proteins localization shows that AtLEA3-3 and AtLEA3-4 are mainly distributed in the ER and that AtLEA3-3 also localizes in the nucleus, and in response to salt, mannitol, cold, or BFA treatments, the localization of AtLEA3-3 and AtLEA3-4 is altered and becomes more condensed. Protein translocalization may be a positive and effective strategy for responding to abiotic stresses. Taken together, these results suggest that AtLEA3-3 has an important function during seed germination and seedling development of Arabidopsis under abiotic stress conditions.
Similar content being viewed by others
References
Ingram J., Bartels D. 1996. The molecular basis of dehydration tolerance in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47, 377–403.
Bartels D., Sunkar R. 2005. Drought and salt tolerance in plants. Crit. Rev. Plant Sci. 24, 23–58.
Dure L., Chlan C. 1981. Developmental biochemistry of cottonseed embryogenesis and germination: 12. Purification and properties of principal storage proteins. Plant Physiol. 68, 180–186.
Battaglia M., Olvera-Carrillo Y., Garciarrubio A., et al. 2008. The enigmatic LEA proteins and other hydrophilins. Plant Physiol. 148, 6–24.
Wise M.J. 2003. LEAping to conclusions: A computational reanalysis of late embryogenesis abundant proteins and their possible roles. BMC Bioinform. 4, 52.
Bies-Ethe’ve N., Gaubier-Comella P., Debures A., et al. 2008. Inventory, evolution and expression profiling diversity of the LEA (late embryogenesis abundant) protein gene family in Arabidopsis thaliana. Plant Mol. Biol. 67, 107–124.
Hundertmark M., Hincha D.K. 2008. LEA (Late Embryogenesis Abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genomics. 9, 118–139.
Dure L. 1993. A repeating 11-mer amino acid motif and plant desiccation. Plant J. 3, 363–369.
Dure L. 2001. Occurrence of a repeating 11-mer amino acid sequence motif in diverse organisms. Protein Pept. Lett. 8, 115–122.
Goyal K., Tisi L., Basran A., et al. 2003. Transition from natively unfolded to folded state induced by desiccation in an anhydrobiotic nematode protein. J. Biol. Chem. 278, 12977–12984.
Wolkers W.F., McCready S., Brandt W.F., et al. 2001. Isolation and characterization of a D-7 LEA protein from pollen that stabilizes glasses in vitro. Biochim. Biophys. Acta. 1544, 196–206.
Tolleter D., Jaquinod M., Mangavel C., et al. 2007. Structure and function of a mitochondrial late embryogenesis abundant protein are revealed by desiccation. Plant Cell. 19, 1580–1589.
Tunnacliffe A., Wise M.J. 2007. The continuing conundrum of LEA proteins. Naturwissenschaften. 94, 791–812.
Bray E.A. 1993. Molecular responses to water deficit. Plant Physiol. 103, 1035–1040.
Cuming A.C. 1999. LEA proteins. In: Seed Proteins. Eds. Casey R., Shewry P.R. Dordrecht: Kluwer, 753–780.
Honjoh K.I., Matsumoto H., Shimizu H., et al. 2000. Cryoprotective activities of Group 3 late embryogenesis abundant proteins from Chlorella vulgaris C-27. Biosci. Biotechnol. Biochem. 64, 1656–1663.
Goyal K., Walton L.J., Tunnacliffe A. 2005. LEA proteins prevent protein aggregation due to water stress. Biochem. J. 388, 151–157.
Grelet J., Benamar A., Teyssier E., et al. 2005. Identification in pea seed mitochondria of a late-embryogenesis abundant protein able to protect enzymes from drying. Plant Physiol. 137, 157–167.
Reyes J.L., Rodrigo M.J., Colmenero-Flores J.M., et al. 2005. Hydrophilins from distant organisms can protect enzymatic activities from water limitation effects in vitro. Plant Cell Environ. 28, 709–718.
Chakrabortee S., Boschetti C., Walton L.J., et al. 2007. Hydrophilic protein associated with desiccation tolerance exhibits broad protein stabilization function. Proc. Natl. Acad. Sci. U. S. A. 104, 18073–18078.
Nakayama K., Okawa K., Kakizaki T., et al. 2007. Arabidopsis Cor15am is a chloroplast stromal protein that has cryoprotective activity and forms oligomers. Plant Physiol. 144, 513–523.
Yu J.N., Zhang J.S., Shan L., Chen S.Y. 2005. Two new group 3 LEA genes of wheat and their functional analysis in yeast. J. Integr. Plant Biol. 47, 1372–1381.
Zhang L., Ohta A., Takagi M., Imai R. 2000. Expression of plant Group 2 and Group 3 LEA genes in Saccharomyces cerevisiae revealed functional divergence among LEA proteins. J. Biochem. 127, 611–616.
Liu Y., Zheng Y. 2005. PM2, a group 3 LEA protein from soybean, and its 22-mer repeating region confer salt tolerance in Escherichia coli. Biochem. Biophys. Res. Commun. 331, 325–332.
Xu D., Duan X., Wang B., et al. 1996. Expression of a late embryogenesis abundant protein gene, HVA1, from barley confers tolerance to water deficit and salt stress in transgenic rice. Plant Physiol. 110, 249–257.
Sivamani E., Bahieldin A., Wraith J.M., et al. 2000. Improved biomass productivity and water use efficiency under water deficit conditions in transgenic wheat constitutively expressing the barley HVA1 gene. Plant Sci. 155, 1–9.
NDong C., Danyluk J., Wilson K.E., et al. 2002. Coldregulated cereal chloroplast late embryogenesis abundant-like proteins: Molecular characterization and functional analyses. Plant Physiol. 129, 1368–1381.
Li Y., Lee K.K., Walsh S., et al. 2006. Establishing glucose- and ABA-regulated transcription networks in Arabidopsis by microarray analysis and promoter classification using relevance vector machine. Genome Res. 16, 414–427.
Dong C.H., Hu X., Tang W., et al. 2006 A putative Arabidopsis nucleoporin, AtNUP160, is critical for RNA export and required for plant tolerance to cold stress. Mol. Cell Biol. 26, 9533–9543.
Sakuma Y., Maruyama K., Osakabe Y., et al. 2006. Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell. 18, 1292–1309.
Sakuma Y., Maruyama K., Qin F., et al. 2006. Dual function of an Arabidopsis transcription factor DREB2A in water-stress-responsive and heat-stress-responsive gene expression. Proc. Natl. Acad. Sci. U. S. A. 103, 18822–18827.
Lee J., He K., Stolc V., et al. 2007. Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell. 19, 731–749.
Davletova S., Schlauch K., Coutu J., et al. 2005. The zinc-finger protein Zat12 plays a central role in reactive oxygen and abiotic stress signaling in Arabidopsis. Plant Physiol. 139, 847–856.
Cao D., Cheng H., Wu W., et al. 2006. Gibberellin mobilizes distinct DELLA-dependent transcriptomes to regulate seed germination and floral development in Arabidopsis. Plant Physiol. 142, 509–525.
Batoko H., Zheng H.Q., Hawes C., Moore I. 2000. A Rab1 GTPase is required for transport between the endoplasmic reticulum and Golgi apparatus and for normal Golgi movement in plants. Plant Cell. 12, 2201–2217.
Dalal M., Tayal D., Chinnusamy V., et al. 2009. Abiotic stress and ABA-inducible Group 4 LEA from Brassica napus plays a key role in salt and drought tolerance. J. Biotechnol. 139, 137–145.
Yoo S.D., Cho Y.H., Sheen J. 2007. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nature Protocols. 2, 1565–1572.
Samalova M., Fricker M., Moore I. 2008. Quantitative and qualitative analysis of plant membrane traffic using fluorescent proteins. Methods Cell Biol. 85, 353–380.
McGuffin L.J., Bryson K., Jones D.T. 2000. The PSIPRED protein structure prediction server. Bioinformatics. 16, 404–405.
Zhang Y., Cao G., Qu L.J., Gu H. 2009. Involvement of an R2R3-MYB transcription factor gene AtMYB118 in embryogenesis in Arabidopsis. Plant Cell Rep. 28, 337–346.
Lopez-Molina L., Mongrand S., Chua N.H. 2001. A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 98, 4782–4787.
Lopez-Molina L., Mongrand S., McLachlin D.T., et al. 2002. ABI5 acts downstream of ABI3 to execute an ABA dependent growth arrest during germination. Plant J. 32, 317–328.
Carles C., Bies-Etheve N., Aspart L., et al. 2002. Regulation of Arabidopsis thaliana Em genes: Role of ABI5. Plant J. 30, 373–383.
Author information
Authors and Affiliations
Corresponding author
Additional information
The article is published in the original.
Rights and permissions
About this article
Cite this article
Zhao, P., Liu, F., Ma, M. et al. Overexpression of AtLEA3-3 confers resistance to cold stress in Escherichia coli and provides enhanced osmotic stress tolerance and ABA sensitivity in Arabidopsis thaliana . Mol Biol 45, 785–796 (2011). https://doi.org/10.1134/S0026893311050165
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0026893311050165